Skin Microbiota: Setting up a Protocol to Evaluate a Correlation between the Microbial Flora and Skin Parameters
Abstract
:1. Introduction
2. Materials and Methods
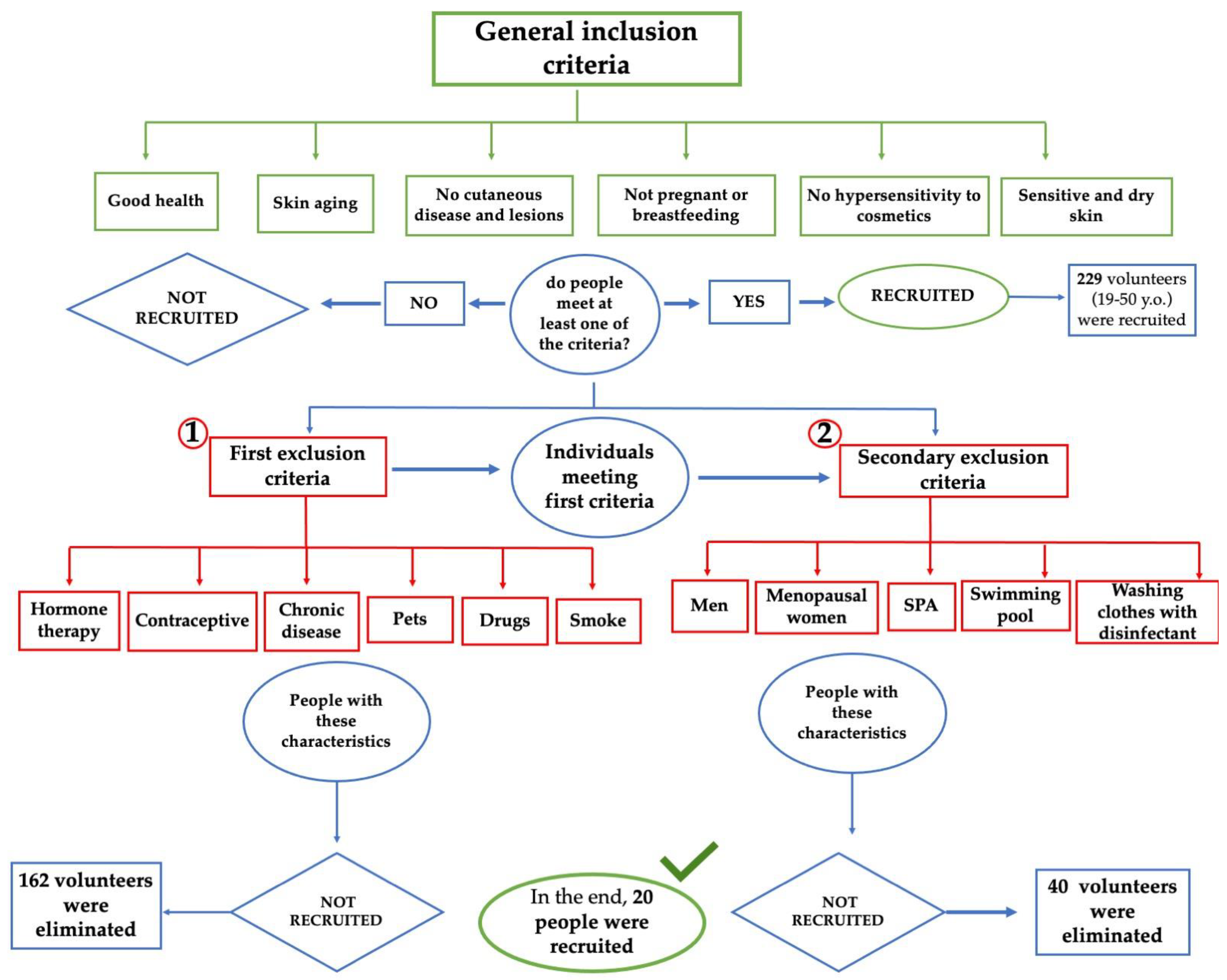
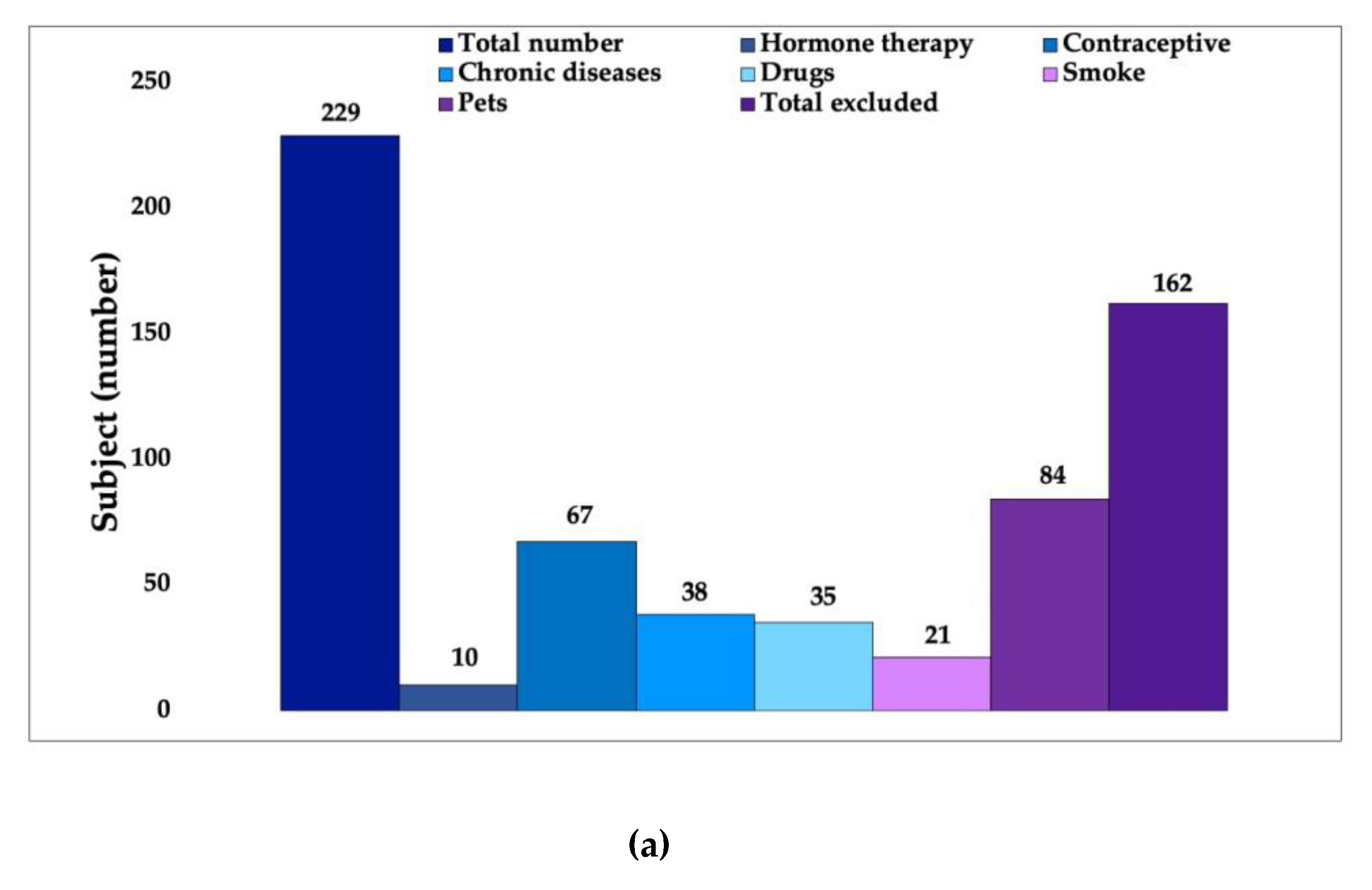
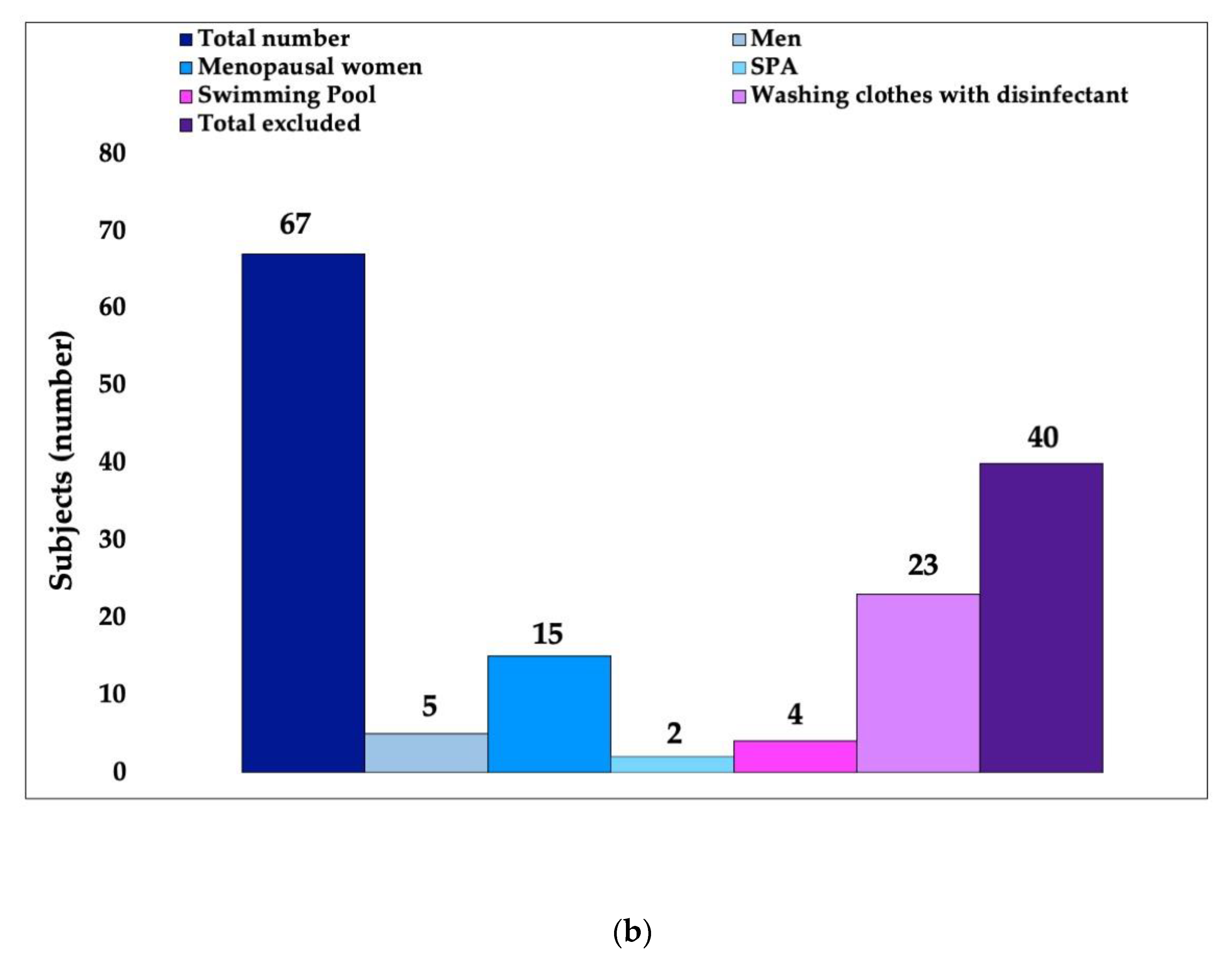
2.1. First Phase: Volunteer Selection
