Maternal Plasma RNA in First Trimester Nullipara for the Prediction of Spontaneous Preterm Birth ≤ 32 Weeks: Validation Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecular Tests
2.2. qRT-PCR Assays
2.3. Statistics
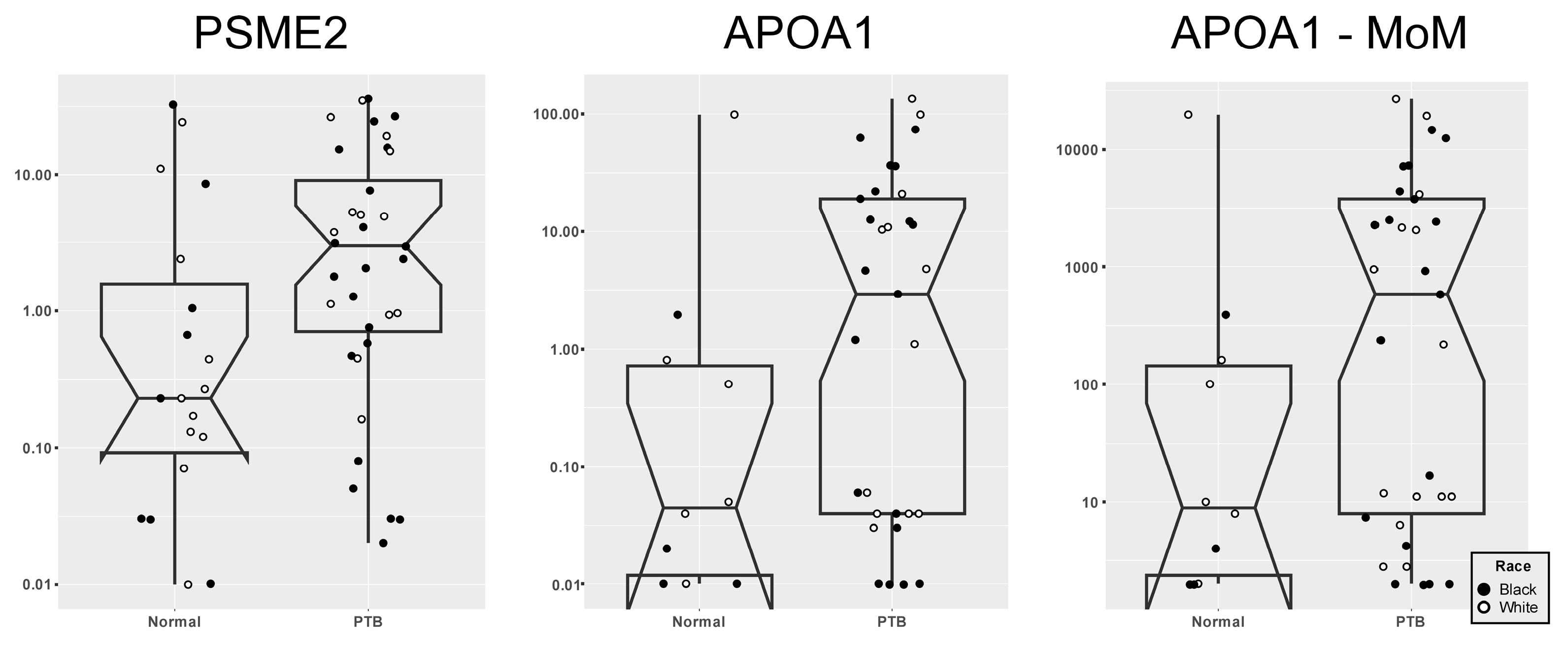
3. Results
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chawanpaiboon, S.; Vogel, J.P.; Moller, A.-B.; Lumbiganon, P.; Petzold, M.; Hogan, D.; Landoulsi, S.; Jampathong, N.; Kongwattanakul, K.; Laopaiboon, M.; et al. Global, regional, and national estimates of levels of preterm birth in 2014: A systematic review and modelling analysis. Lancet Glob. Health 2019, 7, e37–e46. [Google Scholar] [CrossRef] [Green Version]
- Kaempf, J.W.; Tomlinson, M.; Arduza, C.; Anderson, S.; Campbell, B.; Ferguson, L.A.; Zabari, M.; Stewart, V.T. Medical staff guidelines for periviability pregnancy counseling and medical treat. Pediatrics 2006, 117, 22–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, C.J.L.; Vos, T.; Lozano, R.; Naghavi, M.; Flaxman, A.D.; Michaud, C.; Ezzati, M.; Shibuya, K.; Salomon, J.A.; Abdalla, S.; et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2197–2223. [Google Scholar] [CrossRef] [PubMed]
- Koullali, B.; Van Zijl, M.D.; Kazemier, B.M.; Oudijk, M.A.; Mol, B.W.J.; Pajkrt, E.; Ravelli, A.C.J. The association between parity and spontaneous preterm birth: A population based study. BMC Pregnancy Childbirth 2020, 20, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, G.; Shah, I.; White, I.; Pell, J.; Crossley, J.; Dobbie, R.; Smith, G. Maternal and biochemical predictors of antepartum stillbirth among nulliparous women in relation to gestational age of fetal death. BJOG Int. J. Obstet. Gynaecol. 2007, 114, 705–714. [Google Scholar] [CrossRef]
- Van Der Ven, J.; Van Os, M.A.; Kazemier, B.M.; Kleinrouweler, E.; Verhoeven, C.J.; De Miranda, E.; van Wassenaer-Leemhuis, A.G.; Kuiper, P.N.; Porath, M.; Willekes, C.; et al. The capacity of mid-pregnancy cervical length to predict preterm birth in low-risk women: A national cohort study. Acta. Obs. Gynecol. Scand. 2015, 94, 1223–1234. [Google Scholar] [CrossRef] [Green Version]
- Rosenbloom, J.I.; Raghuraman, N.; Temming, L.A.; Stout, M.J.; Tuuli, M.G.; Dicke, J.M.; Macones, G.A.; Cahill, A.G. Predictive Value of Midtrimester Universal Cervical Length Screening Based on Parity. J. Ultrasound Med. 2020, 39, 147–154. [Google Scholar] [CrossRef]
