Oral Microbiota Signatures in the Pathogenesis of Euthyroid Hashimoto’s Thyroiditis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Sample Collection and Storage
2.3. DNA Extraction from Unstimulated Saliva Samples
2.4. PCR Amplification and Sequencing
2.5. Bioinformatics and Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Klubo-Gwiezdzinska, J.; Wartofsky, L. Hashimoto thyroiditis: An evidence-based guide: Etiology, diagnosis and treatment. Pol. Arch. Intern. Med. 2022, 132, 16222. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chen, Y.; Shen, Y.; Tian, R.; Sheng, Y.; Que, H. Global prevalence and epidemiological trends of Hashimoto’s thyroiditis in adults: A systematic review and meta-analysis. Front. Public Health 2022, 10, 1020709. [Google Scholar] [CrossRef] [PubMed]
- Danailova, Y.; Velikova, T.; Nikolaev, G.; Mitova, Z.; Shinkov, A.; Gagov, H.; Konakchieva, R. Nutritional Management of Thyroiditis of Hashimoto. Int. J. Mol. Sci. 2022, 23, 5144. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, F.; Fallahi, P.; Elia, G.; Gonnella, D.; Paparo, S.R.; Giusti, C.; Churilov, L.P.; Ferrari, S.M.; Antonelli, A. Hashimotos’ thyroiditis: Epidemiology, pathogenesis, clinic and therapy. Best Pract. Res. Clin. Endocrinol. Metab. 2019, 33, 101367. [Google Scholar] [CrossRef]
- Pyzik, A.; Grywalska, E.; Matyjaszek-Matuszek, B.; Roliński, J. Immune Disorders in Hashimoto’s Thyroiditis: What Do We Know So Far? J. Immunol. Res. 2015, 2015, 979167. [Google Scholar] [CrossRef]
- Salazar-Viedma, M.; Vergaño-Salazar, J.G.; Pastenes, L.; D’Afonseca, V. Simulation Model for Hashimoto Autoimmune Thyroiditis Disease. Endocrinology 2021, 162, bqab190. [Google Scholar] [CrossRef]
- Deo, P.N.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Y.; Yang, X.; Li, C.; Song, Z. The Oral Microbiota: Community Composition, Influencing Factors, Pathogenesis, and Interventions. Front. Microbiol. 2022, 13, 895537. [Google Scholar] [CrossRef]
- Lu, M.; Xuan, S.; Wang, Z. Oral microbiota: A new view of body health. Food Sci. Hum. Wellness 2019, 8, 8–15. [Google Scholar] [CrossRef]
- Dutzan, N.; Kajikawa, T.; Abusleme, L.; Greenwell-Wild, T.; Zuazo, C.E.; Ikeuchi, T.; Brenchley, L.; Abe, T.; Hurabielle, C.; Martin, C.; et al. A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Sci. Transl. Med. 2018, 10, eaat0797. [Google Scholar] [CrossRef]
