A Simplified and Effective Approach for the Isolation of Small Pluripotent Stem Cells Derived from Human Peripheral Blood
Abstract
1. Introduction
2. Materials and Methods
2.1. Healthy Volunteers
2.2. Isolation of SBSCs
2.3. SBSCs Counting
2.4. Kyoto Probe 1 Staining
2.5. Immunofluorescence
2.6. Hematoxylin and Eosin (H&E) Staining
2.7. Culture of Human Embryonic Stem Cell Line H1
2.8. Isolation of White Blood Cells (WBCs)
2.9. Sample Preparation for Proteomic Analysis
2.10. Proteomic Liquid Chromatography-Tandem Mass Spectrometry
2.11. Database Search and Analysis
3. Results
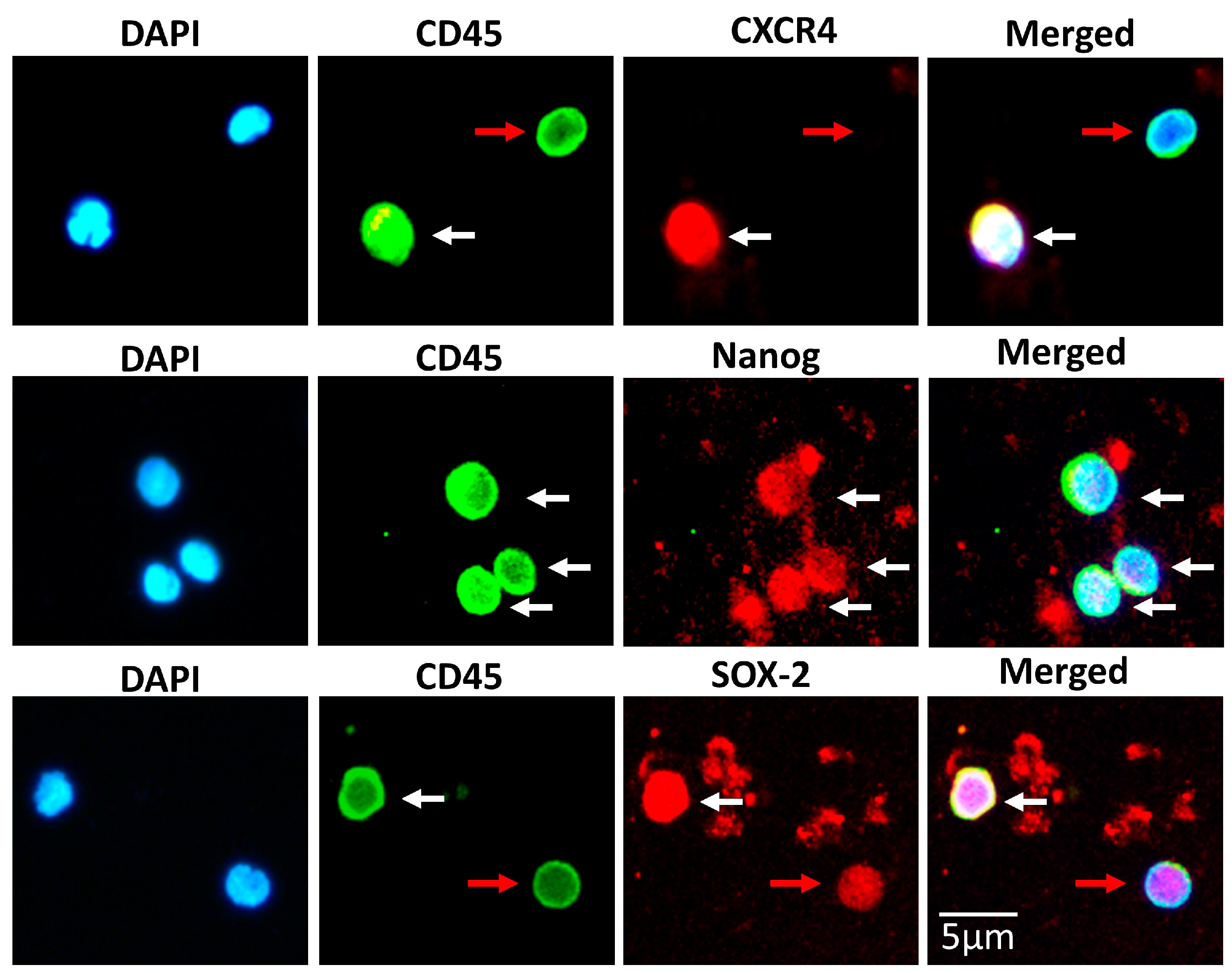
3.1. Isolation of a Rich and Pluripotent Population of SBSCs Using a Simplified Approach
3.2. SBSCs Express Markers of Mesenchymal Origin
3.3. SBSCs Express Certain Hematopoietic Markers
3.4. SBSC Proteomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Suman, S.; Domingues, A.; Ratajczak, J.; Ratajczak, M.Z. Potential Clinical Applications of Stem Cells in Regenerative Medicine. Adv. Exp. Med. Biol. 2019, 1201, 1–22. [Google Scholar] [CrossRef]
- Jarrige, M.; Frank, E.; Herardot, E.; Martineau, S.; Darle, A.; Benabides, M.; Domingues, S.; Chose, O.; Habeler, W.; Lorant, J.; et al. The Future of Regenerative Medicine: Cell Therapy Using Pluripotent Stem Cells and Acellular Therapies Based on Extracellular Vesicles. Cells 2021, 10, 240. [Google Scholar] [CrossRef] [PubMed]
- Bajada, S.; Mazakova, I.; Richardson, J.B.; Ashammakhi, N. Updates on stem cells and their applications in regenerative medicine. J. Tissue Eng. Regen. Med. 2008, 2, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Kolios, G.; Moodley, Y. Introduction to stem cells and regenerative medicine. Respiration 2013, 85, 3–10. [Google Scholar] [CrossRef]
- Ilic, D.; Polak, J.M. Stem cells in regenerative medicine: Introduction. Br. Med. Bull. 2011, 98, 117–126. [Google Scholar] [CrossRef]
- Hipp, J.; Atala, A. Sources of Stem Cells for Regenerative Medicine. Stem Cell Rev. 2008, 4, 3–11. [Google Scholar] [CrossRef]
- Zakrzewski, W.; Dobrzyński, M.; Szymonowicz, M.; Rybak, Z. Stem cells: Past, present, and future. Stem Cell Res. Ther. 2019, 10, 68. [Google Scholar] [CrossRef]
- Triffitt, J.T. Stem cells and the philosopher’s stone. J. Cell. Biochem. Suppl. 2002, 38, 13–19. [Google Scholar] [CrossRef]
- Bongso, A.; Richards, M. History and perspective of stem cell research. Best Pract. Res. Clin. Obstet. Gynaecol. 2004, 18, 827–842. [Google Scholar] [CrossRef]
- Ntege, E.H.; Sunami, H.; Shimizu, Y. Advances in regenerative therapy: A review of the literature and future directions. Regen. Ther. 2020, 14, 136–153. [Google Scholar] [CrossRef]
- Graf, T.; Stadtfeld, M. Heterogeneity of Embryonic and Adult Stem Cells. Cell Stem Cell 2008, 3, 480–483. [Google Scholar] [CrossRef] [PubMed]
- Passier, R.; Mummery, C. Origin and use of embryonic and adult stem cells in differentiation and tissue repair. Cardiovasc. Res. 2003, 58, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Aly, R.M. Current state of stem cell-based therapies: An overview. Stem Cell Investig. 2020, 7, 8. [Google Scholar] [CrossRef]
- Doss, M.X.; Koehler, C.I.; Gissel, C.; Hescheler, J.; Sachinidis, A. Embryonic stem cells: A promising tool for cell replacement therapy. J. Cell. Mol. Med. 2004, 8, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Menendez, P.; Bueno, C.; Wang, L. Human embryonic stem cells: A journey beyond cell replacement therapies. Cytotherapy 2006, 8, 530–541. [Google Scholar] [CrossRef]
- De Luca, M.; Aiuti, A.; Cossu, G.; Parmar, M.; Pellegrini, G.; Robey, P.G. Advances in stem cell research and therapeutic development. Nat. Cell Biol. 2019, 21, 801–811. [Google Scholar] [CrossRef]
- Volarevic, V.; Markovic, B.S.; Gazdic, M.; Volarevic, A.; Jovicic, N.; Arsenijevic, N.; Armstrong, L.; Djonov, V.; Lako, M.; Stojkovic, M. Ethical and Safety Issues of Stem Cell-Based Therapy. Int. J. Med. Sci. 2018, 15, 36–45. [Google Scholar] [CrossRef]
- Brunet, A.; Goodell, M.A.; Rando, T.A. Ageing and rejuvenation of tissue stem cells and their niches. Nat. Rev. Mol. Cell Biol. 2023, 24, 45–62. [Google Scholar] [CrossRef]
- Sobhani, A.; Khanlarkhani, N.; Baazm, M.; Mohammadzadeh, F.; Najafi, A.; Mehdinejadiani, S.; Aval, F.S. Multipotent stem cell and current application. Acta Med. Iran. 2017, 55, 6–23. [Google Scholar]
- Mirzaei, H.; Sahebkar, A.; Sichani, L.S.; Moridikia, A.; Nazari, S.; Sadri Nahand, J.; Salehi, H.; Stenvang, J.; Masoudifar, A.; Mirzaei, H.R.; et al. Therapeutic application of multipotent stem cells. J. Cell. Physiol. 2018, 233, 2815–2823. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induced pluripotent stem cells in medicine and biology. Development 2013, 140, 2457–2461. [Google Scholar] [CrossRef] [PubMed]
- Mostoslavsky, G. Concise Review: The Magic Act of Generating Induced Pluripotent Stem Cells: Many Rabbits in the Hat. Stem Cells 2011, 30, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Maherali, N.; Hochedlinger, K. Guidelines and Techniques for the Generation of Induced Pluripotent Stem Cells. Cell Stem Cell 2008, 3, 595–605. [Google Scholar] [CrossRef]
- Singh, V.K.; Kalsan, M.; Kumar, N.; Saini, A.; Chandra, R. Induced pluripotent stem cells: Applications in regenerative medicine, disease modeling, and drug discovery. Front. Cell Dev. Biol. 2015, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Omole, A.E.; Fakoya, A.O.J. Ten years of progress and promise of induced pluripotent stem cells: Historical origins, characteristics, mechanisms, limitations, and potential applications. PeerJ 2018, 6, e4370. [Google Scholar] [CrossRef]
- Neofytou, E.; O’Brien, C.G.; Couture, L.A.; Wu, J.C. Hurdles to clinical translation of human induced pluripotent stem cells. J. Clin. Investig. 2015, 125, 2551–2557. [Google Scholar] [CrossRef]
- Madrid, M.; Sumen, C.; Aivio, S.; Saklayen, N. Autologous Induced Pluripotent Stem Cell–Based Cell Therapies: Promise, Progress, and Challenges. Curr. Protoc. 2021, 1, e88. [Google Scholar] [CrossRef]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced pluripotent stem cell technology: A decade of progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef]
