Novel Histopathological Biomarkers in Prostate Cancer: Implications and Perspectives
Abstract
:1. Introduction
2. Evidence Acquisition
3. Biomarkers
3.1. Extracellular Biomarkers
3.1.1. CD169
3.1.2. Neuropilin-1 (NRP1)
3.1.3. CD15 (Lewis X/Lex)
3.2. Intracellular Biomarkers
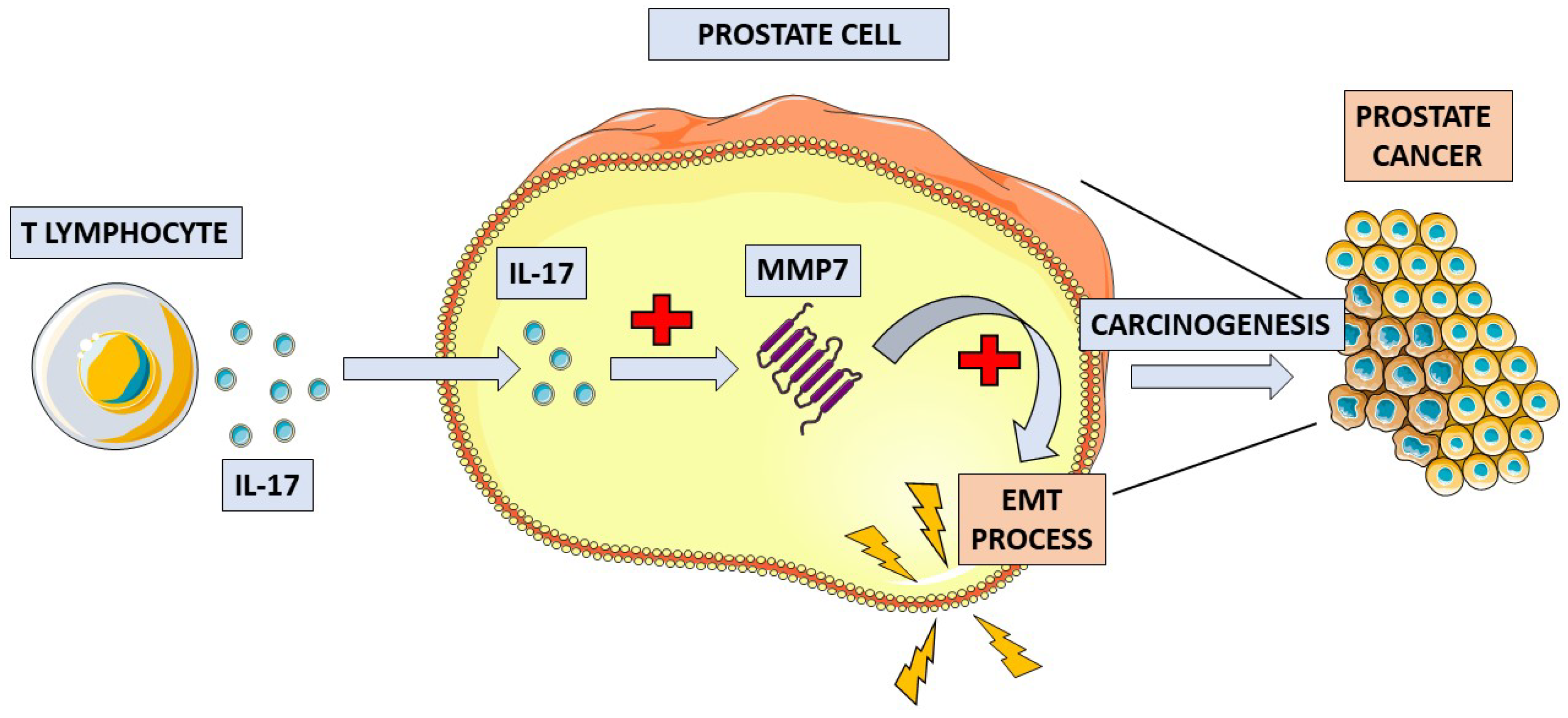
3.2.1. Interleukin 17 (IL-17)
3.2.2. Cofilin-1 (CFL1)
3.2.3. STAT3
3.2.4. LIM Domain Kinase 1 (LIMK1)
3.2.5. AMACR
3.2.6. Prostate-Specific Membrane Antigen (PSMA)
3.2.7. Appl1
3.2.8. Sortilin
3.2.9. Syndecan-1
3.2.10. p63
| Marker | Localization | Function | Clinical Implications |
|---|---|---|---|
| CD169 | Extracellular; surface antigen on macrophages | Tumor immunity [3]. | Improved survival [3,4]. |
| Neuropilin-1 (NRP1) | Extracellular; transmembrane co-receptor | Vascularization and progression of cancers [6]. | Higher Gleason and T scores. Positive nodal status. Progression to mCRPC [11]. |
| CD15 | Extracellular | Adhesion of cancer cells to the blood vessel endothelium [29,30,31,32]. Changes the structure of prostate cells mucins—NK cells cannot detect cancer cells [35]. | Disease aggressiveness. Hormone-refractory type of PCa [24,28]. |
| Cofilin-1 | Intracellular | Monomer binding; cytoskeleton reorganization [67]. | Higher Gleason score [73]. Positive nodal status [74]. Chemoresistance [67,68]. |
| Signal transducer and activator of transcription protein 3 (STAT3) | Intracellular | Transcription activator; promotes tumor cell proliferation [75,83]. | Worse overall and disease-free survival. Progression to hormone-refractory type of PCa [94]. |
| LIMK1 | Intracellular | Reorganization of actin cytoskeleton; intracellular androgen receptor signaling [120,121,122,123]. | Independent risk factor for lymph node metastasis and biochemical recurrence after prostatectomy [128]. |
| IL-17 family | Intracellular | Induces and mediates proinflammatory responses [42,43]. | Unclear, elevated levels of IL-17RC in CRPC [60,61]; higher expression of IL-17F in cancers with higher histological grades [66]. |
| AMACR | Intracellular; localized in peroxisomes and mitochondria | Degradation of branched fatty acids [133]. | Negative marker for benignity of prostate glands [139]. Possible role as a novel non-invasive diagnostic marker [153]. |
| Prostate-specific membrane antigen (PSMA) | Intracellular | Integral membrane protein [155]. | Detection of recurrence or metastases in PET/CT [159]. Radio-guided salvage surgery [161]. High expression linked with poor prognosis [166]. |
| Appl1 | Intracellular; in early endosomes | Controls intracellular transport speed [174]. | Associated with more aggressive PCa [177]. Improves the pathology diagnosis and grade of PCa [178]. |
| Sortilin | Intracellular | Intracellular transport; involved in sugar metabolism [179,180]. | Improves the pathology diagnosis and grade of PCa [178]. Delays progression of CRPC [182]. |
| Syndecan-1 | Intracellular/transmembrane | Cell proliferation, migration [183,184]. | Improves the pathology diagnosis and grade of PCa [178]. Predictor of poor prognosis [186]. |
| p63 | Intracellular; usually nuclei of basal cells | Epithelium development, regulation of proliferation and apoptosis [191]. | Positive marker for benignity of prostate glands [194]. |
4. New Perspectives for Biomarkers in PCa
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | artificial intelligence |
| AMACR | alpha-methylacyl-CoA racemase |
| Appl1 | adaptor protein containing a pleckstrin-homology domain, phosphotyrosine binding domain, and leucine zipper motif 1 |
| AR | androgen receptors |
| BCR | biochemical recurrence |
| BMI | body mass index |
| BPH | benign prostatic hyperplasia |
| CD15s | sialyl form of CD15 |
| CFL1 | cofilin-1 |
| CRPC | castration-resistant prostatę cancer |
| csPCa | clinically significant prostate cancer |
| CSS | cancer-specific survival |
| CT | computed tomography |
| ctDNA | circulating tumor DNA |
| DDR | DNA damage response |
| EMT | epithelial-to-mesenchymal transition |
| GLUT | glucose transporter |
| GS | Gleason score |
| HMW-CK | High Molecular Weight-Cytokeratin |
| HIF | hypoxia-inducible factor |
| ICG | indocyanine green |
| IL-17 | interleukin 17 |
| LB | liquid biopsy |
| LIMK1 | LIM domain kinase 1 |
| LIMKi | LIM domain kinase 1 inhibitor |
| mCRPC | metastatic castration-resistant prostate cancer |
| ML | machine learning |
| MMP7 | matrix metalloproteinase 7 |
| mpMRI | multiparametric magnetic resonance imaging |
| NK cell | natural killer cell |
| NRP1 | neuropilin-1 |
| nsPCa | non-significant prostate cancer |
| PARP | poly(ADP-ribose) polymerase |
| PCa | prostate cancer |
| PET | positron emission tomography |
| PSA | prostate-specific antigen |
| PSAD | prostate-specific antigen density |
| PSMA | prostate-specific membrane antigen |
| pSTAT3 | phosphorylated signal transducer and activator of transcription proteins 3 |
| ROS | reactive oxygen species |
| miRNA | microRNA |
| siRNA | small interference RNA |
| SLN | sentinel lymph node |
| STAT3 | signal transducer and activator of transcription proteins 3 |
| TβRI | transforming growth factor-β type I receptor |
| VEGF | vascular endothelial growth factor |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012: Globocan 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef] [PubMed]
- Komohara, Y.; Ohnishi, K.; Takeya, M. Possible Functions of CD169-Positive Sinus Macrophages in Lymph Nodes in Anti-Tumor Immune Responses. Cancer Sci. 2017, 108, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Strömvall, K.; Sundkvist, K.; Ljungberg, B.; Bergström, S.H.; Bergh, A. Reduced Number of CD169+ Macrophages in Pre-metastatic Regional Lymph Nodes Is Associated with Subsequent Metastatic Disease in an Animal Model and with Poor Outcome in Prostate Cancer Patients. Prostate 2017, 77, 1468–1477. [Google Scholar] [CrossRef]
- Asano, T.; Ohnishi, K.; Shiota, T.; Motoshima, T.; Sugiyama, Y.; Yatsuda, J.; Kamba, T.; Ishizaka, K.; Komohara, Y. CD169-positive Sinus Macrophages in the Lymph Nodes Determine Bladder Cancer Prognosis. Cancer Sci. 2018, 109, 1723–1730. [Google Scholar] [CrossRef]
- Lampropoulou, A.; Ruhrberg, C. Neuropilin Regulation of Angiogenesis. Biochem. Soc. Trans. 2014, 42, 1623–1628. [Google Scholar] [CrossRef]
- Ioannidou, E.; Moschetta, M.; Shah, S.; Parker, J.S.; Ozturk, M.A.; Pappas-Gogos, G.; Sheriff, M.; Rassy, E.; Boussios, S. Angiogenesis and Anti-Angiogenic Treatment in Prostate Cancer: Mechanisms of Action and Molecular Targets. Int. J. Mol. Sci. 2021, 22, 9926. [Google Scholar] [CrossRef]
- Pavlakis, D.; Kampantais, S.; Gkagkalidis, K.; Gourvas, V.; Memmos, D.; Tsionga, A.; Dimitriadis, G.; Vakalopoulos, I. Hypoxia-Inducible Factor 2a Expression Is Positively Correlated with Gleason Score in Prostate Cancer. Technol. Cancer Res. Treat 2021, 20, 1533033821990010. [Google Scholar] [CrossRef]
- Ravi, R.; Mookerjee, B.; Bhujwalla, Z.M.; Sutter, C.H.; Artemov, D.; Zeng, Q.; Dillehay, L.E.; Madan, A.; Semenza, G.L.; Bedi, A. Regulation of Tumor Angiogenesis by P53-Induced Degradation of Hypoxia-Inducible Factor 1alpha. Genes Dev. 2000, 14, 34–44. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia, Clonal Selection, and the Role of HIF-1 in Tumor Progression. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 71–103. [Google Scholar] [CrossRef]
- Bachelder, R.E.; Crago, A.; Chung, J.; Wendt, M.A.; Shaw, L.M.; Robinson, G.; Mercurio, A.M. Vascular Endothelial Growth Factor Is an Autocrine Survival Factor for Neuropilin-Expressing Breast Carcinoma Cells. Cancer Res. 2001, 61, 5736–5740. [Google Scholar]
- Fakhari, M.; Pullirsch, D.; Abraham, D.; Paya, K.; Hofbauer, R.; Holzfeind, P.; Hofmann, M.; Aharinejad, S. Selective Upregulation of Vascular Endothelial Growth Factor Receptors Neuropilin-1 and -2 in Human Neuroblastoma. Cancer 2002, 94, 258–263. [Google Scholar] [CrossRef]
- Parikh, A.A.; Fan, F.; Liu, W.B.; Ahmad, S.A.; Stoeltzing, O.; Reinmuth, N.; Bielenberg, D.; Bucana, C.D.; Klagsbrun, M.; Ellis, L.M. Neuropilin-1 in Human Colon Cancer: Expression, Regulation, and Role in Induction of Angiogenesis. Am. J. Pathol. 2004, 164, 2139–2151. [Google Scholar] [CrossRef]
- Hong, T.M.; Chen, Y.L.; Wu, Y.Y.; Yuan, A.; Chao, Y.C.; Chung, Y.C.; Wu, M.H.; Yang, S.C.; Pan, S.H.; Shih, J.Y.; et al. Targeting Neuropilin 1 as an Antitumor Strategy in Lung Cancer. Clin. Cancer Res. 2007, 13, 4759–4768. [Google Scholar] [CrossRef]
