Evaluation of CDK9 Inhibition by Dinaciclib in Combination with Apoptosis Modulating izTRAIL for the Treatment of Colorectal Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Samples
2.2. Cell Lines
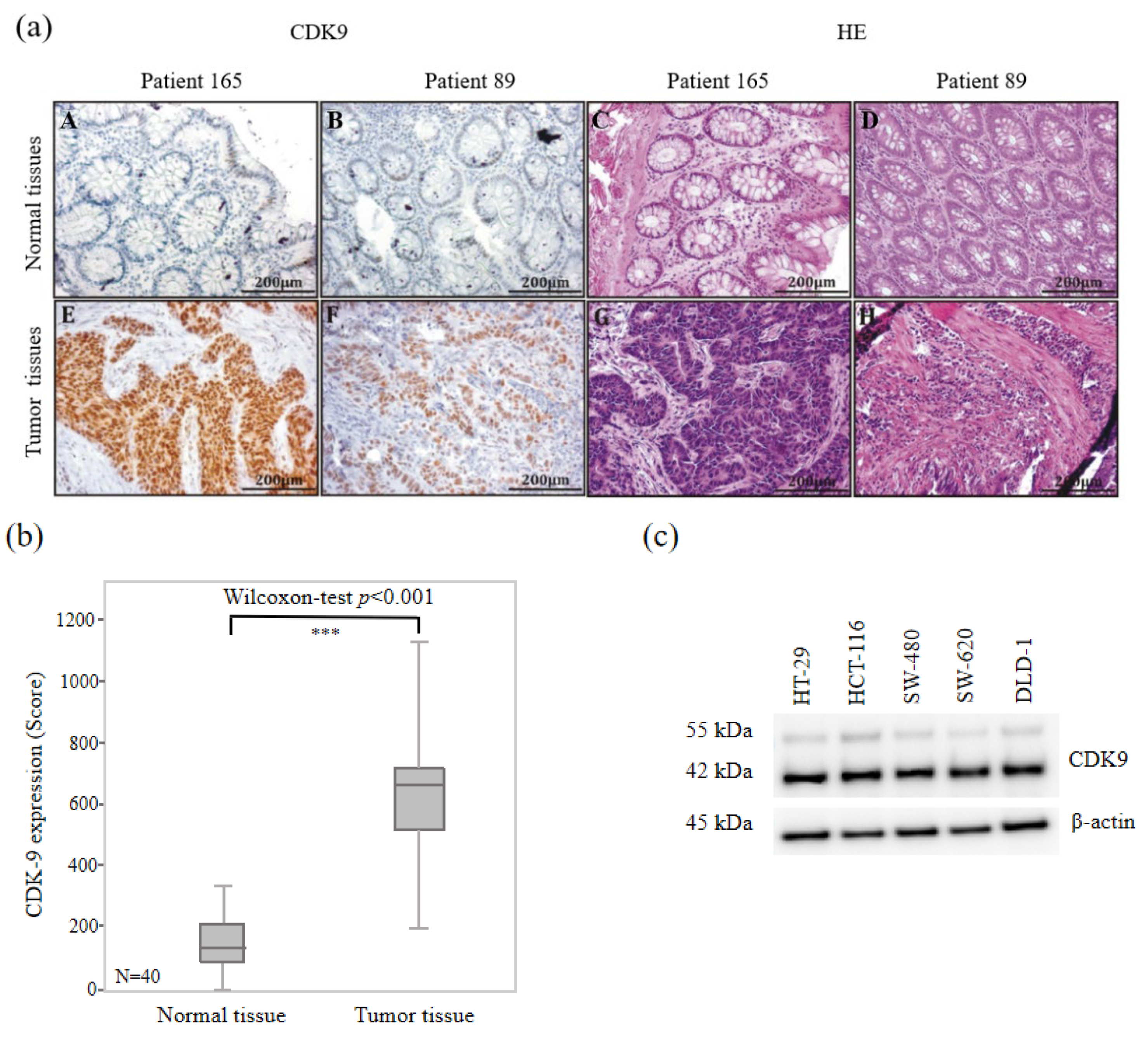
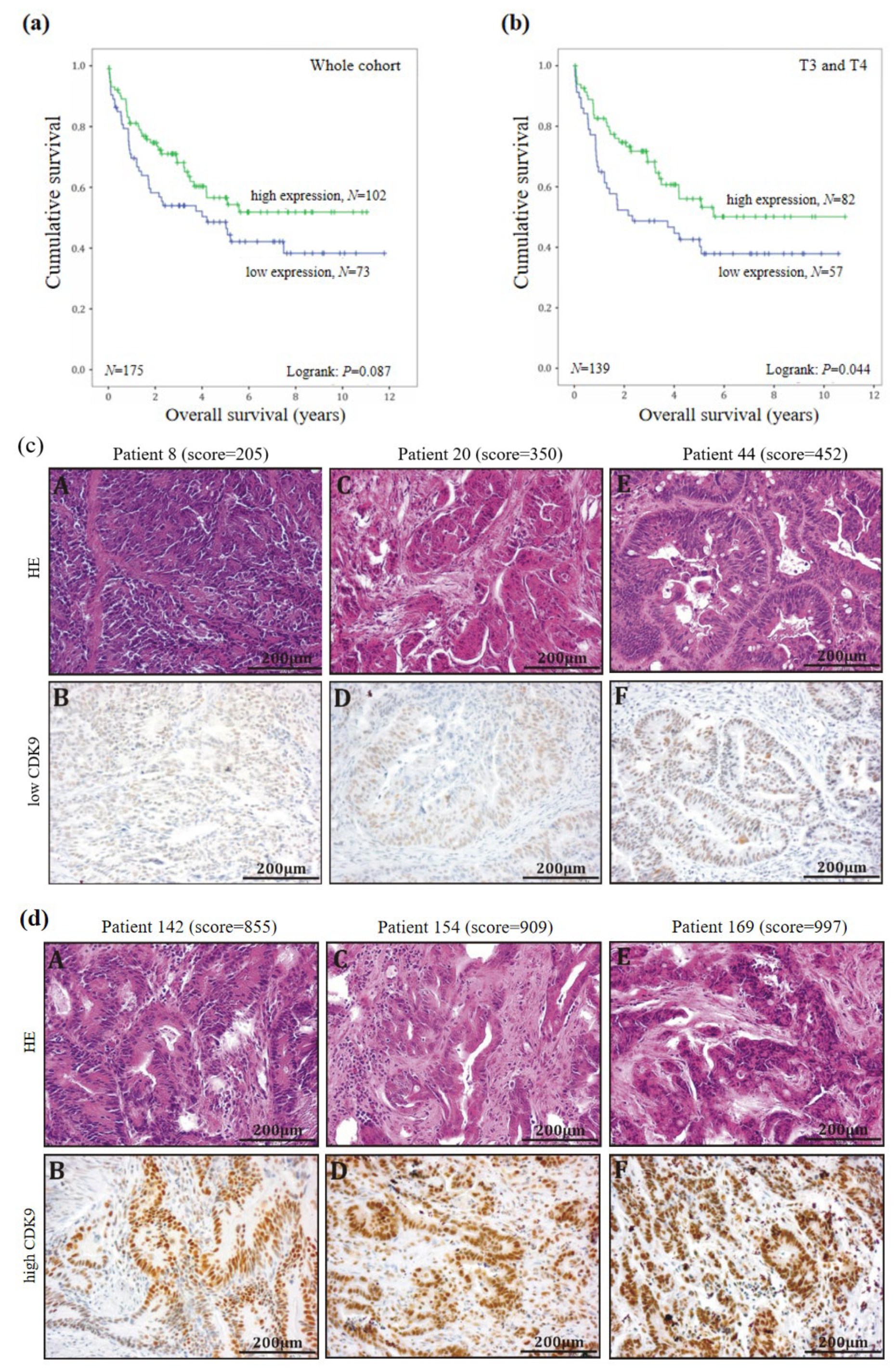
2.3. Immunohistochemistry
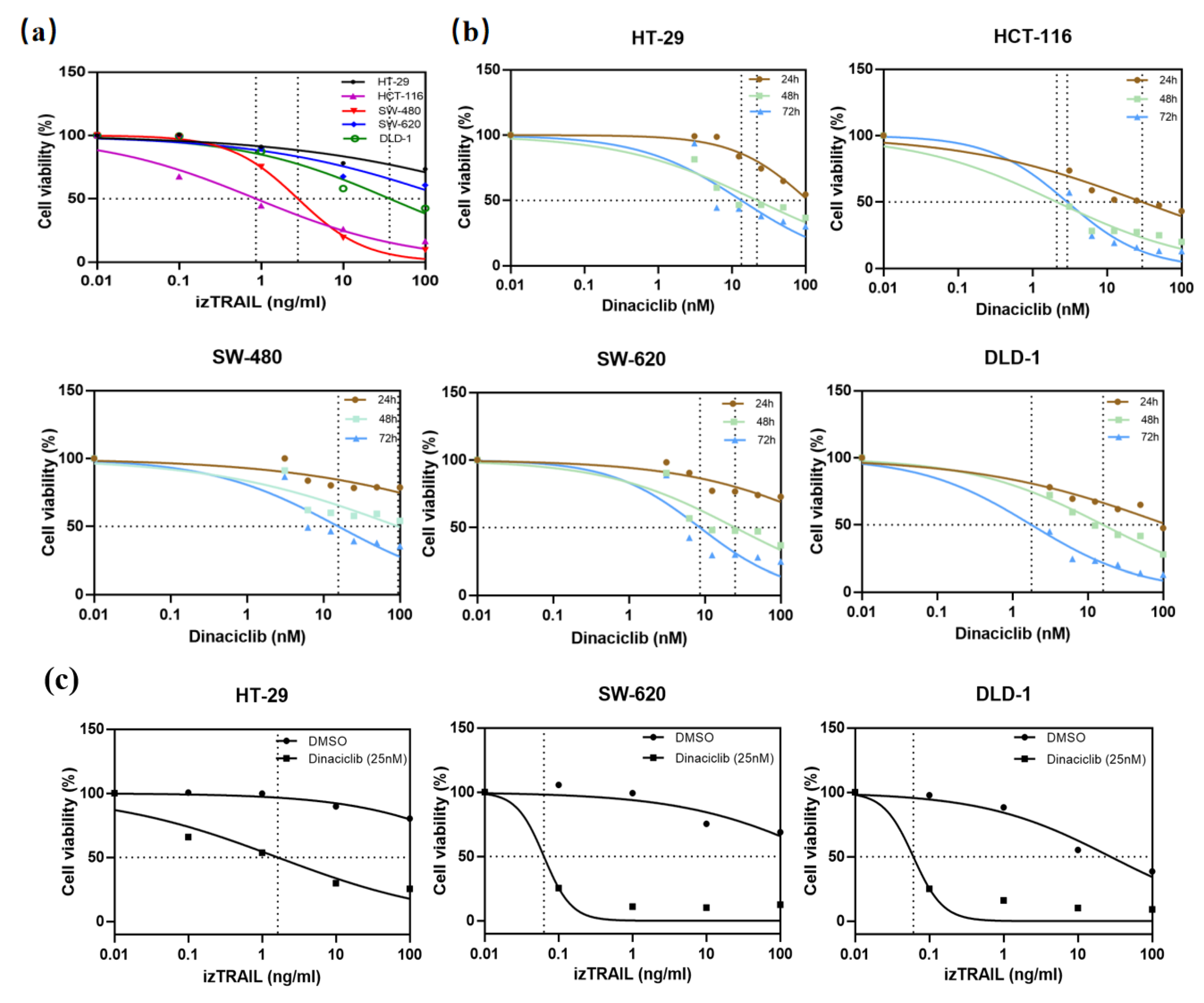
2.4. Cell Viability Assay
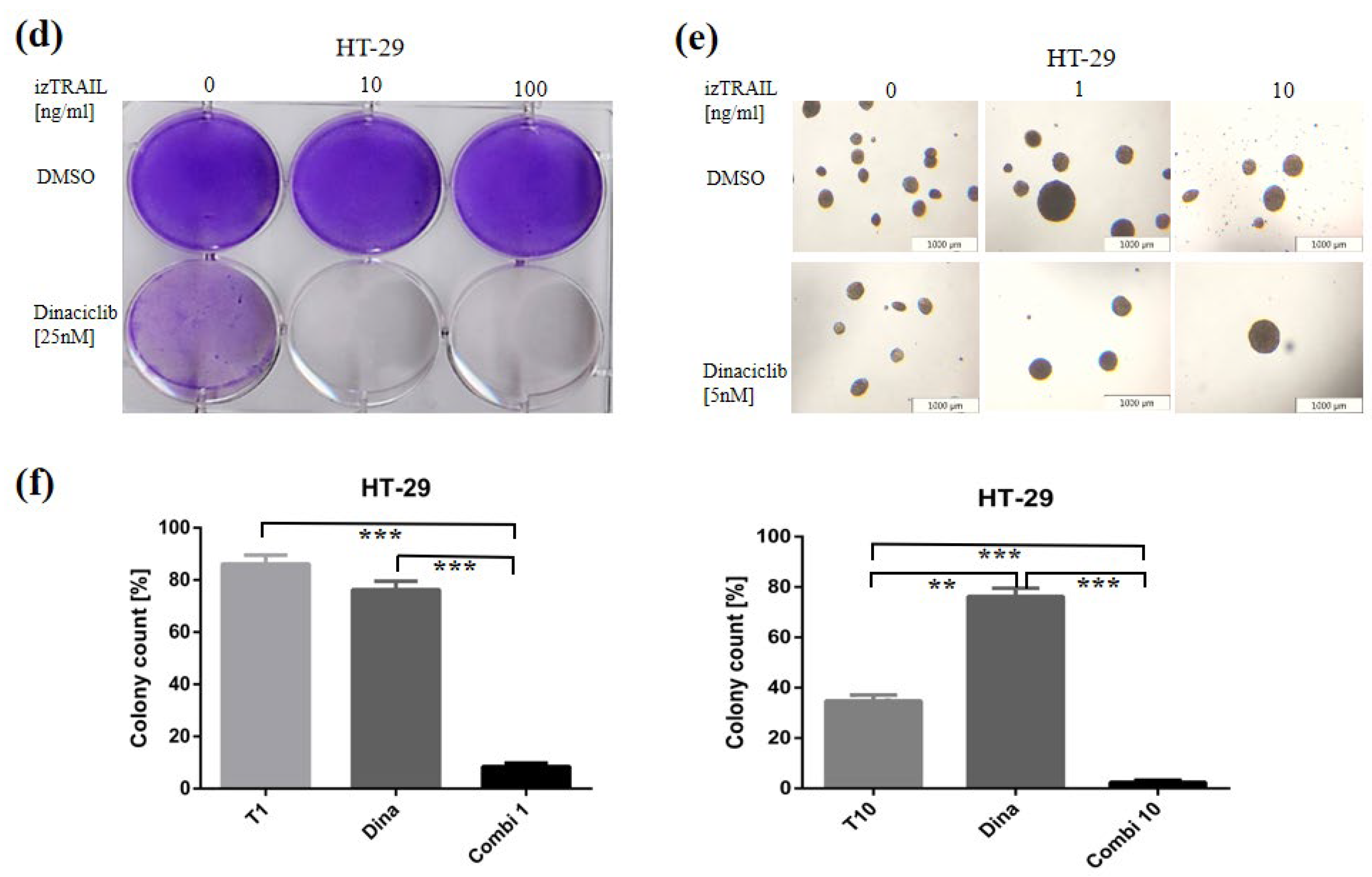
2.5. Analysis of Long-Term Survival and Colony Formation
2.6. Analysis of Cell Cycle and Apoptosis by Flow Cytometry
2.7. Western Blot Analysis
2.8. Statistical Analysis
3. Results
3.1. Patients’ Clinical Characteristics
3.2. High CDK9 Expression in Colorectal Cancer Tissues
3.3. CDK9 as a Positive Prognostic Maker in Colorectal Cancer
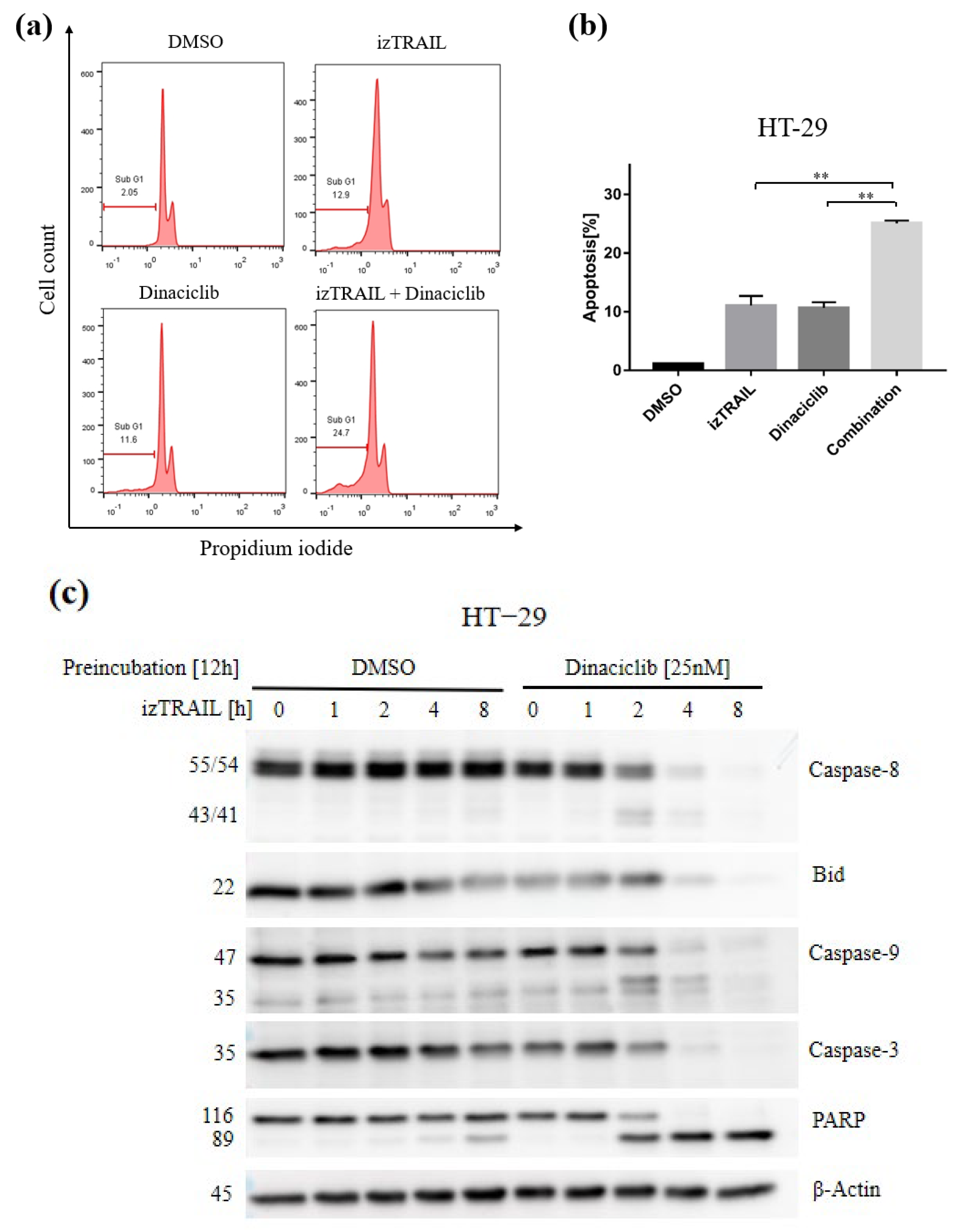
3.4. CDK9 Inhibition by Dinaciclib Sensitizes Colorectal Cancer Cell Lines to izTRAIL Treatment
3.5. The Novel Combination with CDK9 Inhibition Enhances TRAIL-Mediated Cell Apoptosis by Apoptotic Pathways
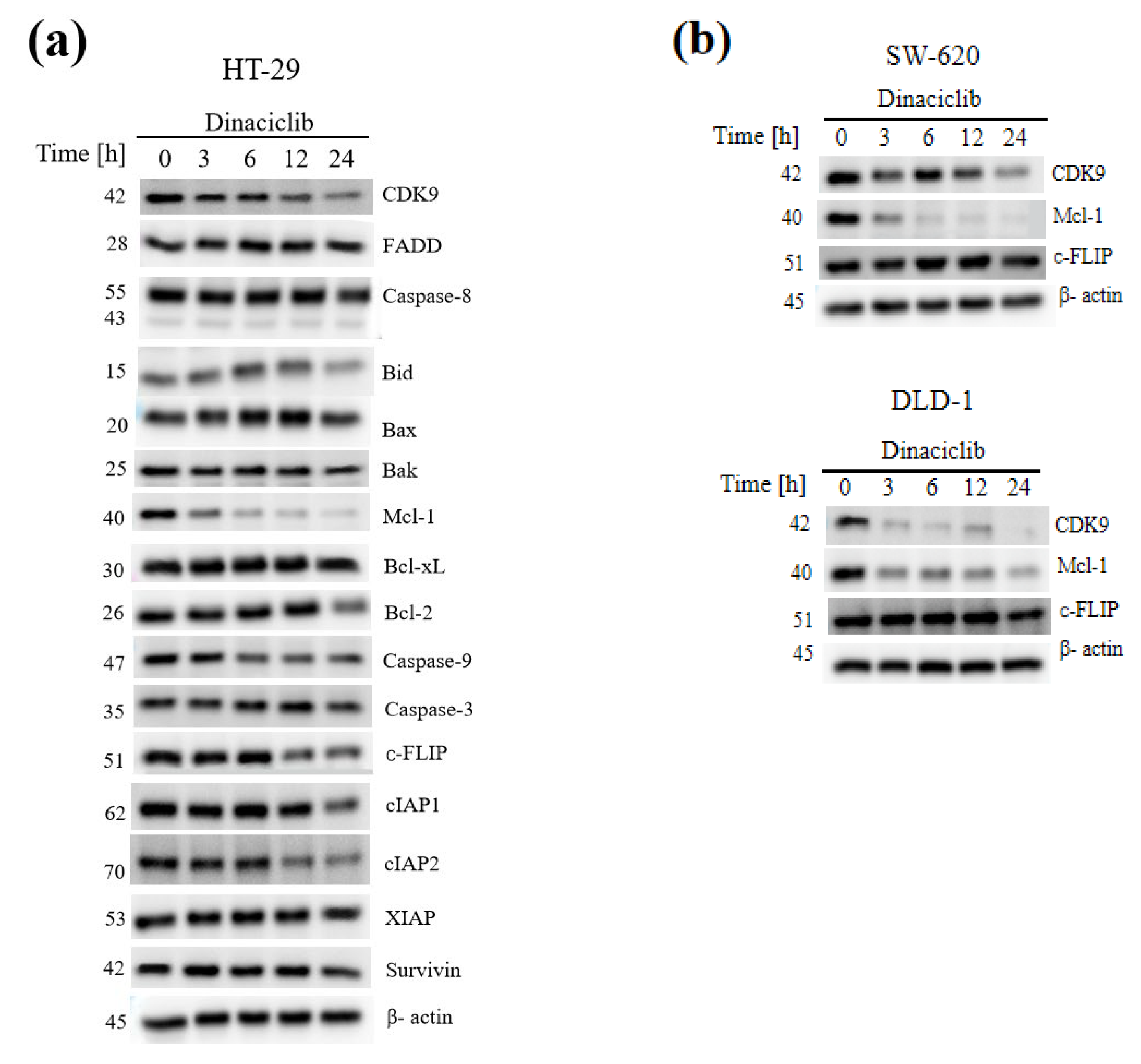
3.6. CDK9 Inhibition Overcomes TRAIL Resistance by Concomitant Downregulation of the Short-Lived Anti-Apoptotic Proteins Mcl-1 and c-FLIP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADP | Adenosine-diphosphate |