- Taylor, C.J.; Bolton, E.M.; Bradley, J.A. Immunological considerations for embryonic and induced pluripotent stem cell banking. Philos. Trans. R. Soc. B: Biol. Sci. 2011, 366, 2312–2322. [Google Scholar] [CrossRef]
- Azuma, K.; Yamanaka, S. Recent policies that support clinical application of induced pluripotent stem cell-based regenerative therapies. Regen. Ther. 2016, 4, 36–47. [Google Scholar] [CrossRef]
- Sullivan, S.; Fairchild, P.J.; Marsh, S.G.E.; Müller, C.R.; Turner, M.L.; Song, J.; Turner, D. Haplobanking induced pluripotent stem cells for clinical use. Stem Cell Res. 2020, 49, 102035. [Google Scholar] [CrossRef]
- Li, C.; Chen, S.; Zhou, Y.; Zhao, Y.; Liu, P.; Cai, J. Application of induced pluripotent stem cell transplants: Autologous or allogeneic? Life Sci. 2018, 212, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Kucia, M.; Reca, R.; Campbell, F.R.; Zuba-Surma, E.; Majka, M.; Ratajczak, J.; Ratajczak, M.Z. A population of very small embryonic-like (VSEL) CXCR4(+)SSEA-1(+)Oct-4+ stem cells identified in adult bone marrow. Leukemia 2006, 20, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.M.; Liu, R.; Wu, W.; Waigel, S.J.; Zacharias, W.; Ratajczak, M.Z.; Kucia, M. Global gene expression analysis of very small embryonic-like stem cells reveals that the Ezh2-dependent bivalent domain mechanism contributes to their pluripotent state. Stem Cells Dev. 2012, 21, 1639–1652. [Google Scholar] [CrossRef]
- Ratajczak, M.Z.; Ratajczak, J.; Suszynska, M.; Miller, D.M.; Kucia, M.; Shin, D.M. A Novel View of the Adult Stem Cell Compartment From the Perspective of a Quiescent Population of Very Small Embryonic-Like Stem Cells. Circ. Res. 2017, 120, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, M.Z.; Ratajczak, J.; Kucia, M. Very small embryonic-like stem cells (VSELs) an update and future directions. Circ. Res. 2019, 124, 208–210. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, M.Z.; Zuba-Surma, E.; Wojakowski, W.; Suszynska, M.; Mierzejewska, K.; Liu, R.; Ratajczak, J.; Shin, D.M.; Kucia, M. Very small embryonic-like stem cells (VSELs) represent a real challenge in stem cell biology: Recent pros and cons in the midst of a lively debate. Leukemia 2014, 28, 473–484. [Google Scholar] [CrossRef]
- Weissman, I.L. Stem cells: Units of development, units of regeneration, and units in evolution. Cell 2000, 100, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Kuruca, S.E.; Çelik, D.D.; Özerkan, D.; Erdemir, G. Characterization and Isolation of Very Small Embryonic-like (VSEL) Stem Cells Obtained from Various Human Hematopoietic Cell Sources. Stem Cell Rev. Rep. 2019, 15, 730–742. [Google Scholar] [CrossRef]
- Gounari, E.; Daniilidis, A.; Tsagias, N.; Michopoulou, A.; Kouzi, K.; Koliakos, G. Isolation of a novel embryonic stem cell cord blood-derived population with in vitro hematopoietic capacity in the presence of Wharton’s jelly-derived mesenchymal stromal cells. Cytotherapy 2019, 21, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Monti, M.; Imberti, B.; Bianchi, N.; Pezzotta, A.; Morigi, M.; Del Fante, C.; Redi, C.A.; Perotti, C. A Novel Method for Isolation of Pluripotent Stem Cells from Human Umbilical Cord Blood. Stem Cells Dev. 2017, 26, 1258–1269. [Google Scholar] [CrossRef]
- Bamias, G.; Filidou, E.; Goukos, D.; Valatas, V.; Arvanitidis, K.; Panagopoulou, M.; Kouklakis, G.; Daikos, G.L.; Ladas, S.D.; Kolios, G. Crohn’s disease-associated mucosal factors regulate the expression of TNF-like cytokine 1A and its receptors in primary subepithelial intestinal myofibroblasts and intestinal epithelial cells. Transl. Res. J. Lab. Clin. Med. 2017, 180, 118–130.e112. [Google Scholar] [CrossRef] [PubMed]
- Filidou, E.; Valatas, V.; Drygiannakis, I.; Arvanitidis, K.; Vradelis, S.; Kouklakis, G.; Kolios, G.; Bamias, G. Cytokine receptor profiling in human colonic subepithelial myofibroblasts: A differential effect of Th polarization associated cytokines in intestinal fibrosis. Inflamm. Bowel Dis. 2018, 24, 2224–2241. [Google Scholar] [CrossRef]
- Kandilogiannakis, L.; Filidou, E.; Drygiannakis, I.; Tarapatzi, G.; Didaskalou, S.; Koffa, M.; Arvanitidis, K.; Bamias, G.; Valatas, V.; Paspaliaris, V.; et al. Development of a Human Intestinal Organoid Model for In Vitro Studies on Gut Inflammation and Fibrosis. Stem Cells Int. 2021, 2021, 9929461. [Google Scholar] [CrossRef]
- Claridge, B.; Rai, A.; Fang, H.; Matsumoto, A.; Luo, J.; McMullen, J.R.; Greening, D.W. Proteome characterisation of extracellular vesicles isolated from heart. Proteomics 2021, 21, e2100026. [Google Scholar] [CrossRef]
- Lozano, J.; Rai, A.; Lees, J.G.; Fang, H.; Claridge, B.; Lim, S.Y.; Greening, D.W. Scalable Generation of Nanovesicles from Human-Induced Pluripotent Stem Cells for Cardiac Repair. Int. J. Mol. Sci. 2022, 23, 14334. [Google Scholar] [CrossRef]
- Hughes, C.S.; Moggridge, S.; Muller, T.; Sorensen, P.H.; Morin, G.B.; Krijgsveld, J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019, 14, 68–85. [Google Scholar] [CrossRef]
- Kompa, A.R.; Greening, D.W.; Kong, A.M.; McMillan, P.J.; Fang, H.; Saxena, R.; Wong, R.C.B.; Lees, J.G.; Sivakumaran, P.; Newcomb, A.E.; et al. Sustained subcutaneous delivery of secretome of human cardiac stem cells promotes cardiac repair following myocardial infarction. Cardiovasc. Res. 2021, 117, 918–929. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Maguire, C.T.; Demarest, B.L.; Hill, J.T.; Palmer, J.D.; Brothman, A.R.; Yost, H.J.; Condic, M.L. Genome-wide analysis reveals the unique stem cell identity of human amniocytes. PLoS ONE 2013, 8, e53372. [Google Scholar] [CrossRef]
- Liebermeister, W.; Noor, E.; Flamholz, A.; Davidi, D.; Bernhardt, J.; Milo, R. Visual account of protein investment in cellular functions. Proc. Natl. Acad. Sci. USA 2014, 111, 8488–8493. [Google Scholar] [CrossRef]
- Tyanova, S.; Cox, J. Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research. Methods Mol. Biol. 2018, 1711, 133–148. [Google Scholar] [CrossRef]
- Hirata, N.; Nakagawa, M.; Fujibayashi, Y.; Yamauchi, K.; Murata, A.; Minami, I.; Tomioka, M.; Kondo, T.; Kuo, T.F.; Endo, H.; et al. A chemical probe that labels human pluripotent stem cells. Cell Rep. 2014, 6, 1165–1174. [Google Scholar] [CrossRef]