- Tse, B.W.C.; Volpert, M.; Ratther, E.; Stylianou, N.; Nouri, M.; McGowan, K.; Lehman, M.L.; McPherson, S.J.; Roshan-Moniri, M.; Butler, M.S.; et al. Neuropilin-1 Is Upregulated in the Adaptive Response of Prostate Tumors to Androgen-Targeted Therapies and Is Prognostic of Metastatic Progression and Patient Mortality. Oncogene 2017, 36, 3417–3427. [Google Scholar] [CrossRef]
- Battaglia, A.; Buzzonetti, A.; Monego, G.; Peri, L.; Ferrandina, G.; Fanfani, F.; Scambia, G.; Fattorossi, A. Neuropilin-1 Expression Identifies a Subset of Regulatory T Cells in Human Lymph Nodes That Is Modulated by Preoperative Chemoradiation Therapy in Cervical Cancer. Immunology 2008, 123, 129–138. [Google Scholar] [CrossRef]
- Snuderl, M.; Batista, A.; Kirkpatrick, N.D.; Ruiz de Almodovar, C.; Riedemann, L.; Walsh, E.C.; Anolik, R.; Huang, Y.; Martin, J.D.; Kamoun, W.; et al. Targeting Placental Growth Factor/Neuropilin 1 Pathway Inhibits Growth and Spread of Medulloblastoma. Cell 2013, 152, 1065–1076. [Google Scholar] [CrossRef]
- Pan, Q.; Chanthery, Y.; Liang, W.-C.; Stawicki, S.; Mak, J.; Rathore, N.; Tong, R.K.; Kowalski, J.; Yee, S.F.; Pacheco, G.; et al. Blocking Neuropilin-1 Function Has an Additive Effect with Anti-VEGF to Inhibit Tumor Growth. Cancer Cell 2007, 11, 53–67. [Google Scholar] [CrossRef]
- Futamura, N.; Nakamura, S.; Tatematsu, M.; Yamamura, Y.; Kannagi, R.; Hirose, H. Clinicopathologic Significance of Sialyl Le(x) Expression in Advanced Gastric Carcinoma. Br. J. Cancer 2000, 83, 1681–1687. [Google Scholar] [CrossRef]
- Schiffmann, L.; Schwarz, F.; Linnebacher, M.; Prall, F.; Pahnke, J.; Krentz, H.; Vollmar, B.; Klar, E. A Novel Sialyl Le(X) Expression Score as a Potential Prognostic Tool in Colorectal Cancer. World J. Surg. Oncol. 2012, 10, 95. [Google Scholar] [CrossRef]
- Torii, A.; Nakayama, A.; Harada, A.; Nakao, A.; Nonami, T.; Sakamoto, J.; Watanabe, T.; Ito, M.; Takagi, H. Expression of the CD15 Antigen in Hepatocellular Carcinoma. Cancer 1993, 71, 3864–3867. [Google Scholar] [CrossRef] [PubMed]
- Croce, M.V. An Introduction to the Relationship between Lewis x and Malignancy Mainly Related to Breast Cancer and Head Neck Squamous Cell Carcinoma (HNSCC). Cancer Investig. 2022, 40, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, I.; Vitagliano, G.; Caputo, A.; Montella, M.; Franco, R.; Ciancia, G.; Selleri, C.; Zeppa, P. CD15, CD30, and PAX5 Evaluation in Hodgkin’s Lymphoma on Fine-Needle Aspiration Cytology Samples. Diagn. Cytopathol. 2020, 48, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, K. Expression of Lewis(x), Sialylated Lewis(x), Lewis(a), and Sialylated Lewis(a) Antigens in Human Lung Carcinoma. Tohoku J. Exp. Med. 1991, 163, 17–30. [Google Scholar] [CrossRef]
- Wang, P.; Gong, S.; Liao, B.; Pan, J.; Wang, J.; Zou, D.; Zhao, L.; Xiong, S.; Deng, Y.; Yan, Q.; et al. HIF1α/HIF2α Induces Glioma Cell Dedifferentiation into Cancer Stem Cells through Sox2 under Hypoxic Conditions. J. Cancer 2022, 13, 1–14. [Google Scholar] [CrossRef]
- Ezeabikwa, B.; Mondal, N.; Antonopoulos, A.; Haslam, S.M.; Matsumoto, Y.; Martin-Caraballo, M.; Lehoux, S.; Mandalasi, M.; Ishaque, A.; Heimburg-Molinaro, J.; et al. Major Differences in Glycosylation and Fucosyltransferase Expression in Low-Grade versus High-Grade Bladder Cancer Cell Lines. Glycobiology 2021, 31, 1444–1463. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Huo, J.-P.; Zhang, X.-K.; Zhang, Y.-J.; Hu, W.-M.; Yang, P.; Lu, J.-B.; Zhang, Z.-L.; Cao, Y. Loss of CD15 Expression in Clear Cell Renal Cell Carcinoma Is Correlated with Worse Prognosis in Chinese Patients. Jpn. J. Clin. Oncol. 2017, 47, 1182–1188. [Google Scholar] [CrossRef]
- Jørgensen, T.; Berner, A.; Kaalhus, O.; Tveter, K.J.; Danielsen, H.E.; Bryne, M. Up-Regulation of the Oligosaccharide Sialyl LewisX: A New Prognostic Parameter in Metastatic Prostate Cancer. Cancer Res. 1995, 55, 1817–1819. [Google Scholar]
- Sheinfeld, J.; Reuter, V.E.; Sarkis, A.S.; Cordon-Cardo, C. Blood Group Antigens in Normal and Neoplastic Urothelium. J. Cell Biochem. Suppl. 1992, 16I, 50–55. [Google Scholar] [CrossRef]
- Szlasa, W.; Wilk, K.; Knecht-Gurwin, K.; Gurwin, A.; Froń, A.; Sauer, N.; Krajewski, W.; Saczko, J.; Szydełko, T.; Kulbacka, J.; et al. Prognostic and Therapeutic Role of CD15 and CD15s in Cancer. Cancers 2022, 14, 2203. [Google Scholar] [CrossRef]
- Mårtensson, S.; Bigler, S.A.; Brown, M.; Lange, P.H.; Brawer, M.K.; Hakomori, S. Sialyl-LewisX and Related Carbohydrate Antigens in the Prostate. Hum. Pathol. 1995, 26, 735–739. [Google Scholar] [CrossRef]
- Satoh, M.; Numahata, K.; Kawamura, S.; Saito, S.; Orikasa, S. Lack of Selectin-Dependent Adhesion in Prostate Cancer Cells Expressing Sialyl Lex. Int. J. Urol. 1998, 5, 86–91. [Google Scholar] [CrossRef]
- Martín-Satué, M.; Marrugat, R.; Cancelas, J.A.; Blanco, J. Enhanced Expression of Alpha(1,3)-Fucosyltransferase Genes Correlates with E-Selectin-Mediated Adhesion and Metastatic Potential of Human Lung Adenocarcinoma Cells. Cancer Res. 1998, 58, 1544–1550. [Google Scholar]
- Takada, A.; Ohmori, K.; Yoneda, T.; Tsuyuoka, K.; Hasegawa, A.; Kiso, M.; Kannagi, R. Contribution of Carbohydrate Antigens Sialyl Lewis A and Sialyl Lewis X to Adhesion of Human Cancer Cells to Vascular Endothelium. Cancer Res. 1993, 53, 354–361. [Google Scholar]
- Numahata, K.; Satoh, M.; Handa, K.; Saito, S.; Ohyama, C.; Ito, A.; Takahashi, T.; Hoshi, S.; Orikasa, S.; Hakomori, S. Sialosyl-Le(x) Expression Defines Invasive and Metastatic Properties of Bladder Carcinoma. Cancer 2002, 94, 673–685. [Google Scholar] [CrossRef]
- Ohyama, C.; Tsuboi, S.; Fukuda, M. Dual Roles of Sialyl Lewis X Oligosaccharides in Tumor Metastasis and Rejection by Natural Killer Cells. EMBO J. 1999, 18, 1516–1525. [Google Scholar] [CrossRef]
- Irimura, T. Cancer Metastasis Determined by Carbohydrate-Mediated Cell Adhesion. Adv. Exp. Med. Biol. 1994, 353, 27–34. [Google Scholar] [CrossRef]
- Patel, T.P.; Goelz, S.E.; Lobb, R.R.; Parekh, R.B. Isolation and Characterization of Natural Protein-Associated Carbohydrate Ligands for E-Selectin. Biochemistry 1994, 33, 14815–14824. [Google Scholar] [CrossRef]
- Okamoto, T.; Yoneyama, M.S.; Hatakeyama, S.; Mori, K.; Yamamoto, H.; Koie, T.; Saitoh, H.; Yamaya, K.; Funyu, T.; Fukuda, M.; et al. Core2 O-Glycan-Expressing Prostate Cancer Cells Are Resistant to NK Cell Immunity. Mol. Med. Rep. 2013, 7, 359–364. [Google Scholar] [CrossRef]
- Mitsuoka, C.; Kawakami-Kimura, N.; Kasugai-Sawada, M.; Hiraiwa, N.; Toda, K.; Ishida, H.; Kiso, M.; Hasegawa, A.; Kannagi, R. Sulfated Sialyl Lewis X, the Putative L-Selectin Ligand, Detected on Endothelial Cells of High Endothelial Venules by a Distinct Set of Anti-Sialyl Lewis X Antibodies. Biochem. Biophys. Res. Commun. 1997, 230, 546–551. [Google Scholar] [CrossRef]
- Munkley, J. Glycosylation Is a Global Target for Androgen Control in Prostate Cancer Cells. Endocr. Relat. Cancer 2017, 24, R49–R64. [Google Scholar] [CrossRef] [PubMed]
- Mastelić, A.; Čulić, V.Č.; Mužinić, N.R.; Vuica-Ross, M.; Barker, D.; Leung, E.Y.; Reynisson, J.; Markotić, A. Glycophenotype of Breast and Prostate Cancer Stem Cells Treated with Thieno [2,3-b]Pyridine Anticancer Compound. Drug Des. Devel. Ther. 2017, 11, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Ideo, H.; Kondo, J.; Nomura, T.; Nonomura, N.; Inoue, M.; Amano, J. Study of Glycosylation of Prostate-Specific Antigen Secreted by Cancer Tissue-Originated Spheroids Reveals New Candidates for Prostate Cancer Detection. Sci. Rep. 2020, 10, 2708. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.H.; Dong, C. A Novel Heterodimeric Cytokine Consisting of IL-17 and IL-17F Regulates Inflammatory Responses. Cell Res. 2007, 17, 435–440. [Google Scholar] [CrossRef]
- Pappu, R.; Ramirez-Carrozzi, V.; Sambandam, A. The Interleukin-17 Cytokine Family: Critical Players in Host Defence and Inflammatory Diseases. Immunology 2011, 134, 8–16. [Google Scholar] [CrossRef]
- De Angulo, A.; Faris, R.; Daniel, B.; Jolly, C.; deGraffenried, L. Age-Related Increase in IL-17 Activates pro-Inflammatory Signaling in Prostate Cells. Prostate 2015, 75, 449–462. [Google Scholar] [CrossRef]
- Song, X.; Qian, Y. IL-17 Family Cytokines Mediated Signaling in the Pathogenesis of Inflammatory Diseases. Cell Signal. 2013, 25, 2335–2347. [Google Scholar] [CrossRef]
- Divekar, R.; Kita, H. Recent Advances in Epithelium-Derived Cytokines (IL-33, IL-25, and Thymic Stromal Lymphopoietin) and Allergic Inflammation. Curr. Opin. Allergy Clin. Immunol. 2015, 15, 98–103. [Google Scholar] [CrossRef]