- Pandiyan, P.; Bhaskaran, N.; Zou, M.; Schneider, E.; Jayaraman, S.; Huehn, J. Microbiome Dependent Regulation of Tregs and Th17 Cells in Mucosa. Front. Immunol. 2019, 10, 426. [Google Scholar] [CrossRef] [PubMed]
- Nagao, J.-I.; Kishikawa, S.; Tanaka, H.; Toyonaga, K.; Narita, Y.; Negoro-Yasumatsu, K.; Tasaki, S.; Arita-Morioka, K.-I.; Nakayama, J.; Tanaka, Y. Pathobiont-responsive Th17 cells in gut-mouth axis provoke inflammatory oral disease and are modulated by intestinal microbiome. Cell Rep. 2022, 40, 111314. [Google Scholar] [CrossRef]
- Liu, J.; Qin, X.; Lin, B.; Cui, J.; Liao, J.; Zhang, F.; Lin, Q. Analysis of gut microbiota diversity in Hashimoto’s thyroiditis patients. BMC Microbiol. 2022, 22, 1–15. [Google Scholar] [CrossRef]
- Ishaq, H.M.; Mohammad, I.S.; Hussain, R.; Parveen, R.; Shirazi, J.H.; Fan, Y.; Shahzad, M.; Hayat, K.; Li, H.; Ihsan, A.; et al. Gut-Thyroid axis: How gut microbial dysbiosis associated with euthyroid thyroid cancer. J. Cancer 2022, 13, 2014–2028. [Google Scholar] [CrossRef] [PubMed]
- Cayres, L.C.d.F.; de Salis, L.V.V.; Rodrigues, G.S.P.; Lengert, A.V.H.; Biondi, A.P.C.; Sargentini, L.D.B.; Brisotti, J.L.; Gomes, E.; de Oliveira, G.L.V. Detection of Alterations in the Gut Microbiota and Intestinal Permeability in Patients With Hashimoto Thyroiditis. Front. Immunol. 2021, 12, 579140. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Zhao, F.; Yuan, K.; Zhu, X.; Wang, N.; Xia, F.; Lu, Y.; Huang, Z. Association Between Serum Thyroid-Stimulating Hormone Levels and Salivary Microbiome Shifts. Front. Cell. Infect. Microbiol. 2021, 11, 603291. [Google Scholar] [CrossRef]
- Kwon, M.; Jeong, Y.; Kwak, J.; Jung, K.; Baek, S. Association between oral health and thyroid disorders: A population-based cross-sectional study. Oral Dis. 2022, 28, 2277–2284. [Google Scholar] [CrossRef]
- Jiao, J.; Zheng, Y.; Zhang, Q.; Xia, D.; Zhang, L.; Ma, N. Saliva microbiome changes in thyroid cancer and thyroid nodules patients. Front. Cell. Infect. Microbiol. 2022, 12, 989188. [Google Scholar] [CrossRef]
- Wang, B.; Xu, Y.; Zhang, M.; Zhang, J.; Hou, X.; Li, J.; Cai, Y.; Sun, Z.; Ban, Y.; Wang, W. Oral and intestinal microbial features in pregnant women with hypothyroidism and their correlations with pregnancy outcomes. Am. J. Physiol. Metab. 2020, 319, E1044–E1052. [Google Scholar] [CrossRef] [PubMed]