- Miller, R.J.; Banisadr, G.; Bhattacharyya, B.J. CXCR4 signaling in the regulation of stem cell migration and development. J. Neuroimmunol. 2008, 198, 31–38. [Google Scholar] [CrossRef]
- Galkowski, D.; Ratajczak, M.Z.; Kocki, J.; Darzynkiewicz, Z. Of Cytometry, Stem Cells and Fountain of Youth. Stem Cell Rev. Rep. 2017, 13, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.J.; Andrews, P.W. Surface marker antigens in the characterization of human embryonic stem cells. Stem Cell Res. 2009, 3, 3–11. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Jordan, N.J.; Kolios, G.; Abbot, S.E.; Sinai, M.A.; Thompson, D.A.; Petraki, K.; Westwick, J. Expression of functional CXCR4 chemokine receptors on human colonic epithelial cells. J. Clin. Investig. 1999, 104, 1061–1069. [Google Scholar] [CrossRef]
- Brown, C.; McKee, C.; Bakshi, S.; Walker, K.; Hakman, E.; Halassy, S.; Svinarich, D.; Dodds, R.; Govind, C.K.; Chaudhry, G.R. Mesenchymal stem cells: Cell therapy and regeneration potential. J. Tissue Eng. Regen. Med. 2019, 13, 1738–1755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kumagai, K.; Saito, T. Effect of parathyroid hormone on early chondrogenic differentiation from mesenchymal stem cells. J. Orthop. Surg. Res. 2014, 9, 68. [Google Scholar] [CrossRef] [PubMed]
- Mo, M.; Wang, S.; Zhou, Y.; Li, H.; Wu, Y. Mesenchymal stem cell subpopulations: Phenotype, property and therapeutic potential. Cell. Mol. Life Sci. 2016, 73, 3311–3321. [Google Scholar] [CrossRef]
- Tarnok, A.; Ulrich, H.; Bocsi, J. Phenotypes of stem cells from diverse origin. Cytom. A 2010, 77, 6–10. [Google Scholar] [CrossRef]
- Chang, Y.J.; Tien, K.E.; Wen, C.H.; Hsieh, T.B.; Hwang, S.M. Recovery of CD45(-)/Lin(-)/SSEA-4(+) very small embryonic-like stem cells by cord blood bank standard operating procedures. Cytotherapy 2014, 16, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Halasa, M.; Baskiewicz-Masiuk, M.; Dabkowska, E.; Machalinski, B. An efficient two-step method to purify very small embryonic-like (VSEL) stem cells from umbilical cord blood (UCB). Folia Histochem. Cytobiol. 2008, 46, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Gonzalez, C.; Duggleby, R.; Vagaska, B.; Querol, S.; Gomez, S.G.; Ferretti, P.; Madrigal, A. Cord blood Lin(-)CD45(-) embryonic-like stem cells are a heterogeneous population that lack self-renewal capacity. PLoS ONE 2013, 8, e67968. [Google Scholar] [CrossRef]
- Shaikh, A.; Nagvenkar, P.; Pethe, P.; Hinduja, I.; Bhartiya, D. Molecular and phenotypic characterization of CD133 and SSEA4 enriched very small embryonic-like stem cells in human cord blood. Leukemia 2015, 29, 1909–1917. [Google Scholar] [CrossRef] [PubMed]
- Bhartiya, D.; Jha, N.; Tripathi, A.; Tripathi, A. Very small embryonic-like stem cells have the potential to win the three-front war on tissue damage, cancer, and aging. Front. Cell Dev. Biol. 2022, 10, 1061022. [Google Scholar] [CrossRef]
- Kuo, T.F.; Mao, D.; Hirata, N.; Khambu, B.; Kimura, Y.; Kawase, E.; Shimogawa, H.; Ojika, M.; Nakatsuji, N.; Ueda, K.; et al. Selective elimination of human pluripotent stem cells by a marine natural product derivative. J. Am. Chem. Soc. 2014, 136, 9798–9801. [Google Scholar] [CrossRef] [PubMed]
- Miyagi-Shiohira, C.; Saitoh, I.; Watanabe, M.; Noguchi, H. Kyoto probe-1 reveals phenotypic differences between mouse ES cells and iTS-P cells. Sci. Rep. 2020, 10, 18084. [Google Scholar] [CrossRef]
- Mao, D.; Ando, S.; Sato, S.I.; Qin, Y.; Hirata, N.; Katsuda, Y.; Kawase, E.; Kuo, T.F.; Minami, I.; Shiba, Y.; et al. A Synthetic Hybrid Molecule for the Selective Removal of Human Pluripotent Stem Cells from Cell Mixtures. Angew. Chem. Int. Ed. Engl. 2017, 56, 1765–1770. [Google Scholar] [CrossRef] [PubMed]
- Wakao, S.; Kitada, M.; Kuroda, Y.; Shigemoto, T.; Matsuse, D.; Akashi, H.; Tanimura, Y.; Tsuchiyama, K.; Kikuchi, T.; Goda, M.; et al. Multilineage-differentiating stress-enduring (Muse) cells are a primary source of induced pluripotent stem cells in human fibroblasts. Proc. Natl. Acad. Sci. USA 2011, 108, 9875–9880. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, Y.; Kitada, M.; Wakao, S.; Nishikawa, K.; Tanimura, Y.; Makinoshima, H.; Goda, M.; Akashi, H.; Inutsuka, A.; Niwa, A.; et al. Unique multipotent cells in adult human mesenchymal cell populations. Proc. Natl. Acad. Sci. USA 2010, 107, 8639–8643. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Ji, X.; Zhang, F.; Li, L.; Ma, L. Embryonic Stem Cell Markers. Molecules 2012, 17, 6196–6236. [Google Scholar] [CrossRef]
- Breimer, M.E.; Säljö, K.; Barone, A.; Teneberg, S. Glycosphingolipids of human embryonic stem cells. Glycoconj. J. 2017, 34, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef]
- Pan, G.; Thomson, J.A. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007, 17, 42–49. [Google Scholar] [CrossRef]
- Cavaleri, F.; Schöler, H.R. Nanog: A New Recruit to the Embryonic Stem Cell Orchestra. Cell 2003, 113, 551–552. [Google Scholar] [CrossRef]
- Blinka, S.; Rao, S. Nanog Expression in Embryonic Stem Cells—An Ideal Model System to Dissect Enhancer Function. Bioessays 2017, 39, 86. [Google Scholar] [CrossRef]
- Lee, J.; Go, Y.; Kang, I.; Han, Y.-M.; Kim, J. Oct-4 controls cell-cycle progression of embryonic stem cells. Biochem. J. 2010, 426, 171–181. [Google Scholar] [CrossRef]
- Pesce, M.; Anastassiadis, K.; Schöler, H.R. Oct-4: Lessons of Totipotency from Embryonic Stem Cells. Cells Tissues Organs 1999, 165, 144–152. [Google Scholar] [CrossRef]
- Zhang, S.; Cui, W. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cells 2014, 6, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, T.; Steiner, R.; Lengerke, C. SOX2 and p53 Expression Control Converges in PI3K/AKT Signaling with Versatile Implications for Stemness and Cancer. Int. J. Mol. Sci. 2020, 21, 4902. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Rao, S.; Chu, J.; Shen, X.; Levasseur, D.N.; Theunissen, T.W.; Orkin, S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature 2006, 444, 364–368. [Google Scholar] [CrossRef]
- Chen, X.; Xu, H.; Yuan, P.; Fang, F.; Huss, M.; Vega, V.B.; Wong, E.; Orlov, Y.L.; Zhang, W.; Jiang, J. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008, 133, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Zhen, S.; Roumiantsev, S.; McDonald, L.T.; Yuan, G.-C.; Orkin, S.H. Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol. Cell. Biol. 2010, 30, 5364–5380. [Google Scholar] [CrossRef]