2.2. Design of Experiment
2.3. Acquisition of Skin Biophysical Parameters
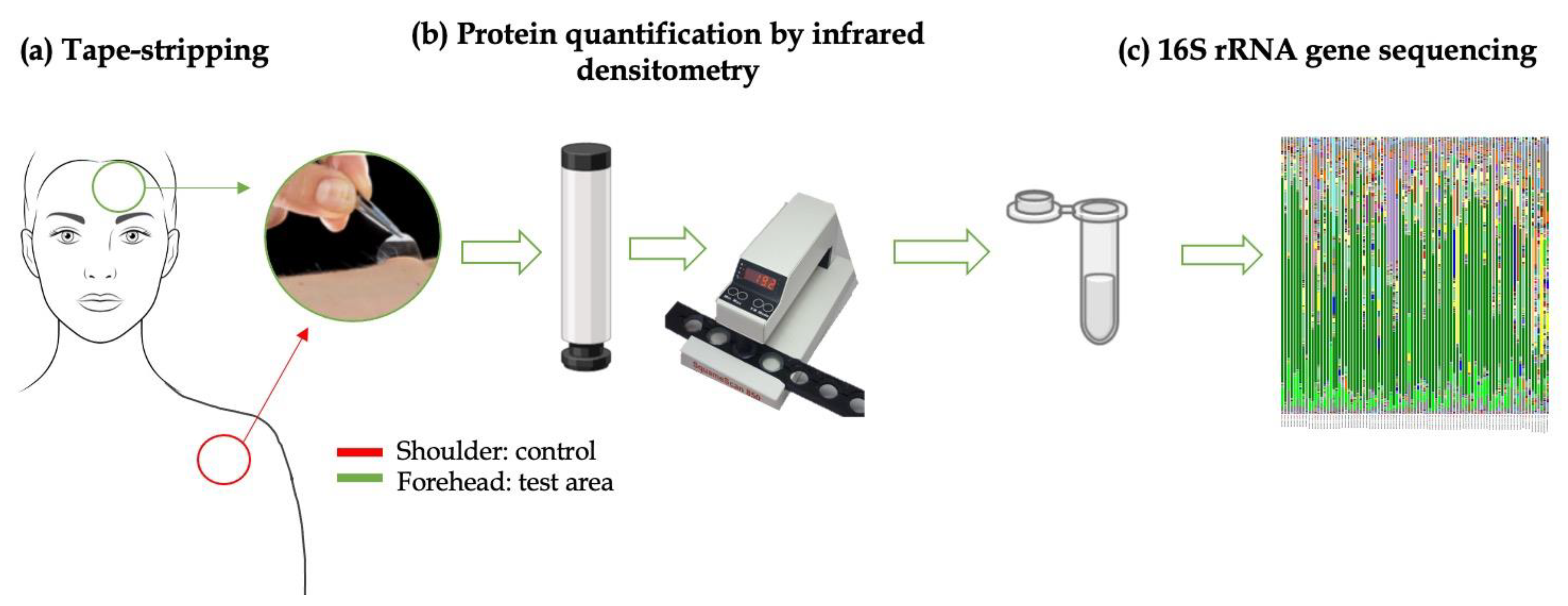
2.4. Skin Flora Collection
- Contamination of the strip: one strip was removed from the film to which it is attached using tweezers. The strip is then inserted directly into a sterile Eppendorf (B12020). This serves to check for possible interference with bacteria present on the tape at purchase.
- Environmental contamination: a second strip, removed from its film as described above, was left in a laboratory environment for 24 h to observe possible interference from bacteria present in the air (B22020).