- Esplin, M.S.; Elovitz, M.; Iams, J.D.; Parker, C.B.; Wapner, R.; Grobman, W.A.; Simhan, H.N.; Wing, D.A.; Haas, D.M.; Silver, R.M.; et al. Predictive Accuracy of Serial Transvaginal Cervical Lengths and Quantitative Vaginal Fetal Fibronectin Levels for Spontaneous Preterm Birth Among Nulliparous Women. JAMA 2017, 317, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Weiner, C.P.; Dong, Y.; Zhou, H.; Cuckle, H.; Ramsey, R.; Egerman, R.; Buhimschi, I.; Buhimschi, C. Early pregnancy prediction of spontaneous preterm birth before 32 completed weeks of pregnancy using plasma RNA: Transcriptome discovery and initial validation of an RNA panel of markers. BJOG 2021, 128, 1870–1880. [Google Scholar] [CrossRef]
- Weiner, C.P.; Cuckle, H.; Weiss, M.L.; Buhimschi, I.A.; Dong, Y.; Zhou, H.; Ramsey, R.; Egerman, R.; Buhimschi, C.S. Evaluation of a Maternal Plasma RNA Panel Predicting Spontaneous Preterm Birth and Its Expansion to the Prediction of Preeclampsia. Diagnostics 2022, 12, 1327. [Google Scholar] [CrossRef]
- Mandrekar, I.N. Biostatistics for Clinicians: Receiver Operating Characteristic Curve in Diagnostic Test Assessment. J. Thoracic. Onc. 2010, 5, 1315–1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Khouli, R.H.; Macura, K.J.; Barker, P.B.; Habba, M.R.; Jacobs, M.A.; Bluemke, D.A. Relationship of temporal resolution to diagnostic performance for dynamic contrast enhanced MRI of the breast. JMRI 2009, 30, 999–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Xu, Y.; Ye, M.; Yan, K.; Gao, Z.; Jin, Q. Learning misclassification costs for imbalanced classification on gene expression data. BMC Bioinformatics. 2019, 20, 681. [Google Scholar] [CrossRef] [PubMed]
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A practical Information-Theoretic Approach, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Luo, W.; Huning, E.Y.-S.; Tran, T.; Phung, D.; Venkatesh, S. Screening for post 32-week preterm birth risk: How helpful is routine perinatal data collection? Heliyon 2016, 2, e00119. [Google Scholar] [CrossRef] [Green Version]
- Dude, C.M.; Levine, L.D.; Schwartz, N. The Impact of Previous Obstetric History on the Risk of Spontaneous Preterm Birth in Women with a Sonographic Short Cervix. Am. J. Perinatol. 2020, 37, 1189–1194. [Google Scholar] [CrossRef]
- Markenson, G.R.; Saade, G.R.; Laurent, L.C.; Heyborne, K.D.; Coonrod, D.V.; Schoen, C.N.; Baxter, J.K.; Haas, D.M.; Longo, S.; Grobman, W.A.; et al. Performance of a proteomic preterm delivery predictor in a large inde-pendent prospective cohort. Am. J. Obstet. Gynecol. MFM 2020, 2, 100140. [Google Scholar] [CrossRef]