| CDK9 | Cyclin-dependent kinase 9 |
| CDKs | Cyclin-dependent kinases |
| c-FLIP | FLICE-inhibitory protein |
| CRC | Colorectal cancer |
| DISC | Death-inducing signaling complex |
| DMSO | Dimethyl sulfoxide |
| DNA | Deoxyribonucleic acid |
| DFS | Disease-free survival |
| DR4/5 | Death receptor 4/5 |
| FADD | Fas-associated death domain |
| FBS | Fetal bovine serum |
| FLICE | FADD-like IL-1-converting enzyme |
| HE | Hematoxylin-eosin |
| IAPs | Inhibitor of Apoptosis family of proteins |
| izTRAIL | Isoleucine-zipper-tagged TRAIL |
| LN | Lymph node |
| Mcl-1 | Myeloid leukemia cell differentiation protein 1 |
| MTT | 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide |
| n.d. | Not determined |
| NSCLC | Non-small cell lung cancer |
| PARP | poly ADP-ribose polymerase |
| PFA | Paraformaldehyde |
| PI | Propidium iodide |
| pTEFb | positive Transcription elongation factor |
| RNA | Ribonucleic acid |
| RNA pol Ⅱ | RNA polymerase Ⅱ |
| SD | Standard deviation |
| TNF | Tumor necrosis factor |
| TRAIL | Tumor necrosis factor-related apoptosis ligand |
| TRAs | TRAIL-receptor agonists |
| UICC | Union for International Cancer Control |
| XIAP | X-linked inhibitor of apoptosis protein |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Gastroenterology 2019, 14, 89–103. [Google Scholar] [CrossRef]