- Ward, S.G.; Bacon, K.; Westwick, J. Chemokines and T lymphocytes: More than an attraction. Immunity 1998, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- McGrath, K.E.; Koniski, A.D.; Maltby, K.M.; McGann, J.K.; Palis, J. Embryonic expression and function of the chemokine SDF-1 and its receptor, CXCR4. Dev. Biol. 1999, 213, 442–456. [Google Scholar] [CrossRef]
- Cencioni, C.; Capogrossi, M.C.; Napolitano, M. The SDF-1/CXCR4 axis in stem cell preconditioning. Cardiovasc. Res. 2012, 94, 400–407. [Google Scholar] [CrossRef]
- Karagiannis, P.; Takahashi, K.; Saito, M.; Yoshida, Y.; Okita, K.; Watanabe, A.; Inoue, H.; Yamashita, J.K.; Todani, M.; Nakagawa, M.; et al. Induced Pluripotent Stem Cells and Their Use in Human Models of Disease and Development. Physiol. Rev. 2019, 99, 79–114. [Google Scholar] [CrossRef] [PubMed]
- Crook, J.M.; Hei, D.; Stacey, G. The International Stem Cell Banking Initiative (ISCBI): Raising standards to bank on. Vitr. Cell. Dev. Biol. Anim. 2010, 46, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Liu, C.L.; Ting, C.Y.; Chiu, Y.T.; Cheng, Y.C.; Nicholson, M.W.; Hsieh, P.C.H. Human iPSC banking: Barriers and opportunities. J. Biomed. Sci. 2019, 26, 87. [Google Scholar] [CrossRef] [PubMed]
- Kolle, G.; Ho, M.; Zhou, Q.; Chy, H.S.; Krishnan, K.; Cloonan, N.; Bertoncello, I.; Laslett, A.L.; Grimmond, S.M. Identification of human embryonic stem cell surface markers by combined membrane-polysome translation state array analysis and immunotranscriptional profiling. Stem Cells 2009, 27, 2446–2456. [Google Scholar] [CrossRef] [PubMed]
- Oka, M.; Tagoku, K.; Russell, T.L.; Nakano, Y.; Hamazaki, T.; Meyer, E.M.; Yokota, T.; Terada, N. CD9 is associated with leukemia inhibitory factor-mediated maintenance of embryonic stem cells. Mol. Biol. Cell 2002, 13, 1274–1281. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Yu, J.M.; Joo, H.J.; Kim, H.K.; Cho, H.H.; Bae, Y.C.; Jung, J.S. Role of CD9 in proliferation and proangiogenic action of human adipose-derived mesenchymal stem cells. Pflug. Arch. 2007, 455, 283–296. [Google Scholar] [CrossRef]
- Prinsloo, E.; Setati, M.M.; Longshaw, V.M.; Blatch, G.L. Chaperoning stem cells: A role for heat shock proteins in the modulation of stem cell self-renewal and differentiation? BioEssays 2009, 31, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.-C. Role of heat shock proteins in stem cell behavior. Prog. Mol. Biol. Transl. Sci. 2012, 111, 305–322. [Google Scholar] [PubMed]
- Held, T.; Barakat, A.Z.; Mohamed, B.A.; Paprotta, I.; Meinhardt, A.; Engel, W.; Adham, I.M. Heat-shock protein HSPA4 is required for progression of spermatogenesis. Reproduction 2011, 142, 133–144. [Google Scholar] [CrossRef]
- Osman, A.M.; van Dartel, D.A.; Zwart, E.; Blokland, M.; Pennings, J.L.; Piersma, A.H. Proteome profiling of mouse embryonic stem cells to define markers for cell differentiation and embryotoxicity. Reprod. Toxicol. 2010, 30, 322–332. [Google Scholar] [CrossRef]
- Battersby, A.; Jones, R.D.; Lilley, K.S.; McFarlane, R.J.; Braig, H.R.; Allen, N.D.; Wakeman, J.A. Comparative proteomic analysis reveals differential expression of Hsp25 following the directed differentiation of mouse embryonic stem cells. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2007, 1773, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Winger, Q.A.; Guttormsen, J.; Gavin, H.; Bhushan, F. Heat shock protein 1 and the mitogen-activated protein kinase 14 pathway are important for mouse trophoblast stem cell differentiation. Biol. Reprod. 2007, 76, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Krebsbach, P.H.; Villa-Diaz, L.G. The Role of Integrin α6 (CD49f) in Stem Cells: More than a Conserved Biomarker. Stem Cells Dev. 2017, 26, 1090–1099. [Google Scholar] [CrossRef]
- Al-Obaide, M.; Ishmakej, A.; Brown, C.; Mazzella, M.; Agosta, P.; Perez-Cruet, M.; Chaudhry, G.R. The potential role of integrin alpha 6 in human mesenchymal stem cells. Front. Genet. 2022, 13, 968228. [Google Scholar] [CrossRef] [PubMed]
- Hakki, S.S.; Kayis, S.A.; Hakki, E.E.; Bozkurt, S.B.; Duruksu, G.; Unal, Z.S.; Turaç, G.; Karaoz, E. Comparison of mesenchymal stem cells isolated from pulp and periodontal ligament. J. Periodontol. 2015, 86, 283–291. [Google Scholar] [CrossRef]
- Meloche, S.; Pouysségur, J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene 2007, 26, 3227–3239. [Google Scholar] [CrossRef]
- Buscà, R.; Pouysségur, J.; Lenormand, P. ERK1 and ERK2 Map Kinases: Specific Roles or Functional Redundancy? Front. Cell Dev. Biol. 2016, 4, 53. [Google Scholar] [CrossRef]
- Zou, J.; Lei, T.; Guo, P.; Yu, J.; Xu, Q.; Luo, Y.; Ke, R.; Huang, D. Mechanisms shaping the role of ERK1/2 in cellular senescence (Review). Mol. Med. Rep. 2019, 19, 759–770. [Google Scholar] [CrossRef]
- Chan, G.; Gu, S.; Neel, B.G. Erk1 and Erk2 are required for maintenance of hematopoietic stem cells and adult hematopoiesis. Blood 2013, 121, 3594–3598. [Google Scholar] [CrossRef]
- Zhang, C.; Lv, J.; He, Q.; Wang, S.; Gao, Y.; Meng, A.; Yang, X.; Liu, F. Inhibition of endothelial ERK signalling by Smad1/5 is essential for haematopoietic stem cell emergence. Nat. Commun. 2014, 5, 3431. [Google Scholar] [CrossRef]
- Saulnier, N.; Guihard, S.; Holy, X.; Decembre, E.; Jurdic, P.; Clay, D.; Feuillet, V.; Pages, G.; Pouysségur, J.; Porteu, F. ERK1 regulates the hematopoietic stem cell niches. PLoS ONE 2012, 7, e30788. [Google Scholar] [CrossRef]
- Kim, M.O.; Kim, S.H.; Cho, Y.Y.; Nadas, J.; Jeong, C.H.; Yao, K.; Kim, D.J.; Yu, D.H.; Keum, Y.S.; Lee, K.Y.; et al. ERK1 and ERK2 regulate embryonic stem cell self-renewal through phosphorylation of Klf4. Nat. Struct. Mol. Biol. 2012, 19, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Hacohen Lev-Ran, A.; Seger, R. Retention of ERK in the cytoplasm mediates the pluripotency of embryonic stem cells. Stem Cell Rep. 2023, 18, 305–318. [Google Scholar] [CrossRef]