- Benatar, T.; Cao, M.Y.; Lee, Y.; Li, H.; Feng, N.; Gu, X.; Lee, V.; Jin, H.; Wang, M.; Der, S.; et al. Virulizin Induces Production of IL-17E to Enhance Antitumor Activity by Recruitment of Eosinophils into Tumors. Cancer Immunol. Immunother. 2008, 57, 1757–1769. [Google Scholar] [CrossRef]
- Wu, S.; Rhee, K.-J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.-R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A Human Colonic Commensal Promotes Colon Tumorigenesis via Activation of T Helper Type 17 T Cell Responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef]
- Chae, W.-J.; Bothwell, A.L.M. IL-17F Deficiency Inhibits Small Intestinal Tumorigenesis in ApcMin/+ Mice. Biochem. Biophys. Res. Commun. 2011, 414, 31–36. [Google Scholar] [CrossRef]
- Chae, W.-J.; Gibson, T.F.; Zelterman, D.; Hao, L.; Henegariu, O.; Bothwell, A.L.M. Ablation of IL-17A Abrogates Progression of Spontaneous Intestinal Tumorigenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 5540–5544. [Google Scholar] [CrossRef]
- Hyun, Y.S.; Han, D.S.; Lee, A.R.; Eun, C.S.; Youn, J.; Kim, H.-Y. Role of IL-17A in the Development of Colitis-Associated Cancer. Carcinogenesis 2012, 33, 931–936. [Google Scholar] [CrossRef]
- Xiao, M.; Wang, C.; Zhang, J.; Li, Z.; Zhao, X.; Qin, Z. IFNgamma Promotes Papilloma Development by Up-Regulating Th17-Associated Inflammation. Cancer Res. 2009, 69, 2010–2017. [Google Scholar] [CrossRef]
- Wang, L.; Yi, T.; Zhang, W.; Pardoll, D.M.; Yu, H. IL-17 Enhances Tumor Development in Carcinogen-Induced Skin Cancer. Cancer Res. 2010, 70, 10112–10120. [Google Scholar] [CrossRef]
- Chang, S.H.; Mirabolfathinejad, S.G.; Katta, H.; Cumpian, A.M.; Gong, L.; Caetano, M.S.; Moghaddam, S.J.; Dong, C. T Helper 17 Cells Play a Critical Pathogenic Role in Lung Cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 5664–5669. [Google Scholar] [CrossRef]
- Xu, B.; Guenther, J.F.; Pociask, D.A.; Wang, Y.; Kolls, J.K.; You, Z.; Chandrasekar, B.; Shan, B.; Sullivan, D.E.; Morris, G.F. Promotion of Lung Tumor Growth by Interleukin-17. Am. J. Physiol. Lung. Cell Mol. Physiol. 2014, 307, L497–L508. [Google Scholar] [CrossRef]
- Novitskiy, S.V.; Pickup, M.W.; Gorska, A.E.; Owens, P.; Chytil, A.; Aakre, M.; Wu, H.; Shyr, Y.; Moses, H.L. TGF-β Receptor II Loss Promotes Mammary Carcinoma Progression by Th17 Dependent Mechanisms. Cancer Discov. 2011, 1, 430–441. [Google Scholar] [CrossRef]
- Jarocki, M.; Karska, J.; Kowalski, S.; Kiełb, P.; Nowak, Ł.; Krajewski, W.; Saczko, J.; Kulbacka, J.; Szydełko, T.; Małkiewicz, B. Interleukin 17 and Its Involvement in Renal Cell Carcinoma. J. Clin. Med. 2022, 11, 4973. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, W.; Zhao, L.; Liang, Z.; Shen, W.; Hou, Q.; Wang, Z.; Jiang, J.; Ying, S. Immune Analysis of Expression of IL-17 Relative Ligands and Their Receptors in Bladder Cancer: Comparison with Polyp and Cystitis. BMC Immunol. 2016, 17, 36. [Google Scholar] [CrossRef]
- Steiner, G.E.; Newman, M.E.; Paikl, D.; Stix, U.; Memaran-Dagda, N.; Lee, C.; Marberger, M.J. Expression and Function of Pro-Inflammatory Interleukin IL-17 and IL-17 Receptor in Normal, Benign Hyperplastic, and Malignant Prostate. Prostate 2003, 56, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Haudenschild, D.; Moseley, T.; Rose, L.; Reddi, A.H. Soluble and Transmembrane Isoforms of Novel Interleukin-17 Receptor-like Protein by RNA Splicing and Expression in Prostate Cancer. J. Biol. Chem. 2002, 277, 4309–4316. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, X.; Sun, X.; Li, Y.; Wang, Z.; Jiang, J.; Han, H.; Shen, W.; Corrigan, C.J.; Sun, Y. Expression of IL-17A, E, and F and Their Receptors in Human Prostatic Cancer: Comparison with Benign Prostatic Hyperplasia. Prostate 2015, 75, 1844–1856. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Dong, Y.; Kong, X.; Zhang, Y.; Vessella, R.L.; Melamed, J. Differential Expression of IL-17RC Isoforms in Androgen-Dependent and Androgen-Independent Prostate Cancers. Neoplasia 2007, 9, 464–470. [Google Scholar] [CrossRef]
- You, Z.; Shi, X.-B.; DuRaine, G.; Haudenschild, D.; Tepper, C.G.; Lo, S.H.; Gandour-Edwards, R.; de Vere White, R.W.; Reddi, A.H. Interleukin-17 Receptor-like Gene Is a Novel Antiapoptotic Gene Highly Expressed in Androgen-Independent Prostate Cancer. Cancer Res. 2006, 66, 175–183. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, S.; Ge, D.; Zhang, Q.; Xue, Y.; Xiong, Z.; Abdel-Mageed, A.B.; Myers, L.; Hill, S.M.; Rowan, B.G.; et al. Interleukin-17 Promotes Formation and Growth of Prostate Adenocarcinoma in Mouse Models. Cancer Res. 2012, 72, 2589–2599. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, S.; Zhang, Q.; Xiong, Z.; Wang, A.R.; Myers, L.; Melamed, J.; Tang, W.W.; You, Z. Interleukin-17 Promotes Development of Castration-Resistant Prostate Cancer Potentially through Creating an Immunotolerant and pro-Angiogenic Tumor Microenvironment. Prostate 2014, 74, 869–879. [Google Scholar] [CrossRef]
- Cunningham, D.; Zhang, Q.; Liu, S.; Parajuli, K.R.; Nie, Q.; Ma, L.; Zhang, A.; Chen, Z.; You, Z. Interleukin-17 Promotes Metastasis in an Immunocompetent Orthotopic Mouse Model of Prostate Cancer. Am. J. Clin. Exp. Urol. 2018, 6, 114–122. [Google Scholar]
- Zhang, Q.; Liu, S.; Parajuli, K.R.; Zhang, W.; Zhang, K.; Mo, Z.; Liu, J.; Chen, Z.; Yang, S.; Wang, A.R.; et al. Interleukin-17 Promotes Prostate Cancer via MMP7-Induced Epithelial-to-Mesenchymal Transition. Oncogene 2017, 36, 687–699. [Google Scholar] [CrossRef]
- Janiczek, M.; Szylberg, Ł.; Antosik, P.; Kasperska, A.; Marszałek, A. Expression Levels of IL-17A, IL-17F, IL-17RA, and IL-17RC in Prostate Cancer with Taking into Account the Histological Grade According to Gleason Scale in Comparison to Benign Prostatic Hyperplasia: In Search of New Therapeutic Options. J. Immunol. Res. 2020, 2020, 4910595. [Google Scholar] [CrossRef]
- Xiao, P.; Ma, T.; Zhou, C.; Xu, Y.; Liu, Y.; Zhang, H. Anticancer Effect of Docetaxel Induces Apoptosis of Prostate Cancer via the Cofilin-1 and Paxillin Signaling Pathway. Mol. Med. Rep. 2016, 13, 4079–4084. [Google Scholar] [CrossRef]
- Pérez-Martínez, F.C.; Carrión, B.; Lucío, M.I.; Rubio, N.; Herrero, M.A.; Vázquez, E.; Ceña, V. Enhanced Docetaxel-Mediated Cytotoxicity in Human Prostate Cancer Cells through Knockdown of Cofilin-1 by Carbon Nanohorn Delivered SiRNA. Biomaterials 2012, 33, 8152–8159. [Google Scholar] [CrossRef]
- Wang, W.; Mouneimne, G.; Sidani, M.; Wyckoff, J.; Chen, X.; Makris, A.; Goswami, S.; Bresnick, A.R.; Condeelis, J.S. The Activity Status of Cofilin Is Directly Related to Invasion, Intravasation, and Metastasis of Mammary Tumors. J. Cell Biol. 2006, 173, 395–404. [Google Scholar] [CrossRef]
- Li, M.; Yin, J.; Mao, N.; Pan, L. Upregulation of Phosphorylated Cofilin 1 Correlates with Taxol Resistance in Human Ovarian Cancer in Vitro and in Vivo. Oncol. Rep. 2013, 29, 58–66. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, Y.; Fei, J.; Zhang, W. Expression of Cofilin 1 Is Positively Correlated with the Differentiation of Human Epithelial Ovarian Cancer. Oncol. Lett. 2012, 4, 1187–1190. [Google Scholar] [CrossRef]
- Mousavi, S.; Safaralizadeh, R.; Hosseinpour-Feizi, M.; Azimzadeh-Isfanjani, A.; Hashemzadeh, S. Study of Cofilin 1 Gene Expression in Colorectal Cancer. J. Gastrointest. Oncol. 2018, 9, 791–796. [Google Scholar] [CrossRef]
- Lu, L.I.; Fu, N.I.; Luo, X.U.; Li, X.-Y.; Li, X.-P. Overexpression of Cofilin 1 in Prostate Cancer and the Corresponding Clinical Implications. Oncol. Lett. 2015, 9, 2757–2761. [Google Scholar] [CrossRef]
- Chen, L.; Cai, J.; Huang, Y.; Tan, X.; Guo, Q.; Lin, X.; Zhu, C.; Zeng, X.; Liu, H.; Wu, X. Identification of Cofilin-1 as a Novel Mediator for the Metastatic Potentials and Chemoresistance of the Prostate Cancer Cells. Eur. J. Pharmacol. 2020, 880, 173100. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in Cancer Inflammation and Immunity: A Leading Role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Bollrath, J.; Phesse, T.J.; von Burstin, V.A.; Putoczki, T.; Bennecke, M.; Bateman, T.; Nebelsiek, T.; Lundgren-May, T.; Canli, O.; Schwitalla, S.; et al. Gp130-Mediated Stat3 Activation in Enterocytes Regulates Cell Survival and Cell-Cycle Progression during Colitis-Associated Tumorigenesis. Cancer Cell 2009, 15, 91–102. [Google Scholar] [CrossRef]