- Device contamination: the third preliminary analysis involved the application of a constant pressure of 10 s with the appropriate cylinder on one other strip lying on the film, which was then placed in a sterile Eppendorf (B32020). This was useful to verify the possible interference with bacteria present on the cylinder used. One more analysis was performed on another strip, placed on its film, pressured for 10 s with the cylinder, and then placed inside the IR instrument to mimic the reading of the amount of keratin. It was subsequently inserted into a sterile Eppendorf (B42020). This procedure allowed us to understand the possible interferences with bacteria using the device to measure the microbial and the keratin content on the same strip.
2.4.1. In Vivo Skin Microbiota Preliminary Tests
- Zone A: Samples were collected separately using 5 strips for each area (from C1FA to -C5FA for the forehead and from C1SA to -C5SA for the shoulder). The objective was to understand if it is possible to obtain a quantity of genetic microbial material to allow the analysis of the microbiota in all 5 first strips of skin.
- Zone B: Samples were collected using 5 strips and examined together in a single analysis. The objective was to understand if it is more effective to analyze the 5 strips together in order to reduce the number of total analyses.
- Zone C: The skin areas were cleaned 2 h before sampling.
2.4.2. In Vivo Skin Microbiota Sample Collection
- Removal of the adhesive disks (tape) from the film on which they were placed using tweezers.
- Application of the adhesive disk on the skin under constant pressure using the cylinder for 10 s.