- Stein, A.; Atanackovic, D.; Bokemeyer, C. Current standards and new trends in the primary treatment of colorectal cancer. Eur. J. Cancer 2011, 47, S312–S314. [Google Scholar] [CrossRef]
- Liao, X.; Morikawa, T.; Lochhead, P.; Imamura, Y.; Kuchiba, A.; Yamauchi, M.; Nosho, K.; Qian, Z.R.; Nishihara, R.; Meyerhardt, J.A. Prognostic role of PIK3CA mutation in colorectal cancer: Cohort study and literature review. Clin. Cancer Res. 2012, 18, 2257–2268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiley, S.R.; Schooley, K.; Smolak, P.J.; Din, W.S.; Huang, C.P.; Nicholl, J.K.; Sutherland, G.R.; Smith, T.D.; Rauch, C.; Smith, C.A.; et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 1995, 3, 673–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitti, R.M.; Marsters, S.A.; Ruppert, S.; Donahue, C.J.; Moore, A.; Ashkenazi, A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J. Biol. Chem. 1996, 271, 12687–12690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walczak, H.; Miller, R.E.; Ariail, K.; Gliniak, B.; Griffith, T.S.; Kubin, M.; Chin, W.; Jones, J.; Woodward, A.L.-T.; Le, T.; et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 1999, 5, 157–163. [Google Scholar] [CrossRef]