- Yalcin, M.M.; Altinova, A.E.; Cavnar, B.; Bolayir, B.; Akturk, M.; Arslan, E.; Ozkan, C.; Cakir, N.; Toruner, F.B. Is thyroid autoimmunity itself associated with psychological well-being in euthyroid Hashimoto’s thyroiditis? Endocr. J. 2017, 64, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Totsika, M.; Morrison, M.; Punyadeera, C. The saliva microbiome profiles are minimally affected by collection method or DNA extraction protocols. Sci. Rep. 2017, 7, 8523. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Kaya, D.; Genc, E.; Genc, M.A.; Aktas, M.; Eroldogan, O.T.; Guroy, D. Biofloc technology in recirculating aquaculture system as a culture model for green tiger shrimp, Penaeus semisulcatus: Effects of different feeding rates and stocking densities. Aquaculture 2020, 528, 735526. [Google Scholar] [CrossRef]
- Wang, S.; Song, F.; Gu, H.; Wei, X.; Zhang, K.; Zhou, Y.; Luo, H. Comparative Evaluation of the Salivary and Buccal Mucosal Microbiota by 16S rRNA Sequencing for Forensic Investigations. Front. Microbiol. 2022, 13, 777882. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Virili, C.; Fallahi, P.; Antonelli, A.; Benvenga, S.; Centanni, M. Gut microbiota and Hashimoto’s thyroiditis. Rev. Endocr. Metab. Disord. 2018, 19, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Olsen, I.; Yamazaki, K. Can oral bacteria affect the microbiome of the gut? J. Oral Microbiol. 2019, 11, 1586422. [Google Scholar] [CrossRef]
- Demirci, M. Could Neisseria in oral microbiota modulate the inflammatory response of COVID-19? Oral Dis. 2022, 28, 2603–2604. [Google Scholar] [CrossRef]
- Gao, L.; Cheng, Z.; Zhu, F.; Bi, C.; Shi, Q.; Chen, X. The Oral Microbiome and Its Role in Systemic Autoimmune Diseases: A Systematic Review of Big Data Analysis. Front. Big Data 2022, 5, 927520. [Google Scholar] [CrossRef]
- Zhao, F.; Feng, J.; Li, J.; Zhao, L.; Liu, Y.; Chen, H.; Jin, Y.; Zhu, B.; Wei, Y. Alterations of the Gut Microbiota in Hashimoto’s Thyroiditis Patients. Thyroid 2018, 28, 175–186. [Google Scholar] [CrossRef]
- Ishaq, H.M.; Mohammad, I.S.; Guo, H.; Shahzad, M.; Hou, Y.J.; Ma, C.; Naseem, Z.; Wu, X.; Shi, P.; Xu, J. Molecular estimation of alteration in intestinal microbial composition in Hashimoto’s thyroiditis patients. Biomed. Pharmacother. 2017, 95, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; An, Y.; Cao, B.; Sun, R.; Ke, J.; Zhao, D. The Composition of Gut Microbiota in Patients Bearing Hashimoto’s Thyroiditis with Euthyroidism and Hypothyroidism. Int. J. Endocrinol. 2020, 2020, 5036959. [Google Scholar] [CrossRef]