- Chiarle, R.; Simmons, W.J.; Cai, H.; Dhall, G.; Zamo, A.; Raz, R.; Karras, J.G.; Levy, D.E.; Inghirami, G. Stat3 Is Required for ALK-Mediated Lymphomagenesis and Provides a Possible Therapeutic Target. Nat. Med. 2005, 11, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, A.; Wang, S.C.; Morris, J.P.; Folias, A.E.; Liou, A.; Kim, G.E.; Akira, S.; Boucher, K.M.; Firpo, M.A.; Mulvihill, S.J.; et al. Stat3 and MMP7 Contribute to Pancreatic Ductal Adenocarcinoma Initiation and Progression. Cancer Cell 2011, 19, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.; Karin, E.; Terzic, J.; Mucida, D.; Yu, G.-Y.; Vallabhapurapu, S.; Scheller, J.; Rose-John, S.; Cheroutre, H.; Eckmann, L.; et al. IL-6 and Stat3 Are Required for Survival of Intestinal Epithelial Cells and Development of Colitis-Associated Cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef]
- Lee, H.; Deng, J.; Kujawski, M.; Yang, C.; Liu, Y.; Herrmann, A.; Kortylewski, M.; Horne, D.; Somlo, G.; Forman, S.; et al. STAT3-Induced S1PR1 Expression Is Crucial for Persistent STAT3 Activation in Tumors. Nat. Med. 2010, 16, 1421–1428. [Google Scholar] [CrossRef] [PubMed]
- Lesina, M.; Kurkowski, M.U.; Ludes, K.; Rose-John, S.; Treiber, M.; Klöppel, G.; Yoshimura, A.; Reindl, W.; Sipos, B.; Akira, S.; et al. Stat3/Socs3 Activation by IL-6 Transsignaling Promotes Progression of Pancreatic Intraepithelial Neoplasia and Development of Pancreatic Cancer. Cancer Cell 2011, 19, 456–469. [Google Scholar] [CrossRef]
- Yu, H.; Kortylewski, M.; Pardoll, D. Crosstalk between Cancer and Immune Cells: Role of STAT3 in the Tumour Microenvironment. Nat. Rev. Immunol. 2007, 7, 41–51. [Google Scholar] [CrossRef]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 Signalling in Cancer: New and Unexpected Biological Functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef]
- Deng, J.; Liu, Y.; Lee, H.; Herrmann, A.; Zhang, W.; Zhang, C.; Shen, S.; Priceman, S.J.; Kujawski, M.; Pal, S.K.; et al. S1PR1-STAT3 Signaling Is Crucial for Myeloid Cell Colonization at Future Metastatic Sites. Cancer Cell 2012, 21, 642–654. [Google Scholar] [CrossRef]
- Gough, D.J.; Corlett, A.; Schlessinger, K.; Wegrzyn, J.; Larner, A.C.; Levy, D.E. Mitochondrial STAT3 Supports Ras-Dependent Oncogenic Transformation. Science 2009, 324, 1713–1716. [Google Scholar] [CrossRef]
- Ernst, M.; Thiem, S.; Nguyen, P.M.; Eissmann, M.; Putoczki, T.L. Epithelial Gp130/Stat3 Functions: An Intestinal Signaling Node in Health and Disease. Semin. Immunol. 2014, 26, 29–37. [Google Scholar] [CrossRef]
- Lee, H.-J.; Zhuang, G.; Cao, Y.; Du, P.; Kim, H.-J.; Settleman, J. Drug Resistance via Feedback Activation of Stat3 in Oncogene-Addicted Cancer Cells. Cancer Cell 2014, 26, 207–221. [Google Scholar] [CrossRef]
- Dai, B.; Meng, J.; Peyton, M.; Girard, L.; Bornmann, W.G.; Ji, L.; Minna, J.D.; Fang, B.; Roth, J.A. STAT3 Mediates Resistance to MEK Inhibitor through MicroRNA MiR-17. Cancer Res 2011, 71, 3658–3668. [Google Scholar] [CrossRef]
- Hedvat, M.; Huszar, D.; Herrmann, A.; Gozgit, J.M.; Schroeder, A.; Sheehy, A.; Buettner, R.; Proia, D.; Kowolik, C.M.; Xin, H.; et al. The JAK2 Inhibitor AZD1480 Potently Blocks Stat3 Signaling and Oncogenesis in Solid Tumors. Cancer Cell 2009, 16, 487–497. [Google Scholar] [CrossRef]
- Sen, M.; Thomas, S.M.; Kim, S.; Yeh, J.I.; Ferris, R.L.; Johnson, J.T.; Duvvuri, U.; Lee, J.; Sahu, N.; Joyce, S.; et al. First-in-Human Trial of a STAT3 Decoy Oligonucleotide in Head and Neck Tumors: Implications for Cancer Therapy. Cancer Discov. 2012, 2, 694–705. [Google Scholar] [CrossRef]
- Hussain, S.F.; Kong, L.-Y.; Jordan, J.; Conrad, C.; Madden, T.; Fokt, I.; Priebe, W.; Heimberger, A.B. A Novel Small Molecule Inhibitor of Signal Transducers and Activators of Transcription 3 Reverses Immune Tolerance in Malignant Glioma Patients. Cancer Res. 2007, 67, 9630–9636. [Google Scholar] [CrossRef]
- Lin, L.; Hutzen, B.; Zuo, M.; Ball, S.; Deangelis, S.; Foust, E.; Pandit, B.; Ihnat, M.A.; Shenoy, S.S.; Kulp, S.; et al. Novel STAT3 Phosphorylation Inhibitors Exhibit Potent Growth-Suppressive Activity in Pancreatic and Breast Cancer Cells. Cancer Res. 2010, 70, 2445–2454. [Google Scholar] [CrossRef]
- Yan, S.; Li, Z.; Thiele, C.J. Inhibition of STAT3 with Orally Active JAK Inhibitor, AZD1480, Decreases Tumor Growth in Neuroblastoma and Pediatric Sarcomas In Vitro and In Vivo. Oncotarget 2013, 4, 433–445. [Google Scholar] [CrossRef]
- Tam, L.; McGlynn, L.M.; Traynor, P.; Mukherjee, R.; Bartlett, J.M.S.; Edwards, J. Expression Levels of the JAK/STAT Pathway in the Transition from Hormone-Sensitive to Hormone-Refractory Prostate Cancer. Br. J. Cancer 2007, 97, 378–383. [Google Scholar] [CrossRef]
- Golus, M.; Bugajski, P.; Chorbińska, J.; Krajewski, W.; Lemiński, A.; Saczko, J.; Kulbacka, J.; Szydełko, T.; Małkiewicz, B. STAT3 and Its Pathways’ Dysregulation—Underestimated Role in Urological Tumors. Cells 2022, 11, 3024. [Google Scholar] [CrossRef]
- Min, H.; Wei-hong, Z. Constitutive Activation of Signal Transducer and Activator of Transcription 3 in Epithelial Ovarian Carcinoma. J. Obstet. Gynaecol. Res. 2009, 35, 918–925. [Google Scholar] [CrossRef]
- Mano, Y.; Aishima, S.; Fujita, N.; Tanaka, Y.; Kubo, Y.; Motomura, T.; Taketomi, A.; Shirabe, K.; Maehara, Y.; Oda, Y. Tumor-Associated Macrophage Promotes Tumor Progression via STAT3 Signaling in Hepatocellular Carcinoma. Pathobiology 2013, 80, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-H.; Xu, G.-L.; Jia, W.-D.; Li, J.-S.; Ma, J.-L.; Ren, W.-H.; Ge, Y.-S.; Yu, J.-H.; Liu, W.-B.; Wang, W. Activation of STAT3 Signal Pathway Correlates with Twist and E-Cadherin Expression in Hepatocellular Carcinoma and Their Clinical Significance. J. Surg. Res. 2012, 174, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Denley, S.M.; Jamieson, N.B.; McCall, P.; Oien, K.A.; Morton, J.P.; Carter, C.R.; Edwards, J.; McKay, C.J. Activation of the IL-6R/Jak/Stat Pathway Is Associated with a Poor Outcome in Resected Pancreatic Ductal Adenocarcinoma. J. Gastrointest. Surg. 2013, 17, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Huang, R.; Chang, W.; Jiang, T.; Huang, K.; Cao, J.; Sun, X.; Qiu, Z. The Expression and Clinical Significance of PSTAT3, VEGF and VEGF-C in Pancreatic Adenocarcinoma. Neoplasma 2012, 59, 52–61. [Google Scholar] [CrossRef]
- Horiguchi, A.; Oya, M.; Shimada, T.; Uchida, A.; Marumo, K.; Murai, M. Activation of Signal Transducer and Activator of Transcription 3 in Renal Cell Carcinoma: A Study of Incidence and Its Association with Pathological Features and Clinical Outcome. J. Urol. 2002, 168, 762–765. [Google Scholar] [CrossRef]
- Kusaba, T.; Nakayama, T.; Yamazumi, K.; Yakata, Y.; Yoshizaki, A.; Inoue, K.; Nagayasu, T.; Sekine, I. Activation of STAT3 Is a Marker of Poor Prognosis in Human Colorectal Cancer. Oncol. Rep. 2006, 15, 1445–1451. [Google Scholar] [CrossRef]
- Gordziel, C.; Bratsch, J.; Moriggl, R.; Knösel, T.; Friedrich, K. Both STAT1 and STAT3 Are Favourable Prognostic Determinants in Colorectal Carcinoma. Br. J. Cancer 2013, 109, 138–146. [Google Scholar] [CrossRef]
- Monnien, F.; Zaki, H.; Borg, C.; Mougin, C.; Bosset, J.-F.; Mercier, M.; Arbez-Gindre, F.; Kantelip, B. Prognostic Value of Phosphorylated STAT3 in Advanced Rectal Cancer: A Study from 104 French Patients Included in the EORTC 22921 Trial. J. Clin. Pathol. 2010, 63, 873–878. [Google Scholar] [CrossRef]
- Li, X.; Yu, Z.; Li, Y.; Liu, S.; Gao, C.; Hou, X.; Yao, R.; Cui, L. The Tumor Suppressor MiR-124 Inhibits Cell Proliferation by Targeting STAT3 and Functions as a Prognostic Marker for Postoperative NSCLC Patients. Int. J. Oncol. 2015, 46, 798–808. [Google Scholar] [CrossRef]
- WANG, M.; CHEN, G.-Y.; SONG, H.-T.; HONG, X.; YANG, Z.-Y.; SUI, G.-J. Significance of CXCR4, Phosphorylated STAT3 and VEGF-A Expression in Resected Non-Small Cell Lung Cancer. Exp. Ther. Med. 2011, 2, 517–522. [Google Scholar] [CrossRef]
- Yu, Y.; Zhao, Q.; Wang, Z.; Liu, X.-Y. Activated STAT3 Correlates with Prognosis of Non-Small Cell Lung Cancer and Indicates New Anticancer Strategies. Cancer Chemother. Pharmacol. 2015, 75, 917–922. [Google Scholar] [CrossRef]
- Zhang, W.; Pal, S.K.; Liu, X.; Yang, C.; Allahabadi, S.; Bhanji, S.; Figlin, R.A.; Yu, H.; Reckamp, K.L. Myeloid Clusters Are Associated with a Pro-Metastatic Environment and Poor Prognosis in Smoking-Related Early Stage Non-Small Cell Lung Cancer. PLoS ONE 2013, 8, e65121. [Google Scholar] [CrossRef]
- Zhao, X.; Sun, X.; Li, X. Expression and Clinical Significance of STAT3, P-STAT3, and VEGF-C in Small Cell Lung Cancer. Asian Pac. J. Cancer Prev. 2012, 13, 2873–2877. [Google Scholar] [CrossRef]
- Deng, J.; Liang, H.; Zhang, R.; Sun, D.; Pan, Y.; Liu, Y.; Zhang, L.; Hao, X. STAT3 Is Associated with Lymph Node Metastasis in Gastric Cancer. Tumour. Biol. 2013, 34, 2791–2800. [Google Scholar] [CrossRef]