- Removal of the adhesive pads from the skin using tweezers.
- Determining the SC protein content with IR.
- Placing the strip into a sterile Eppendorf and then closing the tube.
- This procedure was repeated three times on the same area of analysis.
2.5. Microbiota Analysis by Sequencing:
2.6. Statistical Analysis
3. Results
3.1. In Vivo Study First Phase: Volunteer Selection
3.2. Instrumental Analysis
3.3. Microbiota Analysis
3.3.1. Preliminary Analyses
3.3.2. In Vivo Skin Microbiota Preliminary Tests
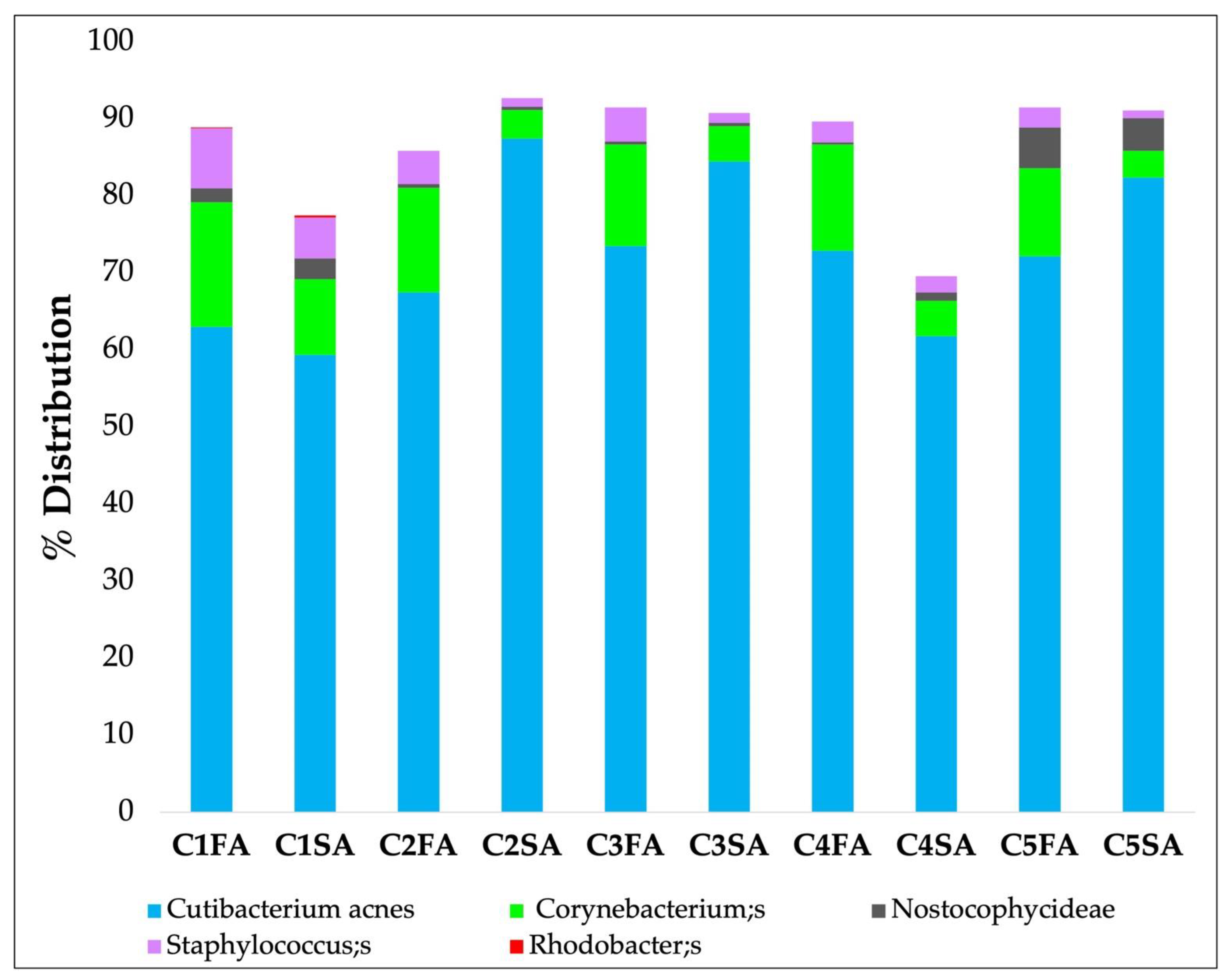
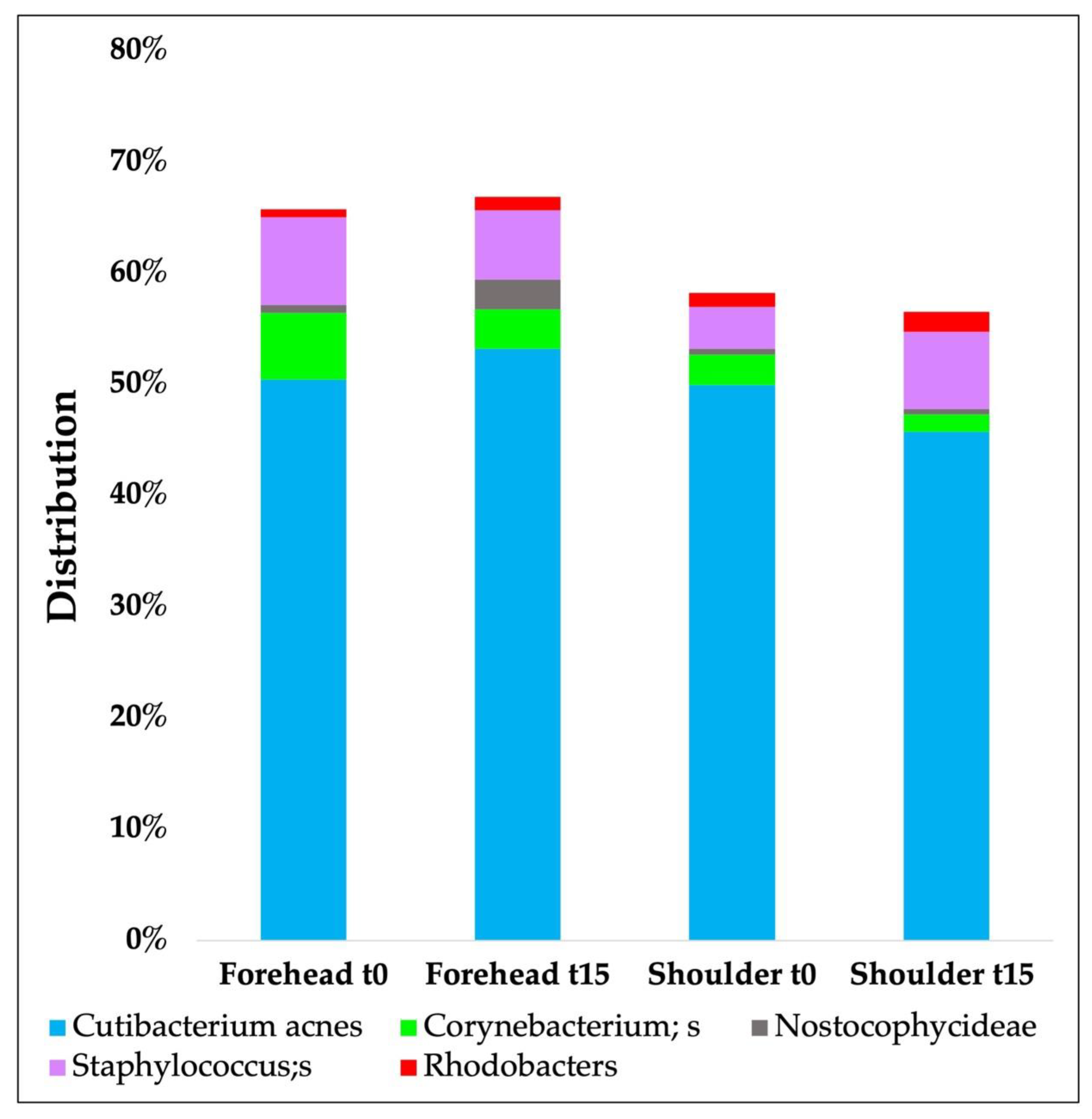
3.3.3. In Vivo Skin Microbiota Evaluation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining thehuman microbiome. Nutr. Rev. 2012, 70, S38–S44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consortium, H.M.P. Structure, function and diversity of the healthy human microbiome. Nature 2012, 207–214. [Google Scholar]
- Dréno, B.; Araviiskaia, E.; Berardesca, E.; Gontijo, G.; Viera, M.S.; Xiang, L.F.; Martin, R.; Bieber, T. Microbiome in healthy skin, update for dermatologist. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 2038–2047. [Google Scholar] [CrossRef] [PubMed]
- Musthaq, S.; Mazuy, A.; Jakus, J. The microbiome in dermatology. Clin. Dermatol. 2018, 36, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.R.; James, W.D. Microbial ecology of the skin. Annu. Rev. Microbiol. 1988, 42, 441–464. [Google Scholar] [CrossRef]
- Lopez-Ojeda, W.; Pandey, A.; Alhajj, M. Anatomy, Skin (Integument); StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online:https://www.ncbi.nlm.nih.gov/books/NBK441980/ (accessed on 17 October 2022).