- Ma, X.; Chen, H.; Chen, L. A dual role of Erk signaling in embryonic stem cells. Exp. Hematol. 2016, 44, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.; Jain, M.; Madhusudhan, N.; Sheppard, N.G.; Strittmatter, L.; Kampf, C.; Huang, J.; Asplund, A.; Mootha, V.K. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014, 5, 3128. [Google Scholar] [CrossRef] [PubMed]
- Sdelci, S.; Rendeiro, A.F.; Rathert, P.; You, W.; Lin, J.-M.G.; Ringler, A.; Hofstätter, G.; Moll, H.P.; Gürtl, B.; Farlik, M.; et al. MTHFD1 interaction with BRD4 links folate metabolism to transcriptional regulation. Nat. Genet. 2019, 51, 990–998. [Google Scholar] [CrossRef]
- Field, M.S.; Kamynina, E.; Agunloye, O.C.; Liebenthal, R.P.; Lamarre, S.G.; Brosnan, M.E.; Brosnan, J.T.; Stover, P.J. Nuclear enrichment of folate cofactors and methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) protect de novo thymidylate biosynthesis during folate deficiency. J. Biol. Chem. 2014, 289, 29642–29650. [Google Scholar] [CrossRef]
- Bhattacharya, B.; Miura, T.; Brandenberger, R.; Mejido, J.; Luo, Y.; Yang, A.X.; Joshi, B.H.; Ginis, I.; Thies, R.S.; Amit, M.; et al. Gene expression in human embryonic stem cell lines: Unique molecular signature. Blood 2004, 103, 2956–2964. [Google Scholar] [CrossRef]
- Krajinovic, M.; Lemieux-Blanchard, E.; Chiasson, S.; Primeau, M.; Costea, I.; Moghrabi, A. Role of polymorphisms in MTHFR and MTHFD1 genes in the outcome of childhood acute lymphoblastic leukemia. Pharm. J. 2004, 4, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Erčulj, N.; Kotnik, B.F.; Debeljak, M.; Jazbec, J.; Dolžan, V. Influence of folate pathway polymorphisms on high-dose methotrexate-related toxicity and survival in childhood acute lymphoblastic leukemia. Leuk. Lymphoma 2012, 53, 1096–1104. [Google Scholar] [CrossRef]
- de Jonge, R.; Hooijberg, J.H.; van Zelst, B.D.; Jansen, G.; van Zantwijk, C.H.; Kaspers, G.J.L.; Peters, F.G.; Ravindranath, Y.; Pieters, R.; Lindemans, J. Effect of polymorphisms in folate-related genes on in vitro methotrexate sensitivity in pediatric acute lymphoblastic leukemia. Blood 2005, 106, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Yin, S.; Zeng, H.; Li, H.; Wan, X. Regulation of Embryonic Stem Cell Self-Renewal. Life 2022, 12, 1151. [Google Scholar] [CrossRef] [PubMed]
- Burdon, T.; Smith, A.; Savatier, P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002, 12, 432–438. [Google Scholar] [CrossRef]
- Kraunsoe, S.; Azami, T.; Pei, Y.; Martello, G.; Jones, K.; Boroviak, T.; Nichols, J. Requirement for STAT3 and its target, TFCP2L1, in self-renewal of naïve pluripotent stem cells in vivo and in vitro. Biol. Open 2023, 12, bio059650. [Google Scholar] [CrossRef] [PubMed]
- Legate, K.R.; Montañez, E.; Kudlacek, O.; Fässler, R. ILK, PINCH and parvin: The tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 2006, 7, 20–31. [Google Scholar] [CrossRef]
- Braun, A.; Bordoy, R.; Stanchi, F.; Moser, M.; ünter Kostka, G.; Ehler, E.; Brandau, O.; Fässler, R. PINCH2 is a new five LIM domain protein, homologous to PINCHand localized to focal adhesions☆. Exp. Cell Res. 2003, 284, 237–248. [Google Scholar] [CrossRef]
- Xu, H.; Cao, H.; Xiao, G. Signaling via PINCH: Functions, binding partners and implications in human diseases. Gene 2016, 594, 10–15. [Google Scholar] [CrossRef]
- Su, J.; Guo, L.; Wu, C. A mechanoresponsive PINCH-1-Notch2 interaction regulates smooth muscle differentiation of human placental mesenchymal stem cells. Stem Cells 2021, 39, 650–668. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Fu, X.; Li, P.; Lin, S.; Yan, Q.; Lai, Y.; Liu, X.; Wang, Y.; Bai, X.; Liu, C.; et al. LIM domain proteins Pinch1/2 regulate chondrogenesis and bone mass in mice. Bone Res. 2020, 8, 37. [Google Scholar] [CrossRef]
- Liao, H.; Wang, F.; Lu, K.; Ma, X.; Yan, J.; Luo, L.; Sun, Y.; Liang, X. Requirement for PINCH in skeletal myoblast differentiation. Cell Tissue Res. 2023, 391, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Radli, M.; Veprintsev, D.B.; Rüdiger, S.G.D. Production and purification of human Hsp90β in Escherichia coli. PLoS ONE 2017, 12, e0180047. [Google Scholar] [CrossRef]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Buchner, J.; Li, J. Structure, function and regulation of the hsp90 machinery. Biomed. J. 2013, 36, 106. [Google Scholar] [CrossRef] [PubMed]
- Jing, R.; Duncan, C.B.; Duncan, S.A. A small-molecule screen reveals that HSP90β promotes the conversion of induced pluripotent stem cell-derived endoderm to a hepatic fate and regulates HNF4A turnover. Development 2017, 144, 1764–1774. [Google Scholar] [CrossRef]
- Nenasheva, V.V.; Kovaleva, G.V.; Khaidarova, N.V.; Novosadova, E.V.; Manuilova, E.S.; Antonov, S.A.; Tarantul, V.Z. Trim14 overexpression causes the same transcriptional changes in mouse embryonic stem cells and human HEK293 cells. Vitr. Cell. Dev. Biol. Anim. 2014, 50, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Schnier, J.; Schwelberger, H.G.; Smit-McBride, Z.; Kang, H.A.; Hershey, J.W. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1991, 11, 3105–3114. [Google Scholar] [CrossRef]
- Jasiulionis, M.G.; Luchessi, A.D.; Moreira, A.G.; Souza, P.P.; Suenaga, A.P.; Correa, M.; Costa, C.A.; Curi, R.; Costa-Neto, C.M. Inhibition of eukaryotic translation initiation factor 5A (eIF5A) hypusination impairs melanoma growth. Cell Biochem. Funct. 2007, 25, 109–114. [Google Scholar] [CrossRef]
- Park, M.H.; Lee, Y.B.; Joe, Y.A. Hypusine is essential for eukaryotic cell proliferation. Biol. Signals 1997, 6, 115–123. [Google Scholar] [CrossRef]
- Dihazi, H.; Dihazi, G.H.; Jahn, O.; Meyer, S.; Nolte, J.; Asif, A.R.; Mueller, G.A.; Engel, W. Multipotent adult germline stem cells and embryonic stem cells functional proteomics revealed an important role of eukaryotic initiation factor 5A (Eif5a) in stem cell differentiation. J. Proteome Res. 2011, 10, 1962–1973. [Google Scholar] [CrossRef]
- Luchessi, A.D.; Cambiaghi, T.D.; Hirabara, S.M.; Lambertucci, R.H.; Silveira, L.R.; Baptista, I.L.; Moriscot, A.S.; Costa-Neto, C.M.; Curi, R. Involvement of eukaryotic translation initiation factor 5A (eIF5A) in skeletal muscle stem cell differentiation. J. Cell. Physiol. 2009, 218, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Ingley, E. Functions of the Lyn tyrosine kinase in health and disease. Cell Commun. Signal. 2012, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Boggon, T.J.; Eck, M.J. Structure and regulation of Src family kinases. Oncogene 2004, 23, 7918–7927. [Google Scholar] [CrossRef] [PubMed]