- Jia, Y.; Liu, D.; Xiao, D.; Ma, X.; Han, S.; Zheng, Y.; Sun, S.; Zhang, M.; Gao, H.; Cui, X.; et al. Expression of AFP and STAT3 Is Involved in Arsenic Trioxide-Induced Apoptosis and Inhibition of Proliferation in AFP-Producing Gastric Cancer Cells. PLoS ONE 2013, 8, e54774. [Google Scholar] [CrossRef]
- Xiong, H.; Du, W.; Wang, J.-L.; Wang, Y.-C.; Tang, J.-T.; Hong, J.; Fang, J.-Y. Constitutive Activation of STAT3 Is Predictive of Poor Prognosis in Human Gastric Cancer. J. Mol. Med. 2012, 90, 1037–1046. [Google Scholar] [CrossRef]
- Woo, S.; Lee, B.L.; Yoon, J.; Cho, S.J.; Baik, T.-K.; Chang, M.S.; Lee, H.E.; Park, J.-W.; Kim, Y.-H.; Kim, W.H. Constitutive Activation of Signal Transducers and Activators of Transcription 3 Correlates with Better Prognosis, Cell Proliferation and Hypoxia-Inducible Factor-1α in Human Gastric Cancer. Pathobiology 2011, 78, 295–301. [Google Scholar] [CrossRef]
- Wu, W.-Y.; Li, J.; Wu, Z.-S.; Zhang, C.-L.; Meng, X.-L.; Lobie, P.E. Prognostic Significance of Phosphorylated Signal Transducer and Activator of Transcription 3 and Suppressor of Cytokine Signaling 3 Expression in Hepatocellular Carcinoma. Exp. Ther. Med. 2011, 2, 647–653. [Google Scholar] [CrossRef]
- Lee, I.; Fox, P.S.; Ferguson, S.D.; Bassett, R.; Kong, L.-Y.; Schacherer, C.W.; Gershenwald, J.E.; Grimm, E.A.; Fuller, G.N.; Heimberger, A.B. The Expression of P-STAT3 in Stage IV Melanoma: Risk of CNS Metastasis and Survival. Oncotarget 2012, 3, 336–344. [Google Scholar] [CrossRef]
- Sonnenblick, A.; Uziely, B.; Nechushtan, H.; Kadouri, L.; Galun, E.; Axelrod, J.H.; Katz, D.; Daum, H.; Hamburger, T.; Maly, B.; et al. Tumor STAT3 Tyrosine Phosphorylation Status, as a Predictor of Benefit from Adjuvant Chemotherapy for Breast Cancer. Breast Cancer Res. Treat. 2013, 138, 407–413. [Google Scholar] [CrossRef]
- Dolled-Filhart, M.; Camp, R.L.; Kowalski, D.P.; Smith, B.L.; Rimm, D.L. Tissue Microarray Analysis of Signal Transducers and Activators of Transcription 3 (Stat3) and Phospho-Stat3 (Tyr705) in Node-Negative Breast Cancer Shows Nuclear Localization Is Associated with a Better Prognosis. Clin. Cancer Res. 2003, 9, 594–600. [Google Scholar]
- Sonnenblick, A.; Shriki, A.; Galun, E.; Axelrod, J.H.; Daum, H.; Rottenberg, Y.; Hamburger, T.; Mali, B.; Peretz, T. Tissue Microarray-Based Study of Patients with Lymph Node-Positive Breast Cancer Shows Tyrosine Phosphorylation of Signal Transducer and Activator of Transcription 3 (Tyrosine705-STAT3) Is a Marker of Good Prognosis. Clin. Transl. Oncol. 2012, 14, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Wu, D.; Zhao, L.; Huang, L.; Shen, G.; Huang, J.; Chai, Y. Prognostic Role of STAT3 in Solid Tumors: A Systematic Review and Meta-Analysis. Oncotarget 2016, 7, 19863–19883. [Google Scholar] [CrossRef] [PubMed]
- McConnell, B.V.; Koto, K.; Gutierrez-Hartmann, A. Nuclear and Cytoplasmic LIMK1 Enhances Human Breast Cancer Progression. Mol. Cancer 2011, 10, 75. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Su, J.; Shi, L.; Liao, Q.; Su, Q. DADS Downregulates the Rac1-ROCK1/PAK1-LIMK1-ADF/Cofilin Signaling Pathway, Inhibiting Cell Migration and Invasion. Oncol. Rep. 2013, 29, 605–612. [Google Scholar] [CrossRef]
- Sousa-Squiavinato, A.C.M.; Vasconcelos, R.I.; Gehren, A.S.; Fernandes, P.V.; de Oliveira, I.M.; Boroni, M.; Morgado-Díaz, J.A. Cofilin-1, LIMK1 and SSH1 Are Differentially Expressed in Locally Advanced Colorectal Cancer and According to Consensus Molecular Subtypes. Cancer Cell Int. 2021, 21, 69. [Google Scholar] [CrossRef]
- Mardilovich, K.; Gabrielsen, M.; McGarry, L.; Orange, C.; Patel, R.; Shanks, E.; Edwards, J.; Olson, M.F. Elevated LIM Kinase 1 in Non-Metastatic Prostate Cancer Reflects Its Role in Facilitating Androgen Receptor Nuclear Translocation. Mol. Cancer Ther. 2015, 14, 246–258. [Google Scholar] [CrossRef]
- Sun, X.; Li, S.; Lin, H. LIMK1 Interacts with STK25 to Regulate EMT and Promote the Proliferation and Metastasis of Colorectal Cancer. J. Oncol. 2022, 2022, 3963883. [Google Scholar] [CrossRef]
- Lu, G.; Zhou, Y.; Zhang, C.; Zhang, Y. Upregulation of LIMK1 Is Correlated with Poor Prognosis and Immune Infiltrates in Lung Adenocarcinoma. Front. Genet. 2021, 12, 671585. [Google Scholar] [CrossRef]
- Li, Z.-F.; Yao, Y.-D.; Zhao, Y.-Y.; Liu, Y.; Liu, Z.-H.; Hu, P.; Zhu, Z.-R. Effects of PAK4/LIMK1/Cofilin-1 Signaling Pathway on Proliferation, Invasion, and Migration of Human Osteosarcoma Cells. J. Clin. Lab. Anal. 2020, 34, e23362. [Google Scholar] [CrossRef]
- Qiao, Y.; Jin, T.; Guan, S.; Cheng, S.; Wen, S.; Zeng, H.; Zhao, M.; Yang, L.; Wan, X.; Qiu, Y.; et al. Long Non-Coding RNA Lnc-408 Promotes Invasion and Metastasis of Breast Cancer Cell by Regulating LIMK1. Oncogene 2021, 40, 4198–4213. [Google Scholar] [CrossRef]
- Huang, J.-B.; Wu, Y.-P.; Lin, Y.-Z.; Cai, H.; Chen, S.-H.; Sun, X.-L.; Li, X.-D.; Wei, Y.; Zheng, Q.-S.; Xu, N.; et al. Up-Regulation of LIMK1 Expression in Prostate Cancer Is Correlated with Poor Pathological Features, Lymph Node Metastases and Biochemical Recurrence. J. Cell Mol. Med. 2020, 24, 4698–4706. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, H.; Pan, X.-W.; Xu, D.-F.; Cui, X.-G.; Chen, J.; Hong, Y.; Gao, Y.; Yin, L.; Ye, J.-Q.; et al. The Prognostic Value of Lymphovascular Invasion in Radical Prostatectomy: A Systematic Review and Meta-Analysis. Asian J. Androl. 2016, 18, 780–785. [Google Scholar] [CrossRef]
- Jiang, W.; Zhang, L.; Wu, B.; Zha, Z.; Zhao, H.; Jun, Y.; Jiang, Y. The Impact of Lymphovascular Invasion in Patients with Prostate Cancer Following Radical Prostatectomy and Its Association with Their Clinicopathological Features: An Updated PRISMA-Compliant Systematic Review and Meta-Analysis. Medicine 2018, 97, e13537. [Google Scholar] [CrossRef]
- Mohler, J.L.; Gregory, C.W.; Ford, O.H.; Kim, D.; Weaver, C.M.; Petrusz, P.; Wilson, E.M.; French, F.S. The Androgen Axis in Recurrent Prostate Cancer. Clin. Cancer Res. 2004, 10, 440–448. [Google Scholar] [CrossRef]
- Lennicke, C.; Rahn, J.; Lichtenfels, R.; Wessjohann, L.A.; Seliger, B. Hydrogen Peroxide—Production, Fate and Role in Redox Signaling of Tumor Cells. Cell Commun. Signal. 2015, 13, 39. [Google Scholar] [CrossRef]
- Esfahani, M.; Ataei, N.; Panjehpour, M. Biomarkers for Evaluation of Prostate Cancer Prognosis. Asian Pac. J. Cancer Prev. 2015, 16, 2601–2611. [Google Scholar] [CrossRef]
- Etheridge, T.; Straus, J.; Ritter, M.A.; Jarrard, D.F.; Huang, W. Semen AMACR Protein as a Novel Method for Detecting Prostate Cancer. Urol. Oncol. 2018, 36, 532.e1–532.e7. [Google Scholar] [CrossRef]
- Eichelberg, C.; Minner, S.; Isbarn, H.; Burandt, E.; Terracciano, L.; Moch, H.; Kell, A.; Heuer, R.; Chun, F.K.; Sauter, G.; et al. Prognostic Value of Alpha-Methyl CoA Racemase (AMACR) Expression in Renal Cell Carcinoma. World J. Urol. 2013, 31, 847–853. [Google Scholar] [CrossRef]
- Nozawa, Y.; Nishikura, K.; Ajioka, Y.; Aoyagi, Y. Relationship between Alpha-Methylacyl-Coenzyme A Racemase Expression and Mucin Phenotype in Gastric Cancer. Hum. Pathol. 2012, 43, 878–887. [Google Scholar] [CrossRef]
- Noske, A.; Zimmermann, A.-K.; Caduff, R.; Varga, Z.; Fink, D.; Moch, H.; Kristiansen, G. Alpha-Methylacyl-CoA Racemase (AMACR) Expression in Epithelial Ovarian Cancer. Virchows Arch. 2011, 459, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-P.; Tsung, A.; Liu, S.; Nalesnick, M.; Geller, D.; Michalopoulos, G.; Luo, J.-H. Detection of Fusion Transcripts in the Serum Samples of Patients with Hepatocellular Carcinoma. Oncotarget 2019, 10, 3352–3360. [Google Scholar] [CrossRef] [PubMed]
- Boran, C.; Kandirali, E.; Yilmaz, F.; Serin, E.; Akyol, M. Reliability of the 34βE12, Keratin 5/6, P63, Bcl-2, and AMACR in the Diagnosis of Prostate Carcinoma. Urol. Oncol. 2011, 29, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Magi-Galluzzi, C.; Luo, J.; Isaacs, W.B.; Hicks, J.L.; de Marzo, A.M.; Epstein, J.I. Alpha-Methylacyl-CoA Racemase: A Variably Sensitive Immunohistochemical Marker for the Diagnosis of Small Prostate Cancer Foci on Needle Biopsy. Am. J. Surg. Pathol. 2003, 27, 1128–1133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Aydin, H.; Kanane, H.; Epstein, J.I. How Often Does Alpha-Methylacyl-CoA-Racemase Contribute to Resolving an Atypical Diagnosis on Prostate Needle Biopsy beyond That Provided by Basal Cell Markers? Am. J. Surg. Pathol. 2004, 28, 239–243. [Google Scholar] [CrossRef]
- Jiang, Z.; Woda, B.A.; Rock, K.L.; Xu, Y.; Savas, L.; Khan, A.; Pihan, G.; Cai, F.; Babcook, J.S.; Rathanaswami, P.; et al. P504S: A New Molecular Marker for the Detection of Prostate Carcinoma. Am. J. Surg. Pathol. 2001, 25, 1397–1404. [Google Scholar] [CrossRef]