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Weyrich, L.S.; Dixit, S.; Farrer, A.G.; Cooper, A.J. The skin microbiome: Associations between altered microbial communities and disease. Australas. J. Derm. 2015, 56, 268–274. [Google Scholar] [CrossRef]
- Cogen, A.L. Skin microbiota: A source of disease or defence. Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef] [Green Version]
- Condrò, G.; Guerini, M.; Castello, M.; Perugini, P. Acne Vulgaris, Atopic Dermatitis and Rosacea: The Role of the Skin Microbiota—A Review. Biomedicines 2022, 10, 2523. [Google Scholar] [CrossRef]
- Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; Deal, C. The NIH human microbiome project. Genome. Res. 2009, 19, 2317–2323. [Google Scholar]
- Capone, K.A.; Dowd, S.E.; Stamatas, G.N.; Nikolovski, J. Diversity of the human skin microbiome early in life. J. Invest. Derm. 2011, 131, 2026–2032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Findley, K.; Grice, E.A. The skin microbiome: A focus on pathogensand their association with skin disease. PLoS Pathog. 2014, 10, e1004436. [Google Scholar] [CrossRef] [PubMed]
- Barnard, E.; Li, H. Shaping of cutaneous function by encounters withcommensals. J. Physiol. 2017, 595, 437–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grice, E.A.; Dawson, T.L., Jr. Host-microbe interactions: Malassezia andhuman skin. Curr. Opin. Microbiol. 2017, 40, 81–87. [Google Scholar] [CrossRef]
- Theelen, B.; Cafarchia, C.; Gaitanis, G.; Bassukas, I.D.; Boekhout, T.; Dawson, T.L., Jr. Malassezia ecology, patho-physiology, and treatment. Med. Mycol. 2018, 56 (Suppl. 1), S10–S25. [Google Scholar] [CrossRef] [Green Version]
- Sanford, J.A.; Gallo, R.L. Functions of the skin microbiota in health and disease. Semin. Immunol. 2013, 25, 370–377. [Google Scholar] [CrossRef] [Green Version]
- Hannigan, G.D.; Meisel, J.S.; Tyldsley, A.S.; Zheng, Q.; Hodkinson, B.P.; SanMiguel, A.J.; Minot, S.; Bushman, F.D.; Grice, E.A. The human skin double-stranded DNA virome: Topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. mBio 2015, 6, e01578-15. [Google Scholar] [CrossRef] [Green Version]
- Blaser, M.J.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Estrada, I.; Gao, Z.; Clemente, J.C.; Costello, E.K.; Knight, R. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. ISME J. 2013, 7, 85–95. [Google Scholar] [CrossRef]
- Staudinger, T.; Pipal, A.; Redl, B. Molecular analysis of the prevalent microbiota of human male and female forehead skin compared to forearm skin and the influence of make up. J. Appl. Microbiol. 2011, 10, 1381–1389. [Google Scholar] [CrossRef]
- Baldwin, H.E.; Bhatia, N.D.; Friedman, A.; Eng, R.M.; Seite, S. The Role of Cutaneous Microbiota Harmony in Maintaining a Functional Skin Barrier. J. Drug Dermatol. 2017, 16, 12–18. [Google Scholar] [CrossRef]
- Maguire, M.; Maguire, G. The role of microbiota, and probiotics and prebiotics in skin health. Arch. Dermatol. Res. 2017, 309, 411–421. [Google Scholar] [CrossRef]
- Tomasoni, D.; Perugini, P. Relationship between Skin Microbiota and Skin Biophysical Parameters in Inflammatory Skin Disease: A Systematic Review. J. Clin. Dermatol. Ther. 2021, 7, 72. [Google Scholar]
- Mukherjee, S.; Mitra, R.; Maitra, A.; Gupta, S.; Kumaran, S.; Chakrabortty, A.; Majumder, P.P. Sebum and Hydration Levels in Specific Regions of Human Face Significantly Predict the Nature and Diversity of Facial Skin Microbiome. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dagnelie, M.A.; Corvec, S.; Saint-Jean, M.; Nguyen, J.-M.; Khammari, A.; Dreno, B. Cutibacterium acnesphylotypes diversity loss: A trigger forskin inflammatory process. JEADV 2019, 33, 2340–2348. [Google Scholar] [PubMed]
- Szepetiuk, G.; Piérard-Franchimont, C.; Quatresooz, P.; Piérard, G.-E. Physico-biological foundation of skin fluorescence—Review. Pathol. Biol. 2012, 60, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Klatte, J.L.; van der Beek, N.; Kemperman, P.M.J.H. 100 years of Wood’s lamp revised. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 842–847. [Google Scholar] [CrossRef]