- Resh, M.D. Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1999, 1451, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Simerly, C.; Hartnett, C.; Schatten, G.; Smithgall, T.E. Src-family tyrosine kinase activities are essential for differentiation of human embryonic stem cells. Stem Cell Res. 2014, 13, 379–389. [Google Scholar] [CrossRef]
- Meyn, M.A., 3rd; Schreiner, S.J.; Dumitrescu, T.P.; Nau, G.J.; Smithgall, T.E. SRC family kinase activity is required for murine embryonic stem cell growth and differentiation. Mol. Pharm. 2005, 68, 1320–1330. [Google Scholar] [CrossRef]
- Alvandi, Z.; Opas, M. c-Src kinase inhibits osteogenic differentiation via enhancing STAT1 stability. PLoS ONE 2020, 15, e0241646. [Google Scholar] [CrossRef]
- Chow, K.H.; Park, H.J.; George, J.; Yamamoto, K.; Gallup, A.D.; Graber, J.H.; Chen, Y.; Jiang, W.; Steindler, D.A.; Neilson, E.G.; et al. S100A4 Is a Biomarker and Regulator of Glioma Stem Cells That Is Critical for Mesenchymal Transition in Glioblastoma. Cancer Res. 2017, 77, 5360–5373. [Google Scholar] [CrossRef]
- Ishikawa, M.; Osaki, M.; Yamagishi, M.; Onuma, K.; Ito, H.; Okada, F.; Endo, H. Correlation of two distinct metastasis-associated proteins, MTA1 and S100A4, in angiogenesis for promoting tumor growth. Oncogene 2019, 38, 4715–4728. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Song, K.; Liu, S.; Zhang, H.; Wang, F.; Ni, C.; Zhai, W.; Liang, J.; Qin, Z.; et al. S100A4 promotes hepatocellular carcinogenesis by intensifying fibrosis-associated cancer cell stemness. Oncoimmunology 2020, 9, 1725355. [Google Scholar] [CrossRef]
- Mishra, S.K.; Siddique, H.R.; Saleem, M. S100A4 calcium-binding protein is key player in tumor progression and metastasis: Preclinical and clinical evidence. Cancer Metastasis Rev. 2012, 31, 163–172. [Google Scholar] [CrossRef]
- Tochimoto, M.; Oguri, Y.; Hashimura, M.; Konno, R.; Matsumoto, T.; Yokoi, A.; Kodera, Y.; Saegusa, M. S100A4/non-muscle myosin II signaling regulates epithelial-mesenchymal transition and stemness in uterine carcinosarcoma. Lab. Invest. 2020, 100, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Stary, M.; Schneider, M.; Sheikh, S.P.; Weitzer, G. Parietal endoderm secreted S100A4 promotes early cardiomyogenesis in embryoid bodies. Biochem. Biophys. Res. Commun. 2006, 343, 555–563. [Google Scholar] [CrossRef]
- Schneider, M.; Hansen, J.L.; Sheikh, S.P. S100A4: A common mediator of epithelial–mesenchymal transition, fibrosis and regeneration in diseases? J. Mol. Med. 2008, 86, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Figeac, N.; Serralbo, O.; Marcelle, C.; Zammit, P.S. ErbB3 binding protein-1 (Ebp1) controls proliferation and myogenic differentiation of muscle stem cells. Dev. Biol. 2014, 386, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Neilson, K.M.; Abbruzzesse, G.; Kenyon, K.; Bartolo, V.; Krohn, P.; Alfandari, D.; Moody, S.A. Pa2G4 is a novel Six1 co-factor that is required for neural crest and otic development. Dev. Biol. 2017, 421, 171–182. [Google Scholar] [CrossRef]
- Yoo, J.; Wang, X.; Rishi, A.; Lessor, T.; Xia, X.; Gustafson, T.; Hamburger, A. Interaction of the PA2G4 (EBP1) protein with ErbB-3 and regulation of this binding by heregulin. Br. J. Cancer 2000, 82, 683–690. [Google Scholar] [CrossRef]
- Somanath, P.; Bush, K.M.; Knoepfler, P.S. ERBB3-Binding Protein 1 (EBP1) Is a Novel Developmental Pluripotency-Associated-4 (DPPA4) Cofactor in Human Pluripotent Cells. Stem Cells 2018, 36, 671–682. [Google Scholar] [CrossRef]
- Song, Y.; Cao, P.; Gu, Z.; Xiao, J.; Lian, M.; Huang, D.; Xing, J.; Zhang, Y.; Feng, X.; Wang, C. The Role of Neuropilin-1-FYN Interaction in Odontoblast Differentiation of Dental Pulp Stem Cells. Cell Reprogram 2018, 20, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Thompson, W.R.; Guilluy, C.; Xie, Z.; Sen, B.; Brobst, K.E.; Yen, S.S.; Uzer, G.; Styner, M.; Case, N.; Burridge, K.; et al. Mechanically activated fyn utilizes mTORC2 to regulate RhoA and adipogenesis in mesenchymal stem cells. Stem Cells 2013, 31, 2528–2537. [Google Scholar] [CrossRef]
- Brzozowski, J.S.; Skelding, K.A. The Multi-Functional Calcium/Calmodulin Stimulated Protein Kinase (CaMK) Family: Emerging Targets for Anti-Cancer Therapeutic Intervention. Pharmaceuticals 2019, 12, 8. [Google Scholar] [CrossRef]
- Takemoto-Kimura, S.; Suzuki, K.; Horigane, S.I.; Kamijo, S.; Inoue, M.; Sakamoto, M.; Fujii, H.; Bito, H. Calmodulin kinases: Essential regulators in health and disease. J. Neurochem. 2017, 141, 808–818. [Google Scholar] [CrossRef]
- Skelding, K.A.; Rostas, J.A.P. Regulation of Multifunctional Calcium/Calmodulin Stimulated Protein Kinases by Molecular Targeting. Adv. Exp. Med. Biol. 2020, 1131, 649–679. [Google Scholar] [CrossRef]
- Boutz, P.L.; Stoilov, P.; Li, Q.; Lin, C.-H.; Chawla, G.; Ostrow, K.; Shiue, L.; Ares, M.; Black, D.L. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007, 21, 1636–1652. [Google Scholar] [CrossRef] [PubMed]
- Coutinho-Mansfield, G.C.; Xue, Y.; Zhang, Y.; Fu, X.D. PTB/nPTB switch: A post-transcriptional mechanism for programming neuronal differentiation. Genes Dev. 2007, 21, 1573–1577. [Google Scholar] [CrossRef]
- Dai, S.; Wang, C.; Zhang, C.; Feng, L.; Zhang, W.; Zhou, X.; He, Y.; Xia, X.; Chen, B.; Song, W. PTB: Not just a polypyrimidine tract-binding protein. J. Cell. Physiol. 2022, 237, 2357–2373. [Google Scholar] [CrossRef] [PubMed]
- Rehn, M.; Wenzel, A.; Frank, A.K.; Schuster, M.B.; Pundhir, S.; Jørgensen, N.; Vitting-Seerup, K.; Ge, Y.; Jendholm, J.; Michaut, M.; et al. PTBP1 promotes hematopoietic stem cell maintenance and red blood cell development by ensuring sufficient availability of ribosomal constituents. Cell Rep. 2022, 39, 110793. [Google Scholar] [CrossRef]
- Iannone, C.; Kainov, Y.; Zhuravskaya, A.; Hamid, F.; Nojima, T.; Makeyev, E.V. PTBP1-activated co-transcriptional splicing controls epigenetic status of pluripotent stem cells. Mol. Cell 2023, 83, 203–218.e209. [Google Scholar] [CrossRef]