- Ngo, T.T.M.; Moufarrej, M.N.; Rasmussen, M.-L.H.; Camunas-Soler, J.; Pan, W.; Okamoto, J.; Neff, N.F.; Liu, K.; Wong, R.J.; Downes, K.; et al. Noninvasive blood tests for fetal development predict gestational age and preterm delivery. Science 2018, 360, 1133–1136. [Google Scholar] [CrossRef] [Green Version]
- Jehan, F.; Sazawal, S.; Baqui, A.H.; Nisar, M.I.; Dhingra, U.; Khanam, R.; Ilyas, M.; Dutta, A.; Mitra, D.K.; Mehmood, U.; et al. Multiomics Characterization of Preterm Birth in Low- and Middle-Income Countries. JAMA Netw. Open 2020, 3, e2029655. [Google Scholar] [CrossRef]
- Camunas-Soler, J.; Gee, E.P.; Reddy, M.; Mi, J.D.; Thao, M.; Brundage, T.; Siddiqui, F.; Hezelgrave, N.L.; Shennan, A.H.; Namsaraev, E.; et al. Predictive RNA profiles for early and very early spontaneous preterm birth. Am. J. Obstet. Gynecol. 2022, 227, 72.e1–72.e16. [Google Scholar] [CrossRef]
- Cook, J.; Bennett, P.R.; Kim, S.H.; Teoh, T.G.; Sykes, L.; Kindinger, L.M.; Garrett, A.; Binkhamis, R.; MacIntyre, D.A.; Terzidou, V. First Trimester Circulating MicroRNA Biomarkers Predictive of Subsequent Preterm Delivery and Cervical Shortening. Sci. Rep. 2019, 9, 5861. [Google Scholar] [CrossRef] [Green Version]
- Hromadnikova, I.; Kotlabova, K.; Krofta, L. First Trimester Prediction of Preterm Delivery in the Absence of Other Pregnan-cy-Related Complications Using Cardiovascular-Disease Associated MicroRNA Biomarkers. Int. J. Mol. Sci. 2022, 23, 3951. [Google Scholar] [CrossRef]
- Del Vecchio, G.; Li, Q.; Li, W.; Thamotharan, S.; Tosevska, A.; Morselli, M.; Sung, K.; Janzen, C.; Zhou, X.; Pellegrini, M.; et al. Cell-free DNA Methylation and Transcriptomic Signature Prediction of Pregnancies with Adverse Outcomes. Epigenetics 2021, 16, 642–661. [Google Scholar] [CrossRef]
- Jain, V.; McDonald, S.D.; Mundle, W.R.; Farine, D. Guideline No. 398: Progesterone for Prevention of Spontaneous Preterm Birth. J. Obstet. Gynaecol. Can. 2020, 42, 806–812. [Google Scholar] [CrossRef]
- National Guideline Alliance Hosted by the Royal College of Obstetricians and Gynaecologists (UK). Preterm Labour and Birth: [A] Evidence Review for Clinical Effectiveness of Prophylactic Progesterone in Preventing Preterm Labour; National Institute for Health and Care Excellence (UK): London, UK, 2019. [Google Scholar]
- Norman, J.E. Progesterone and preterm birth. Int. J. Gynaecol. Obstet. 2020, 150, 24–30. [Google Scholar] [CrossRef]