- Rubin, M.A.; Zhou, M.; Dhanasekaran, S.M.; Varambally, S.; Barrette, T.R.; Sanda, M.G.; Pienta, K.J.; Ghosh, D.; Chinnaiyan, A.M. Alpha-Methylacyl Coenzyme A Racemase as a Tissue Biomarker for Prostate Cancer. JAMA 2002, 287, 1662–1670. [Google Scholar] [CrossRef]
- Luo, J.; Zha, S.; Gage, W.R.; Dunn, T.A.; Hicks, J.L.; Bennett, C.J.; Ewing, C.M.; Platz, E.A.; Ferdinandusse, S.; Wanders, R.J.; et al. Alpha-Methylacyl-CoA Racemase: A New Molecular Marker for Prostate Cancer. Cancer Res. 2002, 62, 2220–2226. [Google Scholar]
- Rathod, S.G.; Jaiswal, D.G.; Bindu, R.S. Diagnostic Utility of Triple Antibody (AMACR, HMWCK and P63) Stain in Prostate Neoplasm. J. Family Med. Prim. Care 2019, 8, 2651–2655. [Google Scholar] [CrossRef]
- Jiang, N.; Zhu, S.; Chen, J.; Niu, Y.; Zhou, L. A-Methylacyl-CoA Racemase (AMACR) and Prostate-Cancer Risk: A Meta-Analysis of 4385 Participants. PLoS ONE 2013, 8, e74386. [Google Scholar] [CrossRef]
- Jain, D.; Gupta, S.; Marwah, N.; Kalra, R.; Gupta, V.; Gill, M.; Jain, N.; Lal, S.; Sen, R. Evaluation of Role of Alpha-Methyl Acyl-Coenzyme A Racemase/P504S and High Molecular Weight Cytokeratin in Diagnosing Prostatic Lesions. J. Cancer Res. Ther. 2017, 13, 21–25. [Google Scholar] [CrossRef]
- Zha, S.; Ferdinandusse, S.; Denis, S.; Wanders, R.J.; Ewing, C.M.; Luo, J.; De Marzo, A.M.; Isaacs, W.B. Alpha-Methylacyl-CoA Racemase as an Androgen-Independent Growth Modifier in Prostate Cancer. Cancer Res. 2003, 63, 7365–7376. [Google Scholar]
- Yevglevskis, M.; Lee, G.L.; Nathubhai, A.; Petrova, Y.D.; James, T.D.; Threadgill, M.D.; Woodman, T.J.; Lloyd, M.D. A Novel Colorimetric Assay for α-Methylacyl-CoA Racemase 1A (AMACR; P504S) Utilizing the Elimination of 2,4-Dinitrophenolate. Chem. Commun. 2017, 53, 5087–5090. [Google Scholar] [CrossRef]
- Carnell, A.J.; Kirk, R.; Smith, M.; McKenna, S.; Lian, L.-Y.; Gibson, R. Inhibition of Human α-Methylacyl CoA Racemase (AMACR): A Target for Prostate Cancer. ChemMedChem 2013, 8, 1643–1647. [Google Scholar] [CrossRef]
- Takahara, K.; Azuma, H.; Sakamoto, T.; Kiyama, S.; Inamoto, T.; Ibuki, N.; Nishida, T.; Nomi, H.; Ubai, T.; Segawa, N.; et al. Conversion of Prostate Cancer from Hormone Independency to Dependency Due to AMACR Inhibition: Involvement of Increased AR Expression and Decreased IGF1 Expression. Anticancer Res. 2009, 29, 2497–2505. [Google Scholar]
- Brahmkhatri, V.P.; Prasanna, C.; Atreya, H.S. Insulin-like Growth Factor System in Cancer: Novel Targeted Therapies. BioMed Res. Int. 2015, 2015, 538019. [Google Scholar] [CrossRef]
- Jin, X.; Ji, J.; Niu, D.; Yang, Y.; Tao, S.; Wan, L.; Xu, B.; Chen, S.; Wang, F.; Chen, M. Urine Exosomal AMACR Is a Novel Biomarker for Prostate Cancer Detection at Initial Biopsy. Front. Oncol. 2022, 12, 904315. [Google Scholar] [CrossRef]
- Kotova, E.S.; Savochkina, Y.A.; Doludin, Y.V.; Vasilyev, A.O.; Prilepskay, E.A.; Potoldykova, N.V.; Babalyan, K.A.; Kanygina, A.V.; Morozov, A.O.; Govorov, A.V.; et al. Identification of Clinically Significant Prostate Cancer by Combined PCA3 and AMACR MRNA Detection in Urine Samples. Res. Rep. Urol. 2020, 12, 403–413. [Google Scholar] [CrossRef]
- Pinto, J.T.; Suffoletto, B.P.; Berzin, T.M.; Qiao, C.H.; Lin, S.; Tong, W.P.; May, F.; Mukherjee, B.; Heston, W.D. Prostate-Specific Membrane Antigen: A Novel Folate Hydrolase in Human Prostatic Carcinoma Cells. Clin. Cancer Res. 1996, 2, 1445–1451. [Google Scholar]
- Morgantetti, G.; Ng, K.L.; Samaratunga, H.; Rhee, H.; Gobe, G.C.; Wood, S.T. Prostate Specific Membrane Antigen (PSMA) Expression in Vena Cava Tumour Thrombi of Clear Cell Renal Cell Carcinoma Suggests a Role for PSMA-Driven Tumour Neoangiogenesis. Transl. Androl. Urol. 2019, 8, S147–S155. [Google Scholar] [CrossRef]
- Holzgreve, A.; Biczok, A.; Ruf, V.C.; Liesche-Starnecker, F.; Steiger, K.; Kirchner, M.A.; Unterrainer, M.; Mittlmeier, L.; Herms, J.; Schlegel, J.; et al. PSMA Expression in Glioblastoma as a Basis for Theranostic Approaches: A Retrospective, Correlational Panel Study Including Immunohistochemistry, Clinical Parameters and PET Imaging. Front. Oncol. 2021, 11, 646387. [Google Scholar] [CrossRef] [PubMed]
- Queisser, A.; Hagedorn, S.A.; Braun, M.; Vogel, W.; Duensing, S.; Perner, S. Comparison of Different Prostatic Markers in Lymph Node and Distant Metastases of Prostate Cancer. Mod. Pathol. 2015, 28, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Woo, S.; Kim, Y.J.; Suh, C.H. Impact of 68Ga-PSMA PET on the Management of Patients with Prostate Cancer: A Systematic Review and Meta-Analysis. Eur. Urol. 2018, 74, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Afshar-Oromieh, A.; Holland-Letz, T.; Giesel, F.L.; Kratochwil, C.; Mier, W.; Haufe, S.; Debus, N.; Eder, M.; Eisenhut, M.; Schäfer, M.; et al. Diagnostic Performance of 68Ga-PSMA-11 (HBED-CC) PET/CT in Patients with Recurrent Prostate Cancer: Evaluation in 1007 Patients. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Maurer, T.; Robu, S.; Schottelius, M.; Schwamborn, K.; Rauscher, I.; van den Berg, N.S.; van Leeuwen, F.W.B.; Haller, B.; Horn, T.; Heck, M.M.; et al. 99mTechnetium-Based Prostate-Specific Membrane Antigen-Radioguided Surgery in Recurrent Prostate Cancer. Eur. Urol. 2019, 75, 659–666. [Google Scholar] [CrossRef]
- Minner, S.; Wittmer, C.; Graefen, M.; Salomon, G.; Steuber, T.; Haese, A.; Huland, H.; Bokemeyer, C.; Yekebas, E.; Dierlamm, J.; et al. High Level PSMA Expression Is Associated with Early PSA Recurrence in Surgically Treated Prostate Cancer. Prostate 2011, 71, 281–288. [Google Scholar] [CrossRef]
- Paschalis, A.; Sheehan, B.; Riisnaes, R.; Rodrigues, D.N.; Gurel, B.; Bertan, C.; Ferreira, A.; Lambros, M.B.K.; Seed, G.; Yuan, W.; et al. Prostate-Specific Membrane Antigen Heterogeneity and DNA Repair Defects in Prostate Cancer. Eur. Urol. 2019, 76, 469–478. [Google Scholar] [CrossRef]
- Ferraro, D.A.; Rüschoff, J.H.; Muehlematter, U.J.; Kranzbühler, B.; Müller, J.; Messerli, M.; Husmann, L.; Hermanns, T.; Eberli, D.; Rupp, N.J.; et al. Immunohistochemical PSMA Expression Patterns of Primary Prostate Cancer Tissue Are Associated with the Detection Rate of Biochemical Recurrence with 68Ga-PSMA-11-PET. Theranostics 2020, 10, 6082–6094. [Google Scholar] [CrossRef]
- Bravaccini, S.; Puccetti, M.; Bocchini, M.; Ravaioli, S.; Celli, M.; Scarpi, E.; De Giorgi, U.; Tumedei, M.M.; Raulli, G.; Cardinale, L.; et al. PSMA Expression: A Potential Ally for the Pathologist in Prostate Cancer Diagnosis. Sci. Rep. 2018, 8, 4254. [Google Scholar] [CrossRef]
- Hupe, M.C.; Philippi, C.; Roth, D.; Kümpers, C.; Ribbat-Idel, J.; Becker, F.; Joerg, V.; Duensing, S.; Lubczyk, V.H.; Kirfel, J.; et al. Expression of Prostate-Specific Membrane Antigen (PSMA) on Biopsies Is an Independent Risk Stratifier of Prostate Cancer Patients at Time of Initial Diagnosis. Front. Oncol. 2018, 8, 623. [Google Scholar] [CrossRef]
- Rüschoff, J.H.; Stratton, S.; Roberts, E.; Clark, S.; Sebastiao, N.; Fankhauser, C.D.; Eberli, D.; Moch, H.; Wild, P.J.; Rupp, N.J. A Novel 5x Multiplex Immunohistochemical Staining Reveals PSMA as a Helpful Marker in Prostate Cancer with Low P504s Expression. Pathol. Res. Pract. 2021, 228, 153667. [Google Scholar] [CrossRef]
- Li, Q.K.; Lih, T.-S.M.; Wang, Y.; Hu, Y.; Höti, N.; Chan, D.W.; Zhang, H. Improving the Detection of Aggressive Prostate Cancer Using Immunohistochemical Staining of Protein Marker Panels. Am. J. Cancer Res. 2022, 12, 1323–1336. [Google Scholar]
- Sharma, S.; Cwiklinski, K.; Sykes, D.E.; Mahajan, S.D.; Chevli, K.; Schwartz, S.A.; Aalinkeel, R. Use of Glycoproteins—Prostate-Specific Membrane Antigen and Galectin-3 as Primary Tumor Markers and Therapeutic Targets in the Management of Metastatic Prostate Cancer. Cancers 2022, 14, 2704. [Google Scholar] [CrossRef]
- Huang, H.; Guma, S.R.; Melamed, J.; Zhou, M.; Lee, P.; Deng, F.-M. NKX3.1 and PSMA Are Sensitive Diagnostic Markers for Prostatic Carcinoma in Bone Metastasis after Decalcification of Specimens. Am. J. Clin. Exp. Urol. 2018, 6, 182–188. [Google Scholar]
- Juzeniene, A.; Stenberg, V.Y.; Bruland, Ø.S.; Larsen, R.H. Preclinical and Clinical Status of PSMA-Targeted Alpha Therapy for Metastatic Castration-Resistant Prostate Cancer. Cancers 2021, 13, 779. [Google Scholar] [CrossRef]
- Czerwińska, M.; Bilewicz, A.; Kruszewski, M.; Wegierek-Ciuk, A.; Lankoff, A. Targeted Radionuclide Therapy of Prostate Cancer—From Basic Research to Clinical Perspectives. Molecules 2020, 25, 1743. [Google Scholar] [CrossRef]