- Wang, D.H.; Chen, J.S.; Hou, R.; Li, Y.; An, J.H.; He, P.; Cai, Z.G.; Liang, X.H.; Liu, Y.L. Comparison of transcriptome profiles of mesenchymal stem cells derived from umbilical cord and bone marrow of giant panda (Ailuropoda melanoleuca). Gene 2022, 845, 146854. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Hai, J.; Liu, W.; Luo, Y.; Chen, K.; Xin, Y.; Pan, J.; Hu, Y.; Gao, Q.; Xiao, F. Gallic Acid Induces Neural Stem Cell Differentiation into Neurons and Proliferation through the MAPK/ERK Pathway. J. Agric. Food Chem. 2021, 69, 12456–12464. [Google Scholar] [CrossRef]
- Maruyama, T.; Jiang, M.; Abbott, A.; Yu, H.I.; Huang, Q.; Chrzanowska-Wodnicka, M.; Chen, E.I.; Hsu, W. Rap1b Is an Effector of Axin2 Regulating Crosstalk of Signaling Pathways During Skeletal Development. J. Bone Min. Res. 2017, 32, 1816–1828. [Google Scholar] [CrossRef]
- Gao, S.; Chen, B.; Zhu, Z.; Du, C.; Zou, J.; Yang, Y.; Huang, W.; Liao, J. PI3K-Akt signaling regulates BMP2-induced osteogenic differentiation of mesenchymal stem cells (MSCs): A transcriptomic landscape analysis. Stem Cell Res. 2023, 66, 103010. [Google Scholar] [CrossRef] [PubMed]
- Halfon, S.; Abramov, N.; Grinblat, B.; Ginis, I. Markers distinguishing mesenchymal stem cells from fibroblasts are downregulated with passaging. Stem Cells Dev. 2011, 20, 53–66. [Google Scholar] [CrossRef]
- Ise, H.; Matsunaga, K.; Shinohara, M.; Sakai, Y. Improved Isolation of Mesenchymal Stem Cells Based on Interactions between N-Acetylglucosamine-Bearing Polymers and Cell-Surface Vimentin. Stem Cells Int. 2019, 2019, 4341286. [Google Scholar] [CrossRef]
- Lee, T.C.; Lee, T.H.; Huang, Y.H.; Chang, N.K.; Lin, Y.J.; Chien, P.W.; Yang, W.H.; Lin, M.H. Comparison of surface markers between human and rabbit mesenchymal stem cells. PLoS ONE 2014, 9, e111390. [Google Scholar] [CrossRef]
- Malekmohammad, K.; Sewell, R.D.E.; Rafieian-Kopaei, M. Antioxidants and Atherosclerosis: Mechanistic Aspects. Biomolecules 2019, 9, 301. [Google Scholar] [CrossRef]
- Yang, J.; Song, T.; Wu, P.; Chen, Y.; Fan, X.; Chen, H.; Zhang, J.; Huang, C. Differentiation potential of human mesenchymal stem cells derived from adipose tissue and bone marrow to sinus node-like cells. Mol. Med. Rep. 2012, 5, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Paspaliaris, V.; Kolios, G. Stem cells in Osteoporosis: From Biology to New Therapeutic Approaches. Stem Cells Int. 2019, 2019, 1730978. [Google Scholar] [CrossRef]
- Ahlström, M.; Pekkinen, M.; Lamberg-Allardt, C. Dexamethasone downregulates the expression of parathyroid hormone-related protein (PTHrP) in mesenchymal stem cells. Steroids 2009, 74, 277–282. [Google Scholar] [CrossRef]
- Lyu, P.; Li, B.; Li, P.; Bi, R.; Cui, C.; Zhao, Z.; Zhou, X.; Fan, Y. Parathyroid Hormone 1 Receptor Signaling in Dental Mesenchymal Stem Cells: Basic and Clinical Implications. Front. Cell Dev. Biol. 2021, 9, 654715. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Hanai, J.I.; Le, P.T.; Bi, R.; Maridas, D.; DeMambro, V.; Figueroa, C.A.; Kir, S.; Zhou, X.; Mannstadt, M.; et al. Parathyroid Hormone Directs Bone Marrow Mesenchymal Cell Fate. Cell Metab. 2017, 25, 661–672. [Google Scholar] [CrossRef]
- Reeve, J.; Meunier, P.J.; Parsons, J.A.; Bernat, M.; Bijvoet, O.; Courpron, P.; Edouard, C.; Klenerman, L.; Neer, R.M.; Renier, J.C. Anabolic effect of human parathyroid hormone fragment on trabecular bone in involutional osteoporosis: A multicentre trial. Br. Med. J. 1980, 280, 1340–1344. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wang, Y.; Hao, Z.; Hu, Y.; Li, J. Parathyroid hormone and its related peptides in bone metabolism. Biochem. Pharmacol. 2021, 192, 114669. [Google Scholar] [CrossRef] [PubMed]
- Sauzay, C.; Voutetakis, K.; Chatziioannou, A.; Chevet, E.; Avril, T. CD90/Thy-1, a Cancer-Associated Cell Surface Signaling Molecule. Front. Cell Dev. Biol. 2019, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Leyton, L.; Díaz, J.; Martínez, S.; Palacios, E.; Pérez, L.A.; Pérez, R.D. Thy-1/CD90 a Bidirectional and Lateral Signaling Scaffold. Front. Cell Dev. Biol. 2019, 7, 132. [Google Scholar] [CrossRef]
- Rege, T.A.; Hagood, J.S. Thy-1 as a regulator of cell-cell and cell-matrix interactions in axon regeneration, apoptosis, adhesion, migration, cancer, and fibrosis. FASEB J. 2006, 20, 1045–1054. [Google Scholar] [CrossRef]
- Horn, P.A.; Tesch, H.; Staib, P.; Kube, D.; Diehl, V.; Voliotis, D. Expression of AC133, a novel hematopoietic precursor antigen, on acute myeloid leukemia cells. Blood 1999, 93, 1435–1437. [Google Scholar] [CrossRef] [PubMed]
- Majka, M.; Ratajczak, J.; Machalinski, B.; Carter, A.; Pizzini, D.; Wasik, M.A.; Gewirtz, A.M.; Ratajczak, M.Z. Expression, regulation and function of AC133, a putative cell surface marker of primitive human haematopoietic cells. Folia Histochem. Cytobiol. 2000, 38, 53–63. [Google Scholar] [PubMed]
- Patel, P.; Abbasian, J.; Mahmud, D.; Mahmud, N.; Horsthemke, P.M.; Chunduri, S.; Rondelli, D. Allostimulatory activity of CD133+ hematopoietic cells. Bone Marrow Transpl. 2013, 48, 742–744. [Google Scholar] [CrossRef] [PubMed]
- Kucia, M.; Halasa, M.; Wysoczynski, M.; Baskiewicz-Masiuk, M.; Moldenhawer, S.; Zuba-Surma, E.; Czajka, R.; Wojakowski, W.; Machalinski, B.; Ratajczak, M.Z. Morphological and molecular characterization of novel population of CXCR4+ SSEA-4+ Oct-4+ very small embryonic-like cells purified from human cord blood: Preliminary report. Leukemia 2007, 21, 297–303. [Google Scholar] [CrossRef]
- Ratajczak, M.Z.; Mierzejewska, K.; Ratajczak, J.; Kucia, M. CD133 Expression Strongly Correlates with the Phenotype of Very Small Embryonic-/Epiblast-Like Stem Cells. Adv. Exp. Med. Biol. 2013, 777, 125–141. [Google Scholar] [CrossRef]
- Virant-Klun, I.; Zech, N.; Rozman, P.; Vogler, A.; Cvjeticanin, B.; Klemenc, P.; Malicev, E.; Meden-Vrtovec, H. Putative stem cells with an embryonic character isolated from the ovarian surface epithelium of women with no naturally present follicles and oocytes. Differentiation 2008, 76, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Wakao, S.; Kushida, Y.; Tatsumi, K.; Kitada, M.; Abe, T.; Niizuma, K.; Tominaga, T.; Kushimoto, S.; Dezawa, M. A Novel Type of Stem Cells Double-Positive for SSEA-3 and CD45 in Human Peripheral Blood. Cell Transpl. 2020, 29, 963689720923574. [Google Scholar] [CrossRef]