- Poon, L.C.; Wright, D.; Rolnik, D.L.; Syngelaki, A.; Delgado, J.L.; Tsokaki, T.; Leipold, G.; Akolekar, R.; Shearing, S.; De Stefani, L.; et al. Aspirin for Evidence-Based Preeclampsia Prevention trial: Effect of aspirin in prevention of preterm preeclampsia in subgroups of women according to their characteristics and medical and obstetrical history. Am. J. Obstet. Gynecol. 2017, 217, 585.e1–585.e5. [Google Scholar] [CrossRef] [Green Version]
- Carlson, S.E.; Gajewski, B.J.; Valentine, C.J.; Kerling, E.H.; Weiner, C.P.; Cackovic, M.; Buhimschi, C.S.; Rogers, L.K.; Sands, S.A.; Brown, A.R.; et al. Higher dose docosahexaenoic acid supplementation during pregnancy reduces early preterm birth: A randomised, double-blind, adaptive-design superiority trial. EClinicalMedicine 2021, 36, 100905. [Google Scholar] [CrossRef]
- Sun, L.; Li, Y.; Xie, W.; Xue, X. Association between omega-3 fatty acid supplementation and lower risk of preterm delivery: A systematic review and meta-analysis. J. Matern. Neonatal Med. 2022, 35, 2294–2303. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, L.A.; Sullivan, T.R.; Skubisz, M.; Middleton, P.F.; Best, K.P.; Yelland, L.N.; Quinlivan, J.; Zhou, S.J.; Liu, G.; McPhee, A.J.; et al. Omega-3 fatty acid supplementation in pregnancy—Baseline omega-3 status and early preterm birth: Exploratory analysis of a randomised controlled trial. BJOG: Int. J. Obstet. Gynaecol. 2020, 127, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.H.; Weiner, C.P. Robust Cost Efficacy of a Novel, Validated Screening Test at 12 to 20 Weeks for the Prediction Preterm Birth (PTB) ≦ 32 Weeks in Singletons. Gynecol. Obstet. Clin. Medicine 2021, 1, 107–111. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [Green Version]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Yoffe, L.; Gilam, A.; Yaron, O.; Polsky, A.; Farberov, L.; Syngelaki, A.; Nicolaides, K.; Hod, M.; Shomron, N. Early Detection of Preeclampsia Using Circulating Small non-coding RNA. Sci. Rep. 2018, 8, 3401. [Google Scholar] [CrossRef] [Green Version]
- Filant, J.; Nejad, P.; Paul, A.; Simonson, B.; Srinivasan, S.; Zhang, X.; Balaj, L.; Das, S.; Gandhi, R.; Laurent, L.C.; et al. Isolation of Extracellular RNA from Serum/Plasma. Methods Mol Biol. 2018, 1740, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, R.L.; Culhane, J.F.; Iams, J.D.; Romero, R. Epidemiology and causes of preterm birth. Lancet 2008, 371, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Esplin, M.S.; O’Brien, E.; Fraser, A.; Kerber, R.A.; Clark, E.; Simonsen, S.E.; Holmgren, C.; Mineau, G.P.; Varner, M.W. Estimating Recurrence of Spontaneous Preterm Delivery. Obstet. Gynecol. 2008, 112, 516–523. [Google Scholar] [CrossRef] [PubMed]