- Sheridan, J.P.; Marsters, S.A.; Pitti, R.M.; Gurney, A.; Skubatch, M.; Baldwin, D.; Ramakrishnan, L.; Gray, C.; Baker, K.; Wood, W.I.; et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science 1997, 277, 818–821. [Google Scholar] [CrossRef]
- Kischkel, F.C.; Lawrence, D.A.; Chuntharapai, A.; Schow, P.; Kim, K.J.; Ashkenazi, A. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity 2000, 12, 611–620. [Google Scholar] [CrossRef] [Green Version]
- Kretz, A.L.; Trauzold, A.; Hillenbrand, A.; Knippschild, U.; Henne-Bruns, D.; von Karstedt, S.; Lemke, J. TRAILblazing Strategies for Cancer Treatment. Cancers 2019, 11, 456. [Google Scholar] [CrossRef] [Green Version]
- Canavese, M.; Santo, L.; Raje, N. Cyclin dependent kinases in cancer: Potential for therapeutic intervention. Cancer Biol. Ther. 2012, 13, 451–457. [Google Scholar] [CrossRef] [Green Version]
- Lapenna, S.; Giordano, A. Cell cycle kinases as therapeutic tar- gets for cancer. Nat. Rev. Drug Discov. 2009, 8, 547–566. [Google Scholar] [CrossRef]
- Diaz-Padilla, I.; Siu, L.L.; Duran, I. Cyclin-dependent kinase inhibitors as potential targeted anticancer agents. Investig. New Drugs 2009, 27, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shi, S.; Lam, F.; Pepper, C.; Fischer, P.M.; Wang, S. CDKI-71, a