- Demirci, M.; Tokman, H.B.; Uysal, H.; Demiryas, S.; Karakullukçu, A.; Saribas, S.; Cokugras, H.C.; Kocazeybek, B. Reduced Akkermansia muciniphila and Faecalibacterium prausnitzii levels in the gut microbiota of children with allergic asthma. Allergol. Et Immunopathol. 2019, 47, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yuan, L.; Zhu, D.; Sun, B.; Du, J.; Wang, J. Alterations and Mechanism of Gut Microbiota in Graves’ Disease and Hashimoto’s Thyroiditis. Pol. J. Microbiol. 2022, 71, 173–189. [Google Scholar] [CrossRef]
- Wang, X.; Pang, K.; Wang, J.; Zhang, B.; Liu, Z.; Lu, S.; Xu, X.; Zhu, L.; Zhou, Z.; Niu, M.; et al. Microbiota dysbiosis in primary Sjögren’s syndrome and the ameliorative effect of hydroxychloroquine. Cell Rep. 2022, 40, 111352. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.T.; Roesch, L.F.W.; Ördberg, M.; Ilonen, J.; Atkinson, M.A.; Schatz, D.A.; Triplett, E.W.; Ludvigsson, J. Genetic risk for autoimmunity is associated with distinct changes in the human gut microbiome. Nat. Commun. 2019, 10, 3621. [Google Scholar] [CrossRef]
- Bruserud, Ø.; Siddiqui, H.; Marthinussen, M.C.; Chen, T.; Jonsson, R.; Oftedal, B.E.; Olsen, I.; Husebye, E.S.; Wolff, A.B. Oral microbiota in autoimmune polyendocrine syndrome type 1. J. Oral Microbiol. 2018, 10, 1442986. [Google Scholar] [CrossRef]
- Bagavant, H.; Araszkiewicz, A.M.; Ingram, J.K.; Cizio, K.; Merrill, J.T.; Arriens, C.; Guthridge, J.M.; James, J.A.; Deshmukh, U.S. Immune Response to Enterococcus gallinarum in Lupus Patients Is Associated With a Subset of Lupus-Associated Autoantibodies. Front. Immunol. 2021, 12, 635072. [Google Scholar] [CrossRef]
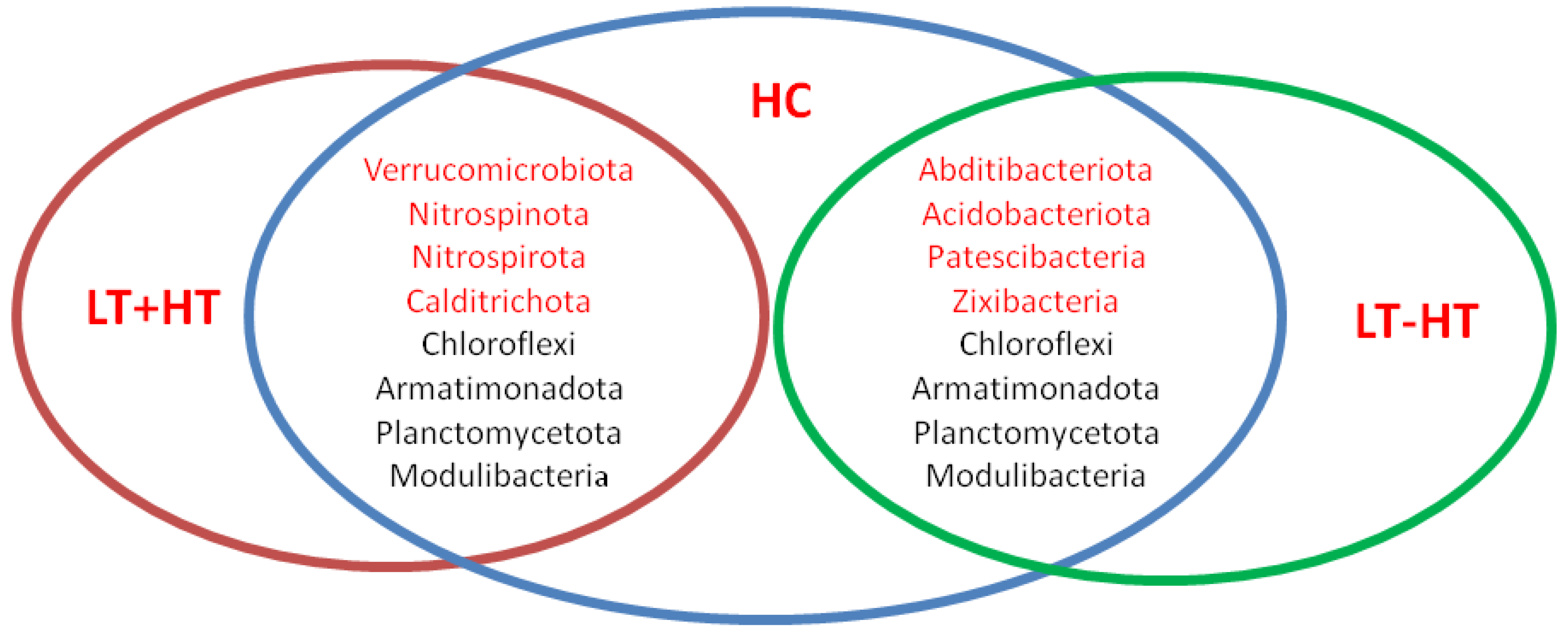



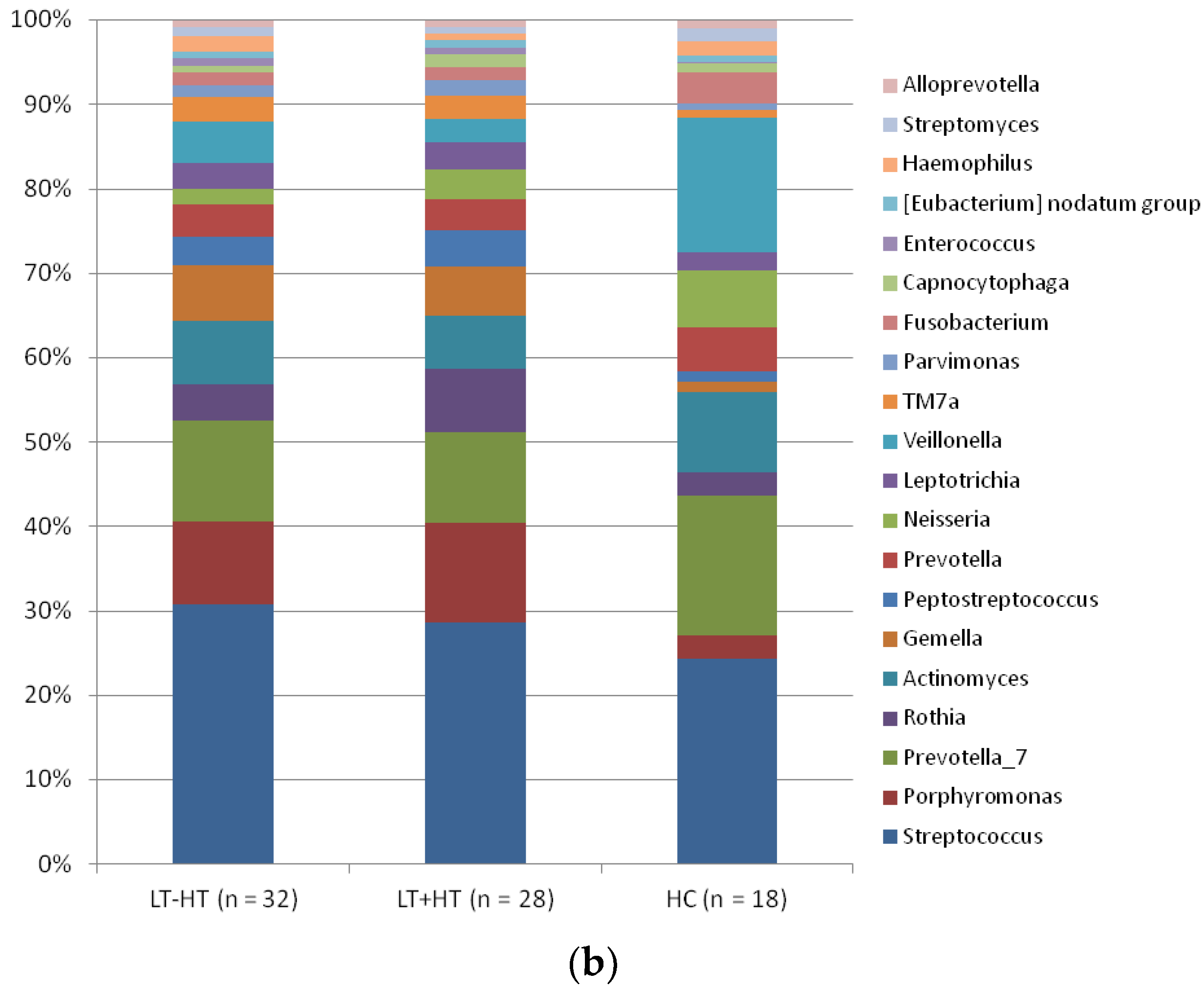
| LT – HT (n = 32) | LT + HT (n = 28) | HC (n = 18) | p ** | |
|---|---|---|---|---|
| Age (years) | 39.4 ± 4.52 | 39.8 ± 6.04 | 39.1 ± 5.8 | 0.893 |
| BMI (kg/m2) | 21.6 ± 2.1 | 22.4 ± 1.9 | 20.8 ± 2.3 | 0.914 |
| TSH (mU/L) | 1.86 ± 1.81 | 2.12 ± 1.94 | 1.61 ± 1.35 | 0.785 |
| TPOAb (IU/mL) | 265 ± 224 | 342 ± 274 | 7.8 ± 4.9 | <0.0001 |