| 12-Week RNA Panel | AUC | p * | 95% CI | Detection Rates of sPTB ≤ 32 Weeks | ||
|---|---|---|---|---|---|---|
| 10% FPR | 20% FPR | 30% FPR | ||||
| PSME2 | 0.65 | 0.05 | 0.50–0.80 | 18 | 35 | 63 |
| PSME2 (MoMs) | 0.65 | 0.06 | 0.50–0.80 | 18 | 38 | 64 |
| PSME2 + MA | 0.69 | <0.02 | 0.54–0.83 | 26 | 45 | 55 |
| PSME2 (MoMs) + MA | 0.67 | <0.02 | 0.53–0.82 | 24 | 40 | 58 |
| PSME2 + CRL, MA, MW, race, tobacco | 0.71 | <0.005 | 0.57–0.85 | 32 | 39 | 58 |
| PSME2 (MoMs) + CRL, MA, MW, race, tobacco | 0.70 | <0.005 | 0.56–0.84 | 32 | 39 | 55 |
| APOA1 | 0.72 | <0.001 | 0.59–0.85 | 45 | 50 | 65 |
| APOA1 (MoMs) | 0.73 | <0.001 | 0.60–0.87 | 45 | 50 | 65 |
| APOA1 + MA | 0.77 | <0.0001 | 0.64–0.90 | 53 | 69 | 70 |
| APOA1 (MoMs) + MA | 0.79 | <0.0001 | 0.66–0.91 | 52 | 61 | 75 |
| APOA1 + CRL, weight, race, tobacco, MA | 0.77 | <0.0001 | 0.64–0.90 | 48 | 72 | 75 |
| APOA1 (MoMs) + CRL, MA, MW, race, tobacco | 0.79 | <0.0001 | 0.66–0.91 | 41 | 61 | 79 |
| PSME2 and APOA1 | 0.72 | <0.002 | 0.58–0.86 | 41 | 50 | 59 |
| PSME2 and APOA1 (MoMs) | 0.73 | <0.002 | 0.59–0.87 | 40 | 48 | 61 |
| PSME2 and APOA1 + MA | 0.78 | <0.0001 | 0.65–0.90 | 50 | 64 | 74 |
| PSME2 and APOA1 (MoMs) + MA | 0.78 | <0.0001 | 0.66–0.91 | 45 | 65 | 78 |
| PSME2 and APOA1 + CRL, MA, MW, race, tobacco | 0.78 | <0.0001 | 0.65–0.90 | 48 | 66 | 72 |
| PSME2 and APOA1 (MoMs) + CRL, MA, MW, race, tobacco | 0.78 | <0.0001 | 0.66–0.91 | 41 | 61 | 78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weiner, C.P.; Zhou, H.; Cuckle, H.; Syngelaki, A.; Nicolaides, K.H.; Weiss, M.L.; Dong, Y. Maternal Plasma RNA in First Trimester Nullipara for the Prediction of Spontaneous Preterm Birth ≤ 32 Weeks: Validation Study. Biomedicines 2023, 11, 1149. https://doi.org/10.3390/biomedicines11041149
Weiner CP, Zhou H, Cuckle H, Syngelaki A, Nicolaides KH, Weiss ML, Dong Y. Maternal Plasma RNA in First Trimester Nullipara for the Prediction of Spontaneous Preterm Birth ≤ 32 Weeks: Validation Study. Biomedicines. 2023; 11(4):1149. https://doi.org/10.3390/biomedicines11041149
Chicago/Turabian StyleWeiner, Carl P., Helen Zhou, Howard Cuckle, Argyro Syngelaki, Kypros H. Nicolaides, Mark L. Weiss, and Yafeng Dong. 2023. "Maternal Plasma RNA in First Trimester Nullipara for the Prediction of Spontaneous Preterm Birth ≤ 32 Weeks: Validation Study" Biomedicines 11, no. 4: 1149. https://doi.org/10.3390/biomedicines11041149