novel CDK9 inhibitor, is preferentially cytotoxic to cancer cells compared to flavopiridol. Int. J. Cancer 2012, 130, 1216–1226. [Google Scholar] [CrossRef]
- Wang, S.; Fischer, P.M. Cyclin-dependent kinase 9: A key transcriptional regulator and potential drug target in oncology, virology and cardiology. Trends Pharmacol. Sci. 2008, 29, 302–313. [Google Scholar] [CrossRef]
- García-Reyes, B.; Kretz, A.L.; Ruff, J.P.; Von Karstedt, S.; Hillenbrand, A.; Knippschild, U.; Henne-Bruns, D.; Lemke, J. The emerging role of cyclin-dependent kinases (CDKs) in pancreatic ductal adenocarcinoma. Int. J. Mol. Sci. 2018, 19, 3219. [Google Scholar] [CrossRef] [Green Version]
- Peterlin, B.M.; Price, D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 2006, 23, 297–305. [Google Scholar] [CrossRef]
- Kretz, A.L.; Schaum, M.; Richter, J.; Kitzig, E.F.; Engler, C.C.; Leithauser, F.; Henne-Bruns, D.; Knippschild, U.; Lemke, J. CDK9 is a prognostic marker and therapeutic target in pancreatic cancer. Tumour Biol. 2017, 39, 1010428317694304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montinaro, A.; Areso Zubiaur, I.; Saggau, J.; Kretz, A.L.; Ferreira, R.M.; Hassan, O.; Walczak, H. Potent pro-apoptotic combination therapy is highly effective in a broad range of cancers. Cell Death Differ. 