Classification of Highly Divergent Viruses from DNA/RNA Sequence Using Transformer-Based Models
Abstract
1. Introduction
2. Literature Review
- The first component involves a pipeline for nucleotide acquisition using the NCBI GenBank database to train a BERT tokenizer.
- The second component is specialized BERT architecture for DNA analysis that learns unsupervised next codons and passes the last hidden state of the CLS token to a classifier to identify relevant features for understanding genotype–phenotype relationships.
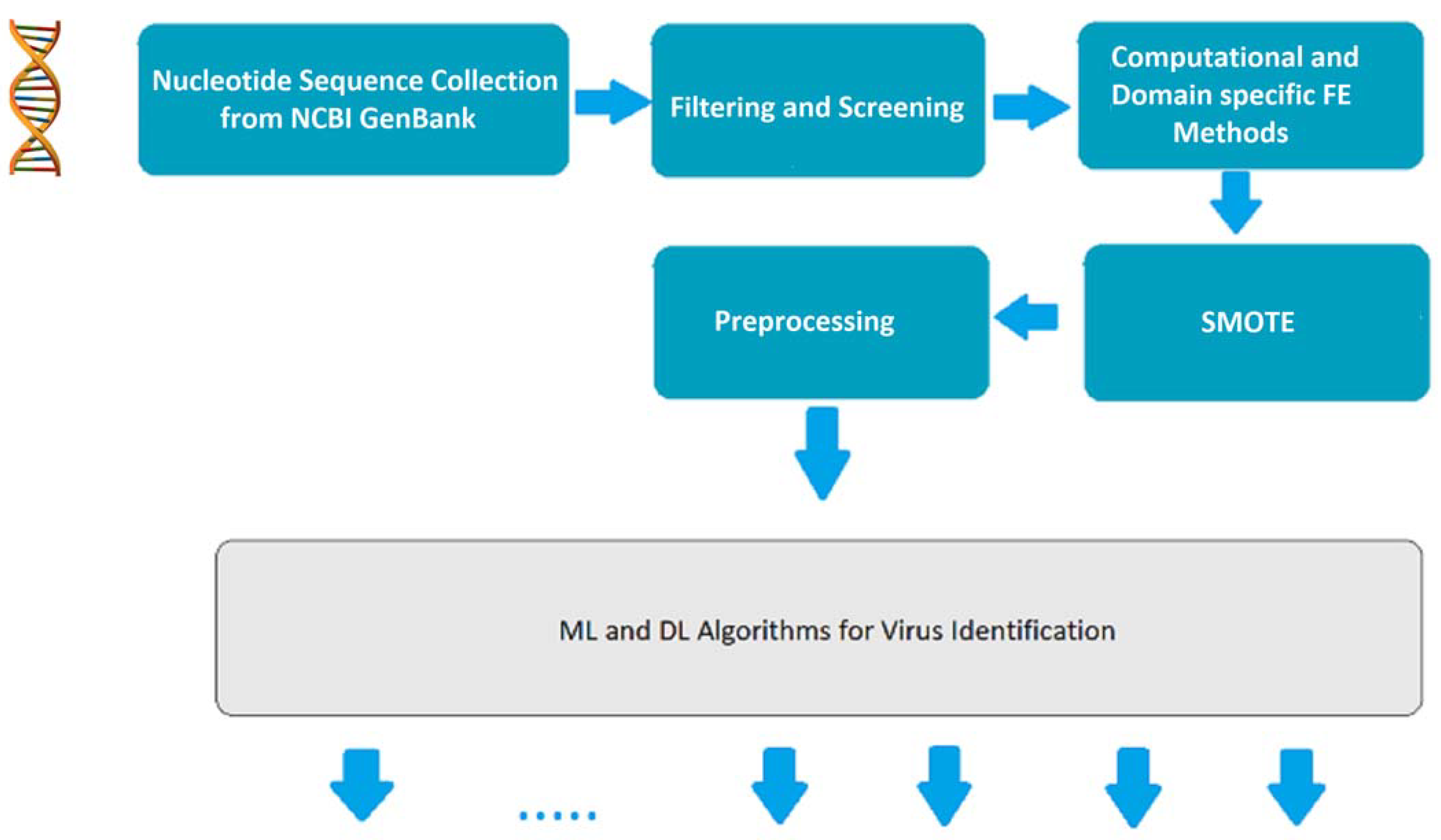
3. Materials and Methods
3.1. Dataset
3.2. Basic Pipeline for Nucleotide Sequence Acquisition
3.2.1. Nucleotide Sequence Collection from GenBank
3.2.2. Filter and Screening
3.2.3. K-Mers for Computational and Domain-Specific Feature Extraction
3.2.4. SMOTE
3.2.5. Additional Preprocessing
3.3. Proposed BERT Model
3.3.1. Proposed DNA/RNA Tokenizer
3.3.2. BERT Padding
3.3.3. Bidirectional Encoder Representation
3.3.4. Classifier
3.4. Evaluation Metrics
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Scimone, C.; Donato, L.; Alafaci, C.; Granata, F.; Rinaldi, C.; Longo, M.; D’Angelo, R.; Sidoti, A. High-throughput sequencing to detect novel likely gene-disrupting variants in pathogenesis of sporadic brain arteriovenous malformations. Front. Genet. 2020, 11, 146. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sadad, T.; Rehman, A.; Hussain, A.; Abbasi, A.A.; Khan, M.Q. A Review on Multi-Organ Cancer Detection Using Advanced Machine Learning Techniques. Curr. Med. Imaging Former. Curr. Med. Imaging Rev. 2020, 17, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2010, 38 (Suppl. S1), 46–51. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Shadab, S.; Alam Khan, M.T.; Neezi, N.A.; Adilina, S.; Shatabda, S. DeepDBP: Deep Neural Networks for Identification of DNA-Binding Proteins. Inf. Med. Unlocked 2020, 19, 100318. [Google Scholar] [CrossRef]
- Saba, T.; Abunadi, I.; Sadad, T.; Khan, A.R.; Bahaj, S.A. Optimizing the transfer-learning with pretrained deep convolutional neural networks for first stage breast tumor diagnosis using breast ultrasound visual images. Microsc. Res. Tech. 2022, 85, 1444–1453. [Google Scholar] [CrossRef]
- Caudai, C.; Galizia, A.; Geraci, F.; Le Pera, L.; Morea, V.; Salerno, E.; Via, A.; Colombo, T. AI Applications in Functional Genomics. Comput. Struct. Biotechnol. J. 2021, 19, 5762–5790. [Google Scholar] [CrossRef]
- Gunasekaran, H.; Ramalakshmi, K.; Arokiaraj, A.R.M.; Kanmani, S.D.; Venkatesan, C.; Dhas, C.S.G. Analysis of DNA sequence classification using CNN and hybrid models. Comput. Math. Methods Med. 2021, 2021, 1835056. [Google Scholar] [CrossRef]
- Mock, F.; Viehweger, A.; Barth, E.; Marz, M. VIDHOP, viral host prediction with Deep Learning. Bioinformatics 2020, 37, 318–325. [Google Scholar] [CrossRef]
- Gałan, W.; Bąk, M.; Jakubowska, M. Host taxon predictor—A tool for predicting the taxon of the host of a newly discovered virus. Sci. Rep. 2019, 9, 3436. [Google Scholar] [CrossRef][Green Version]
- Mock, F.; Kretschmer, F.; Kriese, A.; Böcker, S.; Marz, M. BERTax: Taxonomic classification of DNA sequences with Deep Neural Networks. bioRxiv 2021. Available online: https://www.biorxiv.org/content/10.1101/2021.07.09.451778v1 (accessed on 27 February 2023).