- Sathekge, M.M.; Bruchertseifer, F.; Vorster, M.; Morgenstern, A.; Lawal, I.O. Global Experience with PSMA-Based Alpha Therapy in Prostate Cancer. Eur. J. Nucl. Med. Mol. Imaging 2021, 49, 30–46. [Google Scholar] [CrossRef]
- Allelein, S.; Aerchlimann, K.; Rösch, G.; Khajehamiri, R.; Kölsch, A.; Freese, C.; Kuhlmeier, D. Prostate-Specific Membrane Antigen (PSMA)-Positive Extracellular Vesicles in Urine—A Potential Liquid Biopsy Strategy for Prostate Cancer Diagnosis? Cancers 2022, 14, 2987. [Google Scholar] [CrossRef]
- Diggins, N.L.; Webb, D.J. APPL1 Is a Multi-Functional Endosomal Signaling Adaptor Protein. Biochem. Soc. Trans. 2017, 45, 771. [Google Scholar] [CrossRef]
- Johnson, I.R.D.; Parkinson-Lawrence, E.J.; Keegan, H.; Spillane, C.D.; Barry-O’Crowley, J.; Watson, W.R.; Selemidis, S.; Butler, L.M.; O’Leary, J.J.; Brooks, D.A. Endosomal Gene Expression: A New Indicator for Prostate Cancer Patient Prognosis? Oncotarget 2015, 6, 37919–37929. [Google Scholar] [CrossRef]
- Wu, K.K.L.; Long, K.; Lin, H.; Siu, P.M.F.; Hoo, R.L.C.; Ye, D.; Xu, A.; Cheng, K.K.Y. The APPL1-Rab5 Axis Restricts NLRP3 Inflammasome Activation through Early Endosomal-Dependent Mitophagy in Macrophages. Nat. Commun. 2021, 12, 6637. [Google Scholar] [CrossRef] [PubMed]
- APPL1 Endocytic Adaptor as a Fine Tuner of Dvl2-Induced Transcription. FEBS Lett. 2015, 589, 532–539. [CrossRef] [PubMed]
- Sandsmark, E.; Hansen, A.F.; Selnæs, K.M.; Bertilsson, H.; Bofin, A.M.; Wright, A.J.; Viset, T.; Richardsen, E.; Drabløs, F.; Bathen, T.F.; et al. A Novel Non-Canonical Wnt Signature for Prostate Cancer Aggressiveness. Oncotarget 2016, 8, 9572–9586. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Mu, Y.; Li, C.; Bergh, A.; Miaczynska, M.; Heldin, C.-H.; Landström, M. APPL Proteins Promote TGFβ-Induced Nuclear Transport of the TGFβ Type I Receptor Intracellular Domain. Oncotarget 2015, 7, 279–292. [Google Scholar] [CrossRef]
- Martini, C.; Logan, J.M.; Sorvina, A.; Gordon, C.; Beck, A.R.; Ung, B.S.-Y.; Caruso, M.C.; Moore, C.; Hocking, A.; Johnson, I.R.D.; et al. Aberrant Protein Expression of Appl1, Sortilin and Syndecan-1 during the Biological Progression of Prostate Cancer. Pathology 2022, 55, 40–51. [Google Scholar] [CrossRef]
- Canuel, M.; Korkidakis, A.; Konnyu, K.; Morales, C.R. Sortilin Mediates the Lysosomal Targeting of Cathepsins D and H. Biochem. Biophys. Res. Commun. 2008, 373, 292–297. [Google Scholar] [CrossRef]
- Pan, X.; Zaarur, N.; Singh, M.; Morin, P.; Kandror, K.V. Sortilin and Retromer Mediate Retrograde Transport of Glut4 in 3T3-L1 Adipocytes. Mol. Biol. Cell 2017, 28, 1667–1675. [Google Scholar] [CrossRef]
- Bogan, J.S.; Kandror, K.V. Biogenesis and Regulation of Insulin-Responsive Vesicles Containing GLUT4. Curr. Opin. Cell Biol. 2010, 22, 506–512. [Google Scholar] [CrossRef]
- Tanimoto, R.; Morcavallo, A.; Terracciano, M.; Xu, S.-Q.; Stefanello, M.; Buraschi, S.; Lu, K.G.; Bagley, D.H.; Gomella, L.G.; Scotlandi, K.; et al. Sortilin Regulates Progranulin Action in Castration-Resistant Prostate Cancer Cells. Endocrinology 2015, 156, 58–70. [Google Scholar] [CrossRef]
- Chilosi, M.; Adami, F.; Lestani, M.; Montagna, L.; Cimarosto, L.; Semenzato, G.; Pizzolo, G.; Menestrina, F. CD138/Syndecan-1: A Useful Immunohistochemical Marker of Normal and Neoplastic Plasma Cells on Routine Trephine Bone Marrow Biopsies. Mod. Pathol. 1999, 12, 1101–1106. [Google Scholar]
- Wang, S.; Zhang, X.; Wang, G.; Cao, B.; Yang, H.; Jin, L.; Cui, M.; Mao, Y. Syndecan-1 Suppresses Cell Growth and Migration via Blocking JAK1/STAT3 and Ras/Raf/MEK/ERK Pathways in Human Colorectal Carcinoma Cells. BMC Cancer 2019, 19, 1160. [Google Scholar] [CrossRef]
- Shimada, K.; Anai, S.; Fujii, T.; Tanaka, N.; Fujimoto, K.; Konishi, N. Syndecan-1 (CD138) Contributes to Prostate Cancer Progression by Stabilizing Tumour-Initiating Cells. J. Pathol. 2013, 231, 495–504. [Google Scholar] [CrossRef]
- Kind, S.; Kluth, M.; Hube-Magg, C.; Möller, K.; Makrypidi-Fraune, G.; Lutz, F.; Lennartz, M.; Rico, S.D.; Schlomm, T.; Heinzer, H.; et al. Increased Cytoplasmic CD138 Expression Is Associated with Aggressive Characteristics in Prostate Cancer and Is an Independent Predictor for Biochemical Recurrence. BioMed Res. Int. 2020, 2020, 5845374. [Google Scholar] [CrossRef]
- Santos, N.J.; Barquilha, C.N.; Barbosa, I.C.; Macedo, R.T.; Lima, F.O.; Justulin, L.A.; Barbosa, G.O.; Carvalho, H.F.; Felisbino, S.L. Syndecan Family Gene and Protein Expression and Their Prognostic Values for Prostate Cancer. Int. J. Mol. Sci. 2021, 22, 8669. [Google Scholar] [CrossRef]
- Szarvas, T.; Reis, H.; Vom Dorp, F.; Tschirdewahn, S.; Niedworok, C.; Nyirady, P.; Schmid, K.W.; Rübben, H.; Kovalszky, I. Soluble Syndecan-1 (SDC1) Serum Level as an Independent Pre-Operative Predictor of Cancer-Specific Survival in Prostate Cancer. Prostate 2016, 76, 977–985. [Google Scholar] [CrossRef]
- Szarvas, T.; Sevcenco, S.; Módos, O.; Keresztes, D.; Nyirády, P.; Kubik, A.; Romics, M.; Kovalszky, I.; Reis, H.; Hadaschik, B.; et al. Circulating Syndecan-1 Is Associated with Chemotherapy-Resistance in Castration-Resistant Prostate Cancer. Urol. Oncol. 2018, 36, 312.e9–312.e15. [Google Scholar] [CrossRef]
- Surget, S.; Khoury, M.P.; Bourdon, J.-C. Uncovering the Role of P53 Splice Variants in Human Malignancy: A Clinical Perspective. Onco. Targets Ther. 2013, 7, 57–68. [Google Scholar] [CrossRef]
- Signoretti, S.; Waltregny, D.; Dilks, J.; Isaac, B.; Lin, D.; Garraway, L.; Yang, A.; Montironi, R.; McKeon, F.; Loda, M. P63 Is a Prostate Basal Cell Marker and Is Required for Prostate Development. Am. J. Pathol. 2000, 157, 1769–1775. [Google Scholar] [CrossRef]
- Marchini, S.; Marabese, M.; Marrazzo, E.; Mariani, P.; Cattaneo, D.; Fossati, R.; Compagnoni, A.; Fruscio, R.; Lissoni, A.A.; Broggini, M. DeltaNp63 Expression Is Associated with Poor Survival in Ovarian Cancer. Ann. Oncol. 2008, 19, 501–507. [Google Scholar] [CrossRef]
- Lo Muzio, L.; Santarelli, A.; Caltabiano, R.; Rubini, C.; Pieramici, T.; Trevisiol, L.; Carinci, F.; Leonardi, R.; De Lillo, A.; Lanzafame, S.; et al. P63 Overexpression Associates with Poor Prognosis in Head and Neck Squamous Cell Carcinoma. Hum. Pathol. 2005, 36, 187–194. [Google Scholar] [CrossRef]
- Shiran, M.S.; Tan, G.C.; Sabariah, A.R.; Rampal, L.; Phang, K.S. P63 as a Complimentary Basal Cell Specific Marker to High Molecular Weight-Cytokeratin in Distinguishing Prostatic Carcinoma from Benign Prostatic Lesions. Med. J. Malays. 2007, 62, 36–39. [Google Scholar]
- Brawer, M.K.; Peehl, D.M.; Stamey, T.A.; Bostwick, D.G. Keratin Immunoreactivity in the Benign and Neoplastic Human Prostate. Cancer Res. 1985, 45, 3663–3667. [Google Scholar] [PubMed]
- Hedrick, L.; Epstein, J.I. Use of Keratin 903 as an Adjunct in the Diagnosis of Prostate Carcinoma. Am. J. Surg Pathol. 1989, 13, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.H.-J.; Lapkus, O.; Corbin, M. Comparison of 34betaE12 and P63 in 100 Consecutive Prostate Carcinoma Diagnosed by Needle Biopsies. Appl. Immunohistochem. Mol. Morphol. 2004, 12, 285–289. [Google Scholar] [CrossRef]
- Kalantari, M.R.; Anvari, K.; Jabbari, H.; Tabrizi, F.V. P63 Is More Sensitive and Specific than 34βE12 to Differentiate Adenocarcinoma of Prostate from Cancer Mimickers. Iran. J. Basic Med. Sci. 2014, 17, 497–501. [Google Scholar]
- Srigley, J.R. Benign Mimickers of Prostatic Adenocarcinoma. Mod. Pathol. 2004, 17, 328–348. [Google Scholar] [CrossRef]
- Tan, H.-L.; Haffner, M.C.; Esopi, D.M.; Vaghasia, A.M.; Giannico, G.A.; Ross, H.M.; Ghosh, S.; Hicks, J.L.; Zheng, Q.; Sangoi, A.R.; et al. Prostate Adenocarcinomas Aberrantly Expressing P63 Are Molecularly Distinct from Usual-Type Prostatic Adenocarcinomas. Mod. Pathol. 2015, 28, 446–456. [Google Scholar] [CrossRef]
- Abbas, M.; Habibian, B.; Bettendorf, O. Prostate Adenocarcinoma of Secretory Type with Wide Expression of P63 and Negativity of the Basal Marker Ck5/6: Rare Subtype of Adenocarcinoma of Secretory Origin and to Be Differentiated from Basal Cell Carcinoma. Review of Literature. Rare Tumors 2020, 12, 2036361320971948. [Google Scholar] [CrossRef]
- Sun, X.; Kaufman, P.D. Ki-67: More than a Proliferation Marker. Chromosoma 2018, 127, 175–186. [Google Scholar] [CrossRef]
- Dhillon, P.K.; Barry, M.; Stampfer, M.J.; Perner, S.; Fiorentino, M.; Fornari, A.; Ma, J.; Fleet, J.; Kurth, T.; Rubin, M.A.; et al. Aberrant Cytoplasmic Expression of P63 and Prostate Cancer Mortality. Cancer Epidemiol. Biomark. Prev. 2009, 18, 595–600. [Google Scholar] [CrossRef]