2022, 29, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Parry, D.; Guzi, T.; Shanahan, F.; Davis, N.; Prabhavalkar, D.; Wiswell, D.; Seghezzi, W.; Paruch, K.; Dwyer, M.P.; Doll, R.; et al. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol. Cancer Ther. 2010, 9, 2344–2353. [Google Scholar] [CrossRef] [Green Version]
- Mitri, Z.; Karakas, C.; Wie, C.; Briones, B.; Simmons, H.; Ibrahim, N.; Alvarez, R.; Murray, J.L.; Keyomarsi, K.; Moulder, S. A phase 1 study with dose expansion of the CDK inhibitor dinaciclib (SCH 727965) in combination with epirubicin in patients with metastatic triple negative breast cancer. Investig. New Drugs 2015, 33, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.; Jones, J.; Johnson, A.J.; Andritsos, L.; Maddocks, K.; Jaglowski, S.; Hessler, J.; Grever, M.R.; Im, E.; Zhou, H.; et al. Dinaciclib is a novel cyclin-dependent kinase inhibitor with significant clinical activity in relapsed and refractory chronic lymphocytic leukemia. Leukemia 2015, 29, 1524–1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- U.S. National Library of Medicine. Identifier: NCT01783171. Available online: ClinicalTrials.gov (accessed on 20 September 2018).