- Le, N.Q.K.; Ho, Q.T.; Nguyen, V.N.; Chang, J.S. BERT-Promoter: An improved sequence-based predictor of DNA promoter using BERT pre-trained model and SHAP feature selection. Comput. Biol. Chem. 2022, 99, 107732. [Google Scholar] [CrossRef] [PubMed]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. Proc. Naacl-HLT 2019, 2019, 4171–4186. [Google Scholar]
- Hoarfrost, A.; Aptekmann, A.; Farfañuk, G.; Bromberg, Y. Shedding Light on Microbial Dark Matter with A Universal Language of Life. bioRxiv 2020. [Google Scholar] [CrossRef]
- Busia, A.; Dahl, G.E.; Fannjiang, C.; Alexander, D.H.; Dorfman, E.; Poplin, R.; McLean, C.Y.; Chang, P.-C.; Depristo, M. A Deep Learning Approach to Pattern Recognition for Short DNA Sequences. bioRxiv 2018, 353474. [Google Scholar] [CrossRef][Green Version]
- Rizzo, R.; Fiannaca, A.; La Rosa, M.; Urso, A. A deep learning approach to DNA sequence classification. In Computational Intelligence Methods for Bioinformatics and Biostatistics; CIBB 2015, Lecture Notes in Computer Science; Angelini, C., Rancoita, P., Rovetta, S., Eds.; Springer: Cham, Switzerland, 2016; Volume 9874, pp. 129–140. [Google Scholar]
- Dablain, D.; Krawczyk, B.; Chawla, N.V. DeepSMOTE: Fusing deep learning and SMOTE for imbalanced data. IEEE Trans. Neural Netw. Learn. Syst. 2022. [Google Scholar] [CrossRef] [PubMed]
- Karami, H.; Derakhshani, A.; Ghasemigol, M.; Fereidouni, M.; Miri-Moghaddam, E.; Baradaran, B.; Tabrizi, N.J.; Najafi, S.; Solimando, A.G.; Marsh, L.M.; et al. Weighted gene co-expression network analysis combined with machine learning validation to identify key modules and hub genes associated with SARS-CoV-2 infection. J. Clin. Med. 2021, 10, 3567. [Google Scholar] [CrossRef] [PubMed]
- Le, N.Q.K. Potential of deep representative learning features to interpret the sequence information in proteomics. Proteomics 2021, 22, e2100232. [Google Scholar] [CrossRef]
- Scimone, C.; Donato, L.; Marino, S.; Alafaci, C.; D’Angelo, R.; Sidoti, A. Vis-à-vis: A focus on genetic features of cerebral cavernous malformations and brain arteriovenous malformations pathogenesis. Neurol. Sci. 2019, 40, 243–251. [Google Scholar] [CrossRef]
- Lebatteux, D.; Remita, A.M.; Diallo, A.B. Toward an Alignment-Free Method for Feature Extraction and Accurate Classification of Viral Sequences. J. Comput. Biol. 2019, 26, 519–535. [Google Scholar] [CrossRef][Green Version]
- Ofer, D.; Brandes, N.; Linial, M. The language of proteins: NLP, machine learning & protein sequences. Comput. Struct. Biotechnol. J. 2021, 19, 1750–1758. [Google Scholar]
- Alakus, T.B.; Baykara, M. Comparison of Monkeypox and Wart DNA Sequences with Deep Learning Model. Appl. Sci. 2022, 12, 10216. [Google Scholar] [CrossRef]
- Do, D.T.; Le, N.Q.K. Using extreme gradient boosting to identify origin of replication in Saccharomyces cerevisiae via hybrid features. Genomics 2020, 112, 2445–2451. [Google Scholar] [CrossRef] [PubMed]



| Order | Family | Genus | Species |
|---|---|---|---|
| Articulavirales | Orthomyxoviridae | Alphainfluenzavirus | IAV |
| Articulavirales | Orthomyxoviridae | Betainfluenzavirus | IBV |
| Articulavirales | Orthomyxoviridae | Gammainfluenzavirus | ICV |
| Bunyavirales | Phenuiviridae | Bandavirus | SFTS |
| Flaviviridae | Flaviviridae | Flavivirus | Dengue |
| Picornavirales | Picornaviridae | Enterovirus | Enterovirus A |
| Picornavirales | Picornaviridae | Enterovirus | Enterovirus B |
| Blubervirales | Hepadnaviridae | Orthohepadnavirus | HBV |
| Amarillovirales | Flaviviridae | Hepacivirus | HCV |