- Gai, K.; Nagase, S.; Mukai, K.; Iuchi, T.; Mori, Y.; Matsue, M.; Sugitani, K.; Sugama, J.; Okamoto, S. A Comparison of Techniques for Collecting Skin Microbiome Samples: Swabbing Versus Tape-Stripping. Front. Microbiol. 2018, 9, 2362. [Google Scholar]
- Van Horn, K.G.; Audette, C.D.; Tucker, K.A.; Sebeck, D. Comparison of 3 swab transport systems for direct release and recovery of aerobic and anaerobic bacteria. Diagn. Microbiol. Infect. Dis. 2008, 62, 471–473. [Google Scholar] [CrossRef]
- Ono, S.; Eda, N.; Mori, T.; Otsuka, A.; Nakamura, N.; Inai, Y.; Ota, N.; Akama, T. Tape stripping method is useful for the quantification of antimicrobial peptides on the human skin surface including the stratum corneum. Sci. Rep. 2020, 10, 15259. [Google Scholar] [CrossRef]
- Association, W.M. World Medical Association declaration of Helsinki ethical principles for medical research involving human subjects. JAMA. 2013, 310, 20. [Google Scholar]
- Blichman, C.W.; Serup, J. Assessment of skin moisture. Measurement of electrical conductance, capacitance and transepidermal water loss. Acta. Derm. Venereol. (Stockh) 1988, 68, 284–290. [Google Scholar]
- Lee, C.M.; Maibach, H.I. Bioengineering analysis of water hydration: An overview. Exog Derm. 2002, 1, 269–275. [Google Scholar] [CrossRef]
- Pinnagoda, J.; Tupkek, R.; Agner, T.; Serup, J. Guidelines for transepidermal water loss (TEWL) measurement. Contact Dermat. 1990, 22, 164–178. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Chávez, J.J.; Merino-Sanjuán, V.; López-Cervantes, M.; Urban-Morlan, Z.; Piñón-Segundo, E.; Quintanar-Guerrero, D.; Ganem-Quintanar, A. The tapestripping technique as a method for drug quantification in skin. J. Pharm. Pharm. Sci. 2008, 11, 104–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobrev, H. Fluorescence diagnostic imaging in patients with acne. Photodermatol. Photoimmunol. Photomed. 2010, 26, 285–289. [Google Scholar] [CrossRef]
- Akoglu, H. User’s guide to correlation coefficients. Turk. J. Emerg. Med. 2018, 18, 91–93. [Google Scholar] [CrossRef]
- Rintala, H. Actinobacteria in indoor environments: Exposures and respiratory health effects. Front. Biosci. (Schol. Ed.) 2011, 3, 1273–1284. [Google Scholar]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2015, 80, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Franchitti, E.; Caredda, C.; Anedda, E.; Traversi, D. Aerobiome and Effects on Human Health: A Systematic Review and Missing Evidence. Atmosphere 2022, 13, 1148. [Google Scholar] [CrossRef]
- Moreira, C.; Ramos, V.; Azevedo, J.; Vasconcelos, V. Methods to detect cyanobacteria and their toxins in the environment. Appl. Microbiol. Biotechnol. 2014, 98, 8073–8082. [Google Scholar] [CrossRef]

| Coefficient Value | Strength Interpretation | |
|---|---|---|
| +1 | −1 | Perfect positive or negative correlation |
| +0.9–0.7 | −0.9–0.7 | Very strong correlation |
| +0.6–0.4 | −0.6–0.4 | Strong correlation |
| +0.3 | −0.3 | Moderate correlation |
| +0.2 | −0.2 | Weak correlation |
| +0.1 | −0.1 | Negligible correlation |
| 0 | 0 | No correlation |
| Parameters | Forehead t0 | Forehead t15 | Shoulder t0 | Shoulder t15 | Forehead p t0 vs. t15 | Shoulder p t0 vs. t15 |
|---|---|---|---|---|---|---|
| Hydration (A.U.) | 53.65 | 50.22 | 43.50 | 43.97 | 0.0624 | 0.7984 |
| 6.16 | 5.98 | 7.28 | 5.87 | |||
| TEWL (g/m2h) | 9.74 | 9.85 | 8.46 | 9.31 | 0.9197 | 0.0210 * |
| 1.50 | 1.09 | 1.61 | 1.06 | |||
| pH | 5.16 | 5.24 | 5.53 | 5.18 | 0.4358 | 0.0272 * |
| 0.43 | 0.30 | 0.52 | 0.32 | |||
| Sebum levels (µg/cm2) | 81.70 | 99.48 | 15.85 | 15.80 | 0.0054 ** | 0.3463 |
| 36.35 | 46.93 | 9.16 | 13.59 | |||
| Protein content (µg/cm2) | 15.85 | 14.76 | 15.57 | 14.90 | 0.2196 | 0.7490 |
| 3.41 | 3.64 | 3.79 | 3.09 | |||
| Porphyrin intensity (A.U.) | 186.00 | 174.15 | 168.30 | 170.50 | 0.0894 | 0.6017 |
| 19.07 | 26.28 | 44.81 | 24.38 |