- Feldmann, G.; Mishra, A.; Bisht, S.; Karikari, C.; Garrido-Laguna, I.; Rasheed, Z.; Ottenhof, N.A.; Dadon, T.; Alvarez, H.; Fendrich, V.; et al. Cyclin-dependent kinase inhibitor Dinaciclib (SCH727965) inhibits pancreatic cancer growth and progression in murine xenograft models. Cancer Biol. Ther. 2011, 12, 598–609. [Google Scholar] [CrossRef] [Green Version]
- Sobin, L.H.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumours; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Wu, Y.; Giaisi, M.; Kohler, R.; Chen, W.M.; Krammer, P.H.; Li-Weber, M. Rocaglamide breaks TRAIL-resistance in human multiple myeloma and acute T-cell leukemia in vivo in a mouse xenogtraft model. Cancer Lett. 2017, 389, 70–77. [Google Scholar] [CrossRef] [PubMed]
- MILLIPORE®’s Cell Transformation Detection Assay. Catalog No. ECM570, Merck KGaA: Darmstadt, Germany, 2009–2016.
- Riccardi, C.; Nicoletti, I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat. Protoc. 2006, 1, 1458–1461. [Google Scholar] [CrossRef] [PubMed]
- Boffo, S.; Damato, A.; Alfano, L.; Giordano, A. CDK9 inhibitors in acute myeloid leukemia. J. Exp. Clin. Cancer Res. 2018, 37, 36. [Google Scholar] [CrossRef] [Green Version]
- Gentile, M.; Petrungaro, A.; Uccello, G.; Vigna, E.; Recchia, A.G.; Caruso, N.; Bossio, S.; De Stefano, L.; Palummo, A.; Storino, F.; et al. Venetoclax for the treatment of chronic lymphocytic leukemia. Expert Opin. Investig. Drugs 2017, 26, 1307–1316. [Google Scholar] [CrossRef]
- Malumbres, M.; Harlow, E.; Hunt, T.; Hunter, T.; Lahti, J.M.; Manning, G.; Morgan, D.O.; Tsai, L.H.; Wolgemuth, D.J. Cyclin-dependent kinases: A family portrait. Nat. Cell Biol. 2009, 11, 1275–1276. [Google Scholar] [CrossRef] [Green Version]
- Marshall, N.F.; Peng, J.; Xie, Z.; Price, D.H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 1996, 271, 27176–27183. [Google Scholar] [CrossRef] [Green Version]
- Rahaman, M.H.; Kumarasiri, M.; Mekonnen, L.B.; Yu, M.; Diab, S.; Albrecht, H. endocrine-related cancer thematic review targeting cdk9: A promising therapeutic opportunity in prostate cancer. Endocr. Relet. Cancer. 2019, 23, T211–T226. [Google Scholar]
- Schlafstein, A.J.; Withers, A.E.; Rudra, S.; Danelia, D.; Switchenko, J.M.; Mister, D.; Harari, S.; Zhang, H.; Daddacha, W.; Ehdaivand, S.; et al. CDK9 expression shows role as a potential prognostic biomarker in breast cancer patients who fail to achieve pathologic complete response after neoadjuvant chemotherapy. Int. J. Breast Cancer 2018, 2018, 6945129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Dean, D.C.; Hornicek, F.J.; Shi, H.; Duan, Z. Cyclin-dependent kinase 9 (CDK9) is a novel prognostic marker and therapeutic target in ovarian cancer. J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2019, 33, 5990–6000. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Chatterjee, D.; Deng, D.; Veeranki, O.; Mejia, A.; Ajani, J.A.; Hofstetter, W.; Lin, S.; Guha, S.; Kopetz, S.; et al. Antitumor effects of cyclin dependent kinase 9 inhibition in esophageal adenocarcinoma. Oncotarget 2017, 8, 28696–28710. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.S.; Lin, L.F.; Chen, J.C.; Huang, C.C.; Sheu, J.H.; Chen, W.; Tsao, T.Y.; Hsu, C.W. Increased cyclin T1 expression as a favorable prognostic factor in treating gastric adenocarcinoma. Oncol. Lett. 2015, 10, 3712–3718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puccini, A.; Marshall, J.L.; Salem, M.E. Molecular Variances Between Right- and Left-sided Colon Cancers. Curr. Color. Cancer Rep. 2018, 14, 152–158. [Google Scholar] [CrossRef]