| Herpesvirales | Herpesviridae | Human alphaherpesvirus 1 | HSV-1 |
| Zurhausenvirale | Papillomaviridae | Alphapapillomavirus | HPV |
| Chitovirales | Poxviridae | Orthopoxvirus | MPV |
| Amarillovirales | Flaviviridae | Flavivirus | WNV |
| Amarillovirales | Flaviviridae | Flavivirus | Zika |
| Input: NCBI GenBank nucleotide sequences |
| Output: Biomarkers extracted from genome |
| Let us denote the set of input nucleotide sequences as S, and the set of extracted biomarkers as B. Here is the mathematical representation of the given pseudo-code: |
| ● Collect nucleotide sequences: |
| ● S = {s1, s2, …, sn} |
| ● Filter and screen the sequences: |
| ● S′ = {s|s meets certain criteria} |
| ● Transform the genomes into k-mers: |
| ● K = {k1, k2, …, km}, where ki is a k-mer of a nucleotide sequence s |
| ● Train the BERT tokenizer on the k-mers: |
| ● T = Tokenizer.train (K) |
| ● Use SMOTE to balance imbalanced genomic data samples: |
| ● S″ = SMOTE (S′) |
| ● Perform additional preprocessing steps for the BERT model: |
| ● Convert nucleotide sequences to DNA-specific tokens using T |
| ● Apply necessary transformations to prepare the data for the BERT model |
| ● Preprocess the nucleotide sequence for the custom BERT model: |
| ● Tokenize the nucleotide sequence using the proposed DNA/RNA tokenizer |
| ● Pad any gaps or missing sequence regions with specific tokens |
| ● Encode the input k-mers into a bidirectional representation using the BERT model’s bidirectional encoder: |
| ● E = Encoder.encode (S″) |
| ● Extract specific biomarkers from the genome in an unsupervised manner using the BERT model: |
| ● B = Biomarker.extract (E) |
| ● Pass these biomarkers into a deep neural network-based classifier: |
| ● Classifier.train (B) |
| Disease Name | Count | Disease Name | Count |
|---|---|---|---|
| HBV | 5000 | Gamma Influenza Virus | 1941 |
| Betta Influenza Virus | 5000 | Dengue | 1866 |
| Alpha Influenza Virus | 5000 | Human Alpha Herpes | 1479 |
| Entero Virus B | 4653 | Human Papilloma Virus | 1355 |
| Hepaci Virus | 4619 | West Nile Virus | 371 |
| Entero Virus A | 4527 | Zika Virus | 321 |
| Dabie Banda Virus | 4193 | Monkey Pox | 28 |
| Parameter Name | Details | Parameter Name | Details |
| Maximum position embeddings | 5000 | Number of hidden layers | 2 |
| Number of attention heads | 2 | Hidden size | 768 |
| Training ratio | 80 | Testing ratio | 20 |
| Freeze_bert | False | epsilon value | 0.00000001 |
| Learning Rate | 0.00005 | optimizer | Adam |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadad, T.; Aurangzeb, R.A.; Safran, M.; Imran; Alfarhood, S.; Kim, J. Classification of Highly Divergent Viruses from DNA/RNA Sequence Using Transformer-Based Models. Biomedicines 2023, 11, 1323. https://doi.org/10.3390/biomedicines11051323
Sadad T, Aurangzeb RA, Safran M, Imran, Alfarhood S, Kim J. Classification of Highly Divergent Viruses from DNA/RNA Sequence Using Transformer-Based Models. Biomedicines. 2023; 11(5):1323. https://doi.org/10.3390/biomedicines11051323
Chicago/Turabian StyleSadad, Tariq, Raja Atif Aurangzeb, Mejdl Safran, Imran, Sultan Alfarhood, and Jungsuk Kim. 2023. "Classification of Highly Divergent Viruses from DNA/RNA Sequence Using Transformer-Based Models" Biomedicines 11, no. 5: 1323. https://doi.org/10.3390/biomedicines11051323
APA StyleSadad, T., Aurangzeb, R. A., Safran, M., Imran, Alfarhood, S., & Kim, J. (2023). Classification of Highly Divergent Viruses from DNA/RNA Sequence Using Transformer-Based Models. Biomedicines, 11(5), 1323. https://doi.org/10.3390/biomedicines11051323