| Saliva Samples | LT – HT (n = 32) | LT + HT (n = 28) | HC (n = 18) | p ** |
|---|---|---|---|---|
| Simpson | 0.89 ± 0.04 | 0.91 ± 0.03 | 0.87 ± 0.05 | 0.952 |
| Shannon | 3.71 ± 2.12 | 3.68 ± 1.94 | 3.73 ± 2.03 | 0.874 |
| References | Sample Type | Increased in HT | Decreased in HT |
|---|---|---|---|
| Wang et al. [19] | Saliva and Stool | Gammaproteobacteria and Prevotella | Firmicutes, Leptotrichiace, and Actinobacteria |
| Zhao et al. [30] | Stool | Blautia, Roseburia, Ruminococcus_torques_group, Romboutsia, Dorea, Fusicatenibacter, and Eubacterium_hallii_group | Faecalibacterium, Bacteroides, Prevotella_9, and Lachnoclostridium |
| Ishaq et al. [31] | Stool | Firmicutes, Bacteroidetes, Actinobacteria, Verrucomicrobia, Fusobacteria Escherichia-Shigella, and Parasutterella | Proteobacteria, Prevotella_9 and Dialister |
| Liu et al. [32] | Stool | Bacteroidetes, Actinobacteria, Fusobacteria, Proteobacteria, and Alistipes | Firmicutes, Verrucomicrobia, and Faecalibacterium |
| Zhao et al. [34] | Stool | Proteobacteria, Actinobacteria, Erysipelotrichia, Cyanobacteria, and Ruminococcus_2 | Peptostreptococcaceae, Bacillaceae, and Matophyaceae |
| In this study | Saliva | Firmicutes, Bacteroidota, Actinobacteriota, Patescibacteria, Fusobacteriota, Spirochaetota, Enterococcus, and Staphylococcus | Proteobacteria, Campylobacterota, and Bacillus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erdem, M.G.; Unlu, O.; Ates, F.; Karis, D.; Demirci, M. Oral Microbiota Signatures in the Pathogenesis of Euthyroid Hashimoto’s Thyroiditis. Biomedicines 2023, 11, 1012. https://doi.org/10.3390/biomedicines11041012
Erdem MG, Unlu O, Ates F, Karis D, Demirci M. Oral Microbiota Signatures in the Pathogenesis of Euthyroid Hashimoto’s Thyroiditis. Biomedicines. 2023; 11(4):1012. https://doi.org/10.3390/biomedicines11041012
Chicago/Turabian StyleErdem, Mustafa Genco, Ozge Unlu, Fatma Ates, Denizhan Karis, and Mehmet Demirci. 2023. "Oral Microbiota Signatures in the Pathogenesis of Euthyroid Hashimoto’s Thyroiditis" Biomedicines 11, no. 4: 1012. https://doi.org/10.3390/biomedicines11041012
APA StyleErdem, M. G., Unlu, O., Ates, F., Karis, D., & Demirci, M. (2023). Oral Microbiota Signatures in the Pathogenesis of Euthyroid Hashimoto’s Thyroiditis. Biomedicines, 11(4), 1012. https://doi.org/10.3390/biomedicines11041012