- Dimberg, L.Y.; Anderson, C.K.; Camidge, R.; Behbakht, K.; Thorburn, A.; Ford, H.L. On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene 2013, 32, 1341–1350. [Google Scholar] [CrossRef] [Green Version]
| Variable | N = 175 | % | |
|---|---|---|---|
| Gender | Male | 93 | 53.1 |
| Female | 82 | 46.9 | |
| Differentiation | Low (G1 + G2) | 116 | 66.3 |
| High (G3 + G4) | 56 | 32 | |
| n.d. | 3 | 1.7 | |
| T classification | T1, T2 | 35 | 20 |
| T3, T4 | 139 | 79.4 | |
| n.d. | 1 | 0.6 | |
| LN invasion | N0 | 84 | 48 |
| N1, N2 | 90 | 51.4 | |
| n.d. | 1 | 0.6 | |
| Distant metastasis | M0 | 122 | 69.7 |
| M1 | 52 | 29.7 | |
| n.d. | 1 | 0.6 | |
| Stage (UICC) | I | 30 | 17.1 |
| II | 49 | 28 | |
| III | 43 | 24.6 | |
| IV | 52 | 29.7 | |
| n.d. | 1 | 0.6 | |
| Cancer localization | Left colon | 55 | 31.4 |
| Right colon | 119 | 68 | |
| n.d. | 1 | 0.6 | |
| 5-year survival | 54.1 | ||
| Age (years) | Mean | 69.53 | |
| Range | 29.81–93.98 | ||
| Over survival (months) | Mean Range | 30.57 1.02–153.46 |
| Total N | Low CDK9 Expression [Score ≤ 569] N (Median Score) | High CDK9 Expression [Score > 569] N (Median Score) | Median CDK9 Expression [Score] (Min–Max) | |
|---|---|---|---|---|
| Follow-up | ||||
| Dead | 79 | 40 (394.5) | 39 (767) | 567 (3–997) |
| alive | 96 | 33 (450) | 63 (796) | 643 (143–1138) |
| Gender | ||||
| Male | 93 | 41 (385) | 52 (794.5) | 618 (3–1094) |
| Female | 82 | 32 (467.5) | 50 (766.5) | 623 (143–1138) |
| T classification | ||||
| T1, T2 | 35 | 15 (412) | 20 (850.5) | 651 (6–1080) |
| T3, T4 | 139 | 57 (414) | 82 (756.5) | 618 (3–1138) |
| n.d. | 1 | - | ||
| Localization | ||||
| Left colon | 55 | 21 (390) | 34 (738.5) | 618 (143–1094) |
| Right colon | 119 | 51 (447) | 68 (813.5) | 625 (3–1138) |
| n.d. | 1 | - | ||
| Stage (UICC) | ||||
| I | 30 | 13 (397) | 17 (832) | 620.5 (6–1080) |
| II | 49 | 17 (385) | 32 (797) | 635 (143–1138) |
| III | 43 | 19 (458) | 24 (727) | 604 (151–1094) |
| IV | 52 | 23 (395) | 29 (767) | 614 (3–1070) |
| n.d. | 1 | - | ||
| Total | 175 | 73 (412) | 102 (787) | 618 (3–1138) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Kretz, A.-L.; Schneider, S.; Knippschild, U.; Henne-Bruns, D.; Kornmann, M.; Lemke, J.; Traub, B. Evaluation of CDK9 Inhibition by Dinaciclib in Combination with Apoptosis Modulating izTRAIL for the Treatment of Colorectal Cancer. Biomedicines 2023, 11, 928. https://doi.org/10.3390/biomedicines11030928
Shen X, Kretz A-L, Schneider S, Knippschild U, Henne-Bruns D, Kornmann M, Lemke J, Traub B. Evaluation of CDK9 Inhibition by Dinaciclib in Combination with Apoptosis Modulating izTRAIL for the Treatment of Colorectal Cancer. Biomedicines. 2023; 11(3):928. https://doi.org/10.3390/biomedicines11030928
Chicago/Turabian StyleShen, Xiao, Anna-Laura Kretz, Sandra Schneider, Uwe Knippschild, Doris Henne-Bruns, Marko Kornmann, Johannes Lemke, and Benno Traub. 2023. "Evaluation of CDK9 Inhibition by Dinaciclib in Combination with Apoptosis Modulating izTRAIL for the Treatment of Colorectal Cancer" Biomedicines 11, no. 3: 928. https://doi.org/10.3390/biomedicines11030928