| Parameters | Spearman’s Coefficient | Strength Interpretation |
|---|---|---|
| Forehead | ||
| SCWC vs. protein content | −0.69 | Strong correlation |
| Porphyrin intensity vs. sebum | 0.44 | Strong correlation |
| Porphyrin intensity vs. protein content | −0.43 | Strong correlation |
| Sebum vs. TEWL | 0.43 | Strong correlation |
| Protein content vs. pH | 0.32 | Moderate correlation |
| Porphyrin intensity vs. TEWL | 0.38 | Moderate correlation |
| Shoulder | ||
| SCWC vs. pH | −0.63 | Strong correlation |
| Porphyrin intensity vs. TEWL | 0.40 | Strong correlation |
| Porphyrin intensity vs. sebum | 0.48 | Strong correlation |
| Protein content vs. TEWL | 0.31 | Moderate correlation |
| Forehead vs. shoulder | ||
| Porphyrin intensity | 0.61 | Strong correlation |
| pH | 0.46 | Strong correlation |
| TEWL | 0.47 | Strong correlation |
| Taxonomy | B12020 | B22020 | B32020 | B42020 |
|---|---|---|---|---|
| Bacteria; Firmicutes | 9.00 | 6.20 | 11.90 | 15.30 |
| Bacteria; Actinobacteria | 70.00 | 10.90 | 39.10 | 26.70 |
| Bacteria; Cyanobacteria | 1.60 | 54.00 | 0.40 | 1.00 |
| Bacteria; Proteobacteria | 14.40 | 27.30 | 41.80 | 53.40 |
| Bacteria; Bacteroidetes | 3.60 | 1.20 | 3.90 | 2.30 |
| Taxonomy | C2- 3 FA | C2- 3 SA |
|---|---|---|
| Cutibacterium acnes | 70.5 | 86.05 |
| Corynebacterium; s | 13.4 | 4.15 |
| Nostocophycideae | <0.5 | <0.5 |
| Staphylococcus; s | 4.35 | 1.2 |
| Rhodobacter; s | <0.5 | <0.5 |
| Phyla | Forehead t0 | Forehead t15d | Shoulder t0 | Shoulder t15d |
|---|---|---|---|---|
| Actinobacteria | 62.55 (±20.81) | 64.75 (±19.06) | 59.30 (±0.21) | 59.83 (±27.6) |
| Proteobacteria | 19.62 (±19.52) | 16.75 (±12.72) | 23.97 (±0.17) | 24.53 (±15.56) |
| Bacteroidetes | 2.04 (±2.36) | 1.66 (±1.57) | 2.44 (±0.02) | 2.03 (±1.74) |
| Cyanobacteria | 0.89 (±1.59) | 2.86 (±9.33) | 1.79 (±0.03) | 0.53 (±0.15) |
| Firmicutes | 13.98 (±8.25) | 13.03 (±6.42) | 11.66 (±0.10) | 12.07 (±9.54) |
| Microbial Species | Spearman’s Coefficient | Strength Interpretation |
|---|---|---|
| Forehead | ||
| Porphyrin intensity vs. Cutibacterium acnes | 0.57 | Strong correlation |
| Porphyrin intensity vs. Staphylococcus sp. | −0.33 | Moderate correlation |
| Sebum vs. Corynebacterium sp. | 0.48 | Strong correlation |
| Sebum vs. Cutibacterium acnes | 0.40 | Strong correlation |
| pH vs. Cutibacterium acnes | 0.26 | Weak correlation |
| Protein content vs. Corynebacterium sp. | 0.45 | Strong correlation |
| Protein content vs. Staphylococcus sp. | 0.57 | Strong correlation |
| Shoulder | ||
| Sebum vs. Cutibacterium acnes | 0.41 | Strong correlation |
| pH vs. Cutibacterium acnes | 0.55 | Strong correlation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perugini, P.; Grignani, C.; Condrò, G.; van der Hoeven, H.; Ratti, A.; Mondelli, A.; Colpani, A.; Bleve, M. Skin Microbiota: Setting up a Protocol to Evaluate a Correlation between the Microbial Flora and Skin Parameters. Biomedicines 2023, 11, 966. https://doi.org/10.3390/biomedicines11030966
Perugini P, Grignani C, Condrò G, van der Hoeven H, Ratti A, Mondelli A, Colpani A, Bleve M. Skin Microbiota: Setting up a Protocol to Evaluate a Correlation between the Microbial Flora and Skin Parameters. Biomedicines. 2023; 11(3):966. https://doi.org/10.3390/biomedicines11030966
Chicago/Turabian StylePerugini, Paola, Camilla Grignani, Giorgia Condrò, Harald van der Hoeven, Annamaria Ratti, Antonella Mondelli, Antonio Colpani, and Mariella Bleve. 2023. "Skin Microbiota: Setting up a Protocol to Evaluate a Correlation between the Microbial Flora and Skin Parameters" Biomedicines 11, no. 3: 966. https://doi.org/10.3390/biomedicines11030966
APA StylePerugini, P., Grignani, C., Condrò, G., van der Hoeven, H., Ratti, A., Mondelli, A., Colpani, A., & Bleve, M. (2023). Skin Microbiota: Setting up a Protocol to Evaluate a Correlation between the Microbial Flora and Skin Parameters. Biomedicines, 11(3), 966. https://doi.org/10.3390/biomedicines11030966