- Parte, S.; Bhartiya, D.; Telang, J.; Daithankar, V.; Salvi, V.; Zaveri, K.; Hinduja, I. Detection, characterization, and spontaneous differentiation in vitro of very small embryonic-like putative stem cells in adult mammalian ovary. Stem Cells Dev. 2011, 20, 1451–1464. [Google Scholar] [CrossRef] [PubMed]
- Dzierzak, E.; de Pater, E. Regulation of Blood Stem Cell Development. Curr. Top. Dev. Biol. 2016, 118, 1–20. [Google Scholar] [CrossRef]
- Dzierzak, E.; Speck, N.A. Of lineage and legacy: The development of mammalian hematopoietic stem cells. Nat. Immunol. 2008, 9, 129–136. [Google Scholar] [CrossRef]
- Orkin, S.H.; Zon, L.I. Hematopoiesis: An evolving paradigm for stem cell biology. Cell 2008, 132, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Boltze, J.; Arnold, A.; Walczak, P.; Jolkkonen, J.; Cui, L.; Wagner, D.C. The Dark Side of the Force—Constraints and Complications of Cell Therapies for Stroke. Front. Neurol. 2015, 6, 155. [Google Scholar] [CrossRef] [PubMed]
- Lengefeld, J.; Amon, A. 3022—CELL SIZE IS A DETERMINANT OF STEM CELL POTENTIAL DURING AGING. Exp. Hematol. 2021, 100, S54. [Google Scholar] [CrossRef]
- Gao, D.Y.; Chang, Q.; Liu, C.; Farris, K.; Harvey, K.; McGann, L.E.; English, D.; Jansen, J.; Critser, J.K. Fundamental cryobiology of human hematopoietic progenitor cells. I: Osmotic characteristics and volume distribution. Cryobiology 1998, 36, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Bhartiya, D.; Shaikh, A.; Nagvenkar, P.; Kasiviswanathan, S.; Pethe, P.; Pawani, H.; Mohanty, S.; Rao, S.G.; Zaveri, K.; Hinduja, I. Very small embryonic-like stem cells with maximum regenerative potential get discarded during cord blood banking and bone marrow processing for autologous stem cell therapy. Stem Cells Dev. 2012, 21, 1–6. [Google Scholar] [CrossRef]


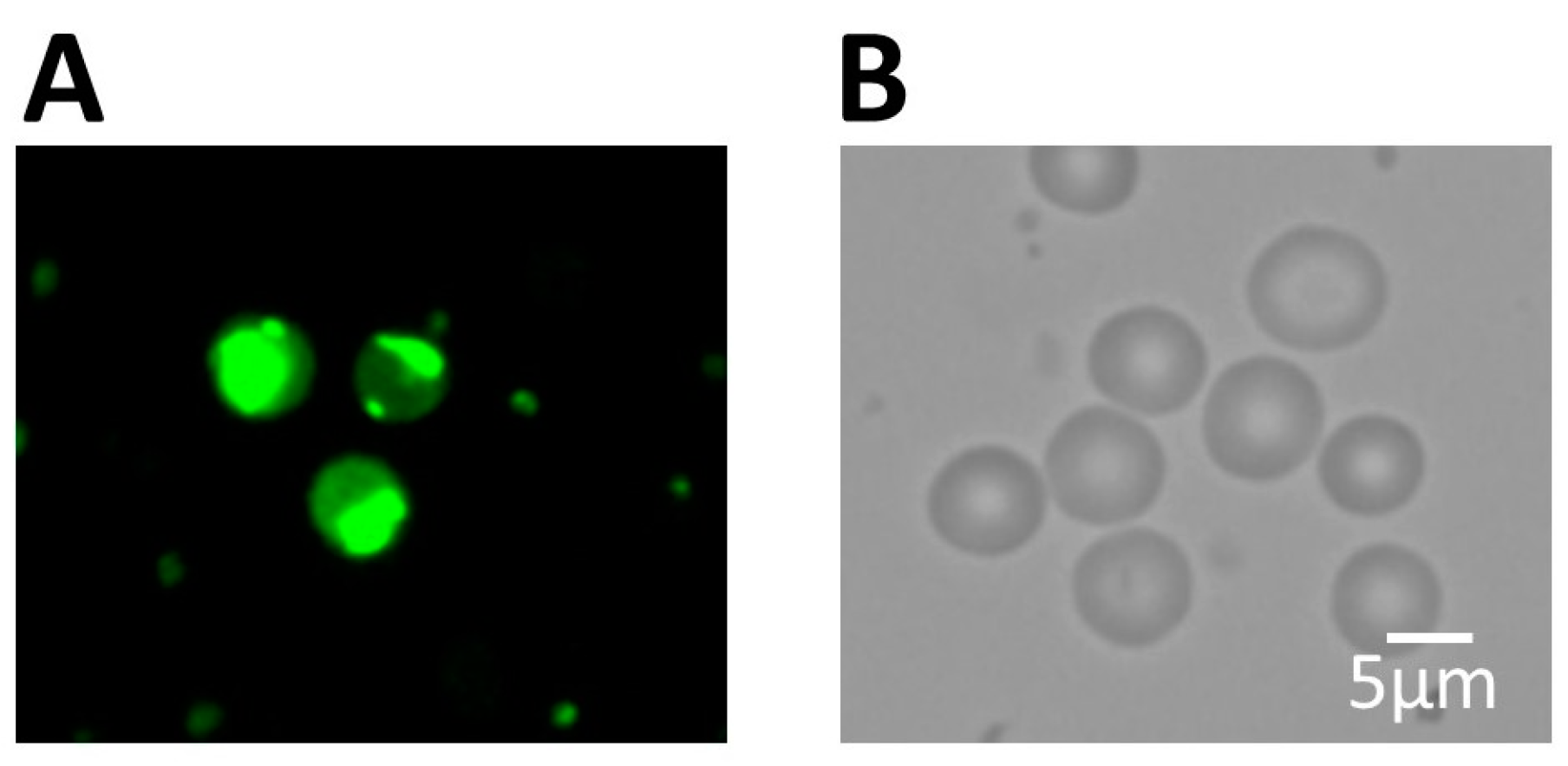
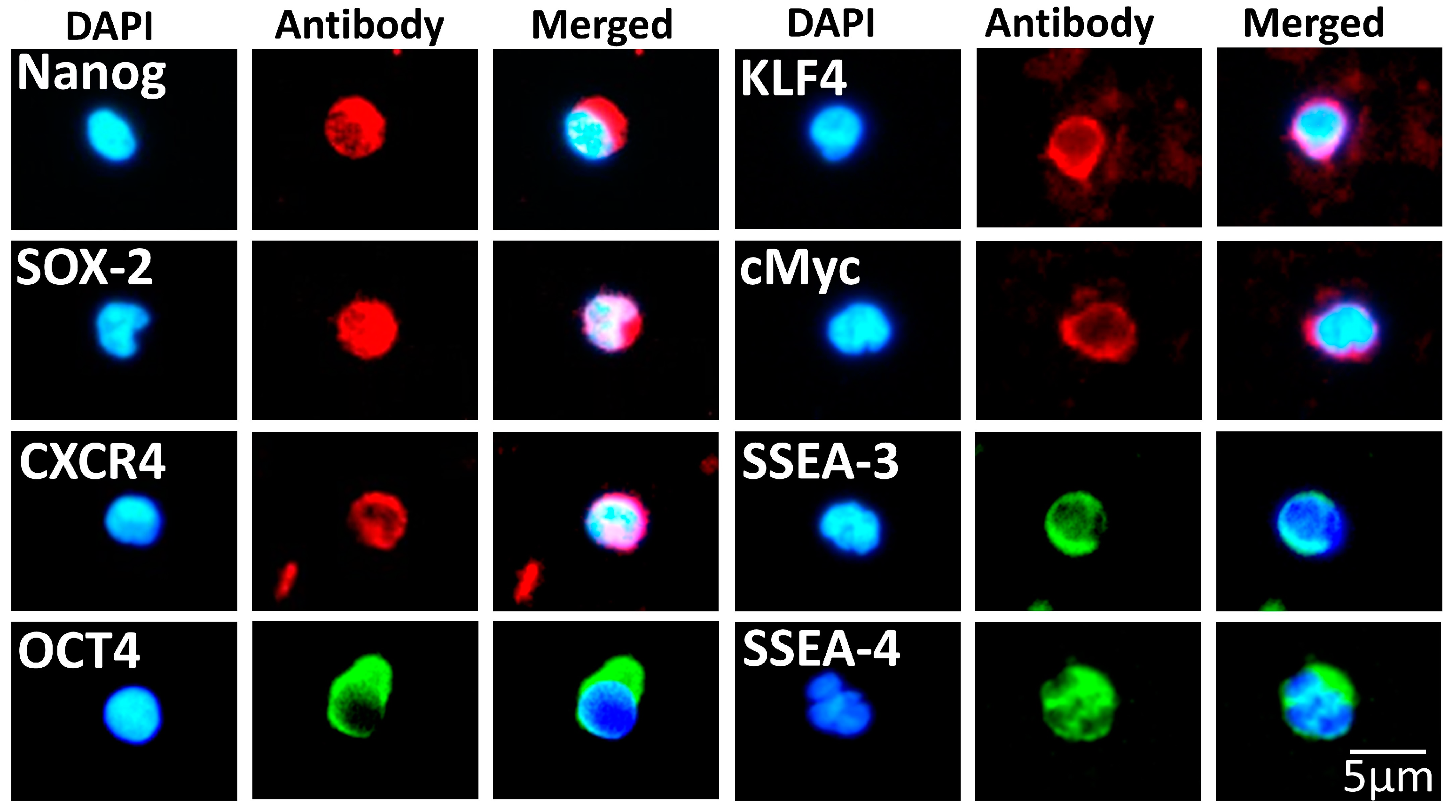
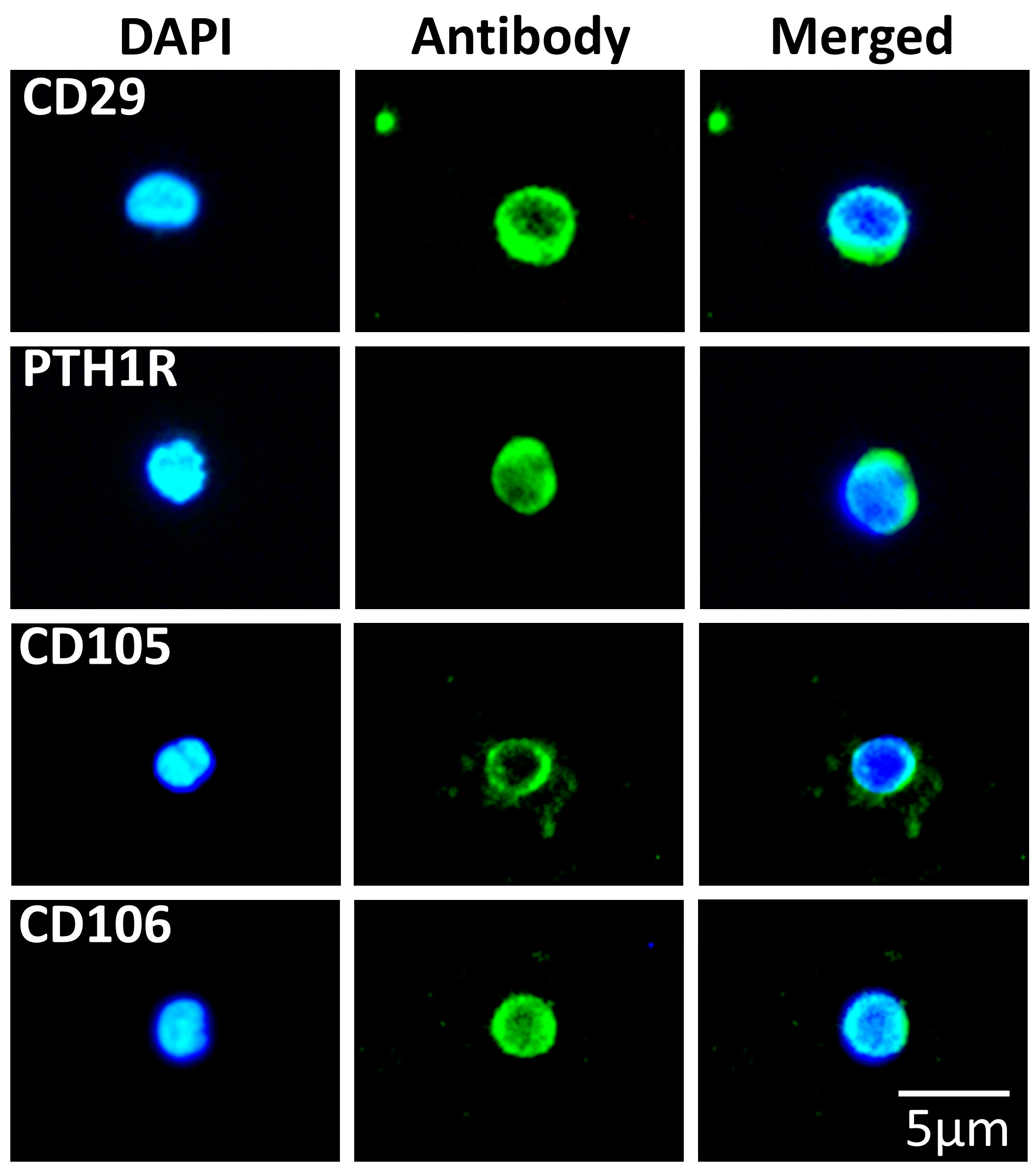
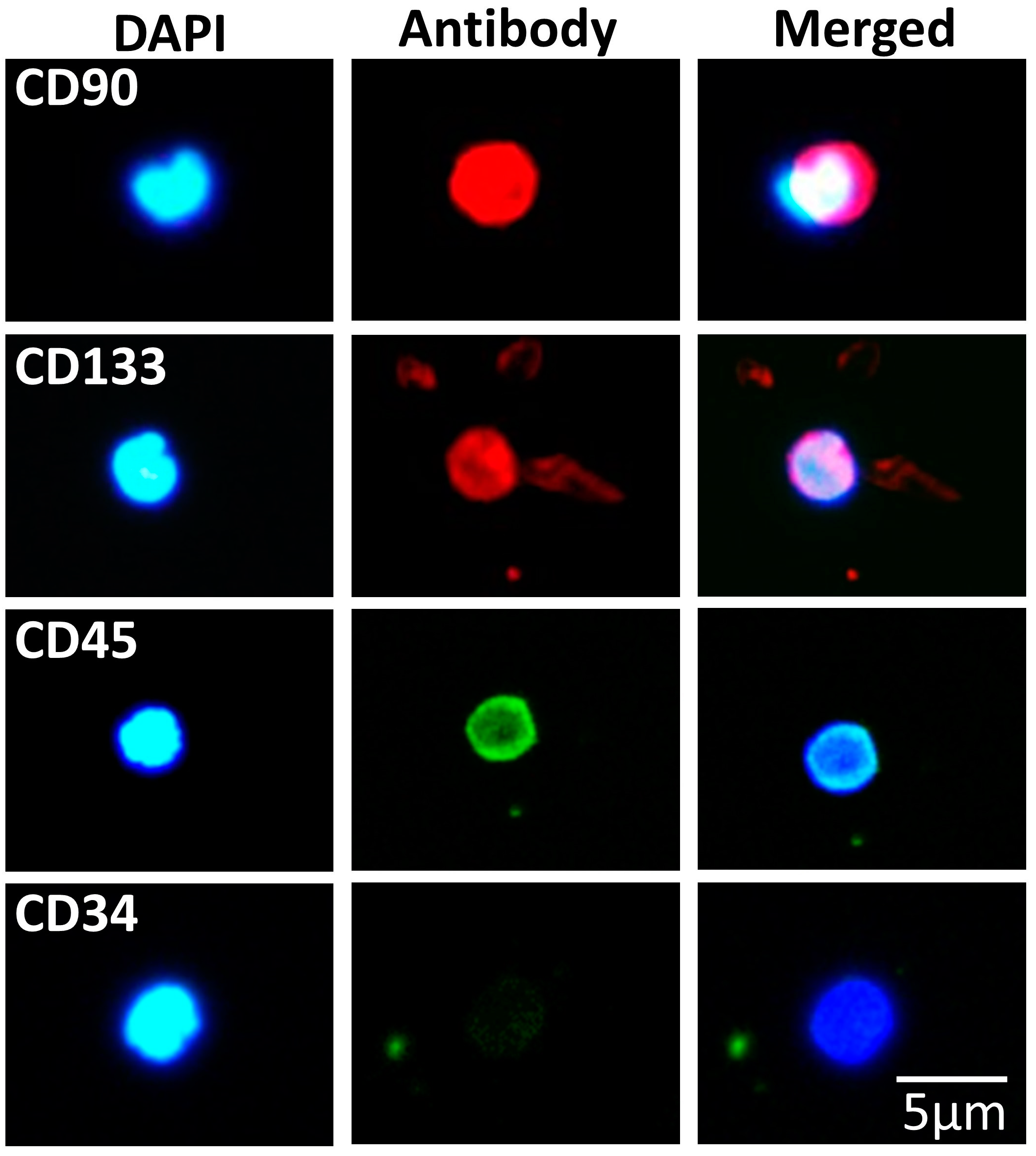


| Pluripotency | Antigen |
|---|---|
| Embryonic | Nanog 1, Oct4 1, CXCR4 1, SOX2 1, KLF4 1, cMyc 1, SSEA-3 1 and SSEA-4 2 |
| Mesenchymal | CD29 3, PTH1R 1, CD105 1 and CD106 3 |
| Hematopoietic | CD45 3, CD34 1, CD90 1 and CD133 1 |
| Protein | Functional Role | Role in Stem Cell Differentiation |
|---|---|---|
| LIMS1 | Adaptor protein that binds to integrin-linked kinase and has a vital role in the processes of cell adhesion [125,126,127] | Promotes chondrogenesis, differentiation of placental mesenchymal cells to smooth muscle cells and plays a crucial part in skeletal myogenic differentiation [128,129,130] |
| HSP90AB1 | Molecular chaperone involved in protein degradation [131,132,133] | Plays a role in the endoderm differentiation towards hepatic cells [134] and in mesodermal differentiation [135] |
| EIF5A | Associated with protein translation and cell proliferation and viability [136,137,138] | Supports embryonic stem cell differentiation and promotes skeletal muscle stem cell differentiation [139,140] |
| LYN | Belongs to the Src-family of tyrosine kinases and it is a nonreceptor cytoplasmic protein. It has a pivotal role in various cellular processes such as migration, cell growth, apoptosis, adhesion, differentiation, metabolism and immune response [141,142,143] | It has significant role in the differentiation process of embryonic stem cells [144,145,146] |
| S100A4 | Belongs to the S100 family of calcium-binding proteins and is involved in various processes, such as angiogenesis, cell growth, motility, differentiation, apoptosis and invasion [147,148,149,150,151] | Promotes the differentiation of endoderm towards cardiomyocytes [152] and has a central role in the epithelial-to-mesenchymal transition [153] |
| PA2G4 | It is a protein regulator of the ErbB3 signaling pathway, and it plays a role in cell growth, apoptosis and differentiation [154,155,156] | It plays a significant role in the differentiation of muscle stem cells and pluripotency of stem cells [154,157] |
| FYN | Belongs to the Src-family of tyrosine kinases and it is involved in various processes, such as cell growth, survival, adhesion, motility and tumor metastasis [142,143,158] | It is implicated in the differentiation process of mesenchymal stem cells [159] and has a significant role in the self-renewal of murine embryonic stem cells [145] |
| CAMK1 | It is a protein kinase, primarily expressed in nerve cells, with various functions [160,161,162] | Unknown |
| PTBP1 | Belongs to the heterogeneous nuclear ribonucleoprotein family and has a pivotal role in neuronal development [163,164,165] | It plays a significant role in the self-renewal of hematopoietic stem cells [166] and in the neuronal development [167] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filidou, E.; Kandilogiannakis, L.; Tarapatzi, G.; Spathakis, M.; Su, C.; Rai, A.; Greening, D.W.; Arvanitidis, K.; Paspaliaris, V.; Kolios, G. A Simplified and Effective Approach for the Isolation of Small Pluripotent Stem Cells Derived from Human Peripheral Blood. Biomedicines 2023, 11, 787. https://doi.org/10.3390/biomedicines11030787
Filidou E, Kandilogiannakis L, Tarapatzi G, Spathakis M, Su C, Rai A, Greening DW, Arvanitidis K, Paspaliaris V, Kolios G. A Simplified and Effective Approach for the Isolation of Small Pluripotent Stem Cells Derived from Human Peripheral Blood. Biomedicines. 2023; 11(3):787. https://doi.org/10.3390/biomedicines11030787
Chicago/Turabian StyleFilidou, Eirini, Leonidas Kandilogiannakis, Gesthimani Tarapatzi, Michail Spathakis, Colin Su, Alin Rai, David W. Greening, Konstantinos Arvanitidis, Vasilis Paspaliaris, and George Kolios. 2023. "A Simplified and Effective Approach for the Isolation of Small Pluripotent Stem Cells Derived from Human Peripheral Blood" Biomedicines 11, no. 3: 787. https://doi.org/10.3390/biomedicines11030787
APA StyleFilidou, E., Kandilogiannakis, L., Tarapatzi, G., Spathakis, M., Su, C., Rai, A., Greening, D. W., Arvanitidis, K., Paspaliaris, V., & Kolios, G. (2023). A Simplified and Effective Approach for the Isolation of Small Pluripotent Stem Cells Derived from Human Peripheral Blood. Biomedicines, 11(3), 787. https://doi.org/10.3390/biomedicines11030787