- Narahashi, T.; Niki, T.; Wang, T.; Goto, A.; Matsubara, D.; Funata, N.; Fukayama, M. Cytoplasmic Localization of P63 Is Associated with Poor Patient Survival in Lung Adenocarcinoma. Histopathology 2006, 49, 349–357. [Google Scholar] [CrossRef]
- Shah, S.; Rachmat, R.; Enyioma, S.; Ghose, A.; Revythis, A.; Boussios, S. BRCA Mutations in Prostate Cancer: Assessment, Implications and Treatment Considerations. Int. J. Mol. Sci. 2021, 22, 12628. [Google Scholar] [CrossRef]
- Nombela, P.; Lozano, R.; Aytes, A.; Mateo, J.; Olmos, D.; Castro, E. BRCA2 and Other DDR Genes in Prostate Cancer. Cancers 2019, 11, 352. [Google Scholar] [CrossRef]
- Castro, E.; Romero-Laorden, N.; Del Pozo, A.; Lozano, R.; Medina, A.; Puente, J.; Piulats, J.M.; Lorente, D.; Saez, M.I.; Morales-Barrera, R.; et al. PROREPAIR-B: A Prospective Cohort Study of the Impact of Germline DNA Repair Mutations on the Outcomes of Patients with Metastatic Castration-Resistant Prostate Cancer. J. Clin. Oncol. 2019, 37, 490–503. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Mateo, J.; Walsh, M.F.; De Sarkar, N.; Abida, W.; Beltran, H.; Garofalo, A.; Gulati, R.; Carreira, S.; Eeles, R.; et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N. Engl. J. Med. 2016, 375, 443–453. [Google Scholar] [CrossRef]
- The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015, 163, 1011–1025. [CrossRef]
- Virtanen, V.; Paunu, K.; Ahlskog, J.K.; Varnai, R.; Sipeky, C.; Sundvall, M. PARP Inhibitors in Prostate Cancer—The Preclinical Rationale and Current Clinical Development. Genes 2019, 10, 565. [Google Scholar] [CrossRef]
- Fossati, N.; Willemse, P.-P.M.; Van den Broeck, T.; van den Bergh, R.C.N.; Yuan, C.Y.; Briers, E.; Bellmunt, J.; Bolla, M.; Cornford, P.; De Santis, M.; et al. The Benefits and Harms of Different Extents of Lymph Node Dissection During Radical Prostatectomy for Prostate Cancer: A Systematic Review. Eur. Urol. 2017, 72, 84–109. [Google Scholar] [CrossRef]
- Murata, Y.; Tatsugami, K.; Yoshikawa, M.; Hamaguchi, M.; Yamada, S.; Hayakawa, Y.; Ueda, K.; Momosaki, S.; Sakamoto, N. Predictive Factors of Biochemical Recurrence after Radical Prostatectomy for High-Risk Prostate Cancer. Int. J. Urol. 2018, 25, 284–289. [Google Scholar] [CrossRef]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef]
- Małkiewicz, B.; Kiełb, P.; Kobylański, M.; Karwacki, J.; Poterek, A.; Krajewski, W.; Zdrojowy, R.; Szydełko, T. Sentinel Lymph Node Techniques in Urologic Oncology: Current Knowledge and Application. Cancers 2023, 15, 2495. [Google Scholar] [CrossRef] [PubMed]
- Dell’Oglio, P.; Meershoek, P.; Maurer, T.; Wit, E.M.K.; van Leeuwen, P.J.; van der Poel, H.G.; van Leeuwen, F.W.B.; van Oosterom, M.N. A DROP-IN Gamma Probe for Robot-Assisted Radioguided Surgery of Lymph Nodes During Radical Prostatectomy. Eur. Urol. 2021, 79, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Mazzone, E.; Dell’Oglio, P.; Grivas, N.; Wit, E.; Donswijk, M.; Briganti, A.; Leeuwen, F.V.; Poel, H. van der Diagnostic Value, Oncologic Outcomes, and Safety Profile of Image-Guided Surgery Technologies During Robot-Assisted Lymph Node Dissection with Sentinel Node Biopsy for Prostate Cancer. J. Nucl. Med. 2021, 62, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Claps, F.; de Pablos-Rodríguez, P.; Gómez-Ferrer, Á.; Mascarós, J.M.; Marenco, J.; Serra, A.C.; Ramón-Borja, J.C.; Fons, A.C.; Trombetta, C.; Rubio-Briones, J.; et al. Free-Indocyanine Green-Guided Pelvic Lymph Node Dissection during Radical Prostatectomy. Urol. Oncol. 2022, 40, 489.e19–489.e26. [Google Scholar] [CrossRef]
- Ionescu, F.; Zhang, J.; Wang, L. Clinical Applications of Liquid Biopsy in Prostate Cancer: From Screening to Predictive Biomarker. Cancers 2022, 14, 1728. [Google Scholar] [CrossRef]
- Trujillo, B.; Wu, A.; Wetterskog, D.; Attard, G. Blood-Based Liquid Biopsies for Prostate Cancer: Clinical Opportunities and Challenges. Br. J. Cancer 2022, 127, 1394–1402. [Google Scholar] [CrossRef]
- Saxby, H.; Mikropoulos, C.; Boussios, S. An Update on the Prognostic and Predictive Serum Biomarkers in Metastatic Prostate Cancer. Diagnostics 2020, 10, 549. [Google Scholar] [CrossRef]
- Chu, T.N.; Wong, E.Y.; Ma, R.; Yang, C.H.; Dalieh, I.S.; Hung, A.J. Exploring the Use of Artificial Intelligence in the Management of Prostate Cancer. Curr. Urol. Rep. 2023, 24, 231–240. [Google Scholar] [CrossRef]
- Boehm, K.M.; Khosravi, P.; Vanguri, R.; Gao, J.; Shah, S.P. Harnessing Multimodal Data Integration to Advance Precision Oncology. Nat. Rev. Cancer 2022, 22, 114–126. [Google Scholar] [CrossRef]
- Woźnicki, P.; Westhoff, N.; Huber, T.; Riffel, P.; Froelich, M.F.; Gresser, E.; von Hardenberg, J.; Mühlberg, A.; Michel, M.S.; Schoenberg, S.O.; et al. Multiparametric MRI for Prostate Cancer Characterization: Combined Use of Radiomics Model with PI-RADS and Clinical Parameters. Cancers 2020, 12, 1767. [Google Scholar] [CrossRef]
- Antonelli, M.; Johnston, E.W.; Dikaios, N.; Cheung, K.K.; Sidhu, H.S.; Appayya, M.B.; Giganti, F.; Simmons, L.A.M.; Freeman, A.; Allen, C.; et al. Machine Learning Classifiers Can Predict Gleason Pattern 4 Prostate Cancer with Greater Accuracy than Experienced Radiologists. Eur. Radiol. 2019, 29, 4754–4764. [Google Scholar] [CrossRef]
- Fehr, D.; Veeraraghavan, H.; Wibmer, A.; Gondo, T.; Matsumoto, K.; Vargas, H.A.; Sala, E.; Hricak, H.; Deasy, J.O. Automatic Classification of Prostate Cancer Gleason Scores from Multiparametric Magnetic Resonance Images. Proc. Natl. Acad. Sci. USA 2015, 112, E6265–E6273. [Google Scholar] [CrossRef]
- Akatsuka, J.; Numata, Y.; Morikawa, H.; Sekine, T.; Kayama, S.; Mikami, H.; Yanagi, M.; Endo, Y.; Takeda, H.; Toyama, Y.; et al. A Data-Driven Ultrasound Approach Discriminates Pathological High Grade Prostate Cancer. Sci. Rep. 2022, 12, 860. [Google Scholar] [CrossRef]
- Bulten, W.; Pinckaers, H.; van Boven, H.; Vink, R.; de Bel, T.; van Ginneken, B.; van der Laak, J.; Hulsbergen-van de Kaa, C.; Litjens, G. Automated Deep-Learning System for Gleason Grading of Prostate Cancer Using Biopsies: A Diagnostic Study. Lancet Oncol. 2020, 21, 233–241. [Google Scholar] [CrossRef]
- Kott, O.; Linsley, D.; Amin, A.; Karagounis, A.; Jeffers, C.; Golijanin, D.; Serre, T.; Gershman, B. Development of a Deep Learning Algorithm for the Histopathologic Diagnosis and Gleason Grading of Prostate Cancer Biopsies: A Pilot Study. Eur. Urol. Focus 2021, 7, 347–351. [Google Scholar] [CrossRef]
- Marginean, F.; Arvidsson, I.; Simoulis, A.; Overgaard, N.C.; Åström, K.; Heyden, A.; Bjartell, A.; Krzyzanowska, A. An Artificial Intelligence-Based Support Tool for Automation and Standardisation of Gleason Grading in Prostate Biopsies. Eur. Urol. Focus 2021, 7, 995–1001. [Google Scholar] [CrossRef]
- Da Silva, L.M.; Pereira, E.M.; Salles, P.G.; Godrich, R.; Ceballos, R.; Kunz, J.D.; Casson, A.; Viret, J.; Chandarlapaty, S.; Ferreira, C.G.; et al. Independent Real-World Application of a Clinical-Grade Automated Prostate Cancer Detection System. J. Pathol. 2021, 254, 147–158. [Google Scholar] [CrossRef]
- Bibault, J.-E.; Hancock, S.; Buyyounouski, M.K.; Bagshaw, H.; Leppert, J.T.; Liao, J.C.; Xing, L. Development and Validation of an Interpretable Artificial Intelligence Model to Predict 10-Year Prostate Cancer Mortality. Cancers 2021, 13, 3064. [Google Scholar] [CrossRef] [PubMed]
- Koo, K.C.; Lee, K.S.; Kim, S.; Min, C.; Min, G.R.; Lee, Y.H.; Han, W.K.; Rha, K.H.; Hong, S.J.; Yang, S.C.; et al. Long Short-Term Memory Artificial Neural Network Model for Prediction of Prostate Cancer Survival Outcomes According to Initial Treatment Strategy: Development of an Online Decision-Making Support System. World J. Urol. 2020, 38, 2469–2476. [Google Scholar] [CrossRef]
- Tan, Y.G.; Fang, A.H.S.; Lim, J.K.S.; Khalid, F.; Chen, K.; Ho, H.S.S.; Yuen, J.S.P.; Huang, H.H.; Tay, K.J. Incorporating Artificial Intelligence in Urology: Supervised Machine Learning Algorithms Demonstrate Comparative Advantage over Nomograms in Predicting Biochemical Recurrence after Prostatectomy. Prostate 2022, 82, 298–305. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiełb, P.; Kowalczyk, K.; Gurwin, A.; Nowak, Ł.; Krajewski, W.; Sosnowski, R.; Szydełko, T.; Małkiewicz, B. Novel Histopathological Biomarkers in Prostate Cancer: Implications and Perspectives. Biomedicines 2023, 11, 1552. https://doi.org/10.3390/biomedicines11061552
Kiełb P, Kowalczyk K, Gurwin A, Nowak Ł, Krajewski W, Sosnowski R, Szydełko T, Małkiewicz B. Novel Histopathological Biomarkers in Prostate Cancer: Implications and Perspectives. Biomedicines. 2023; 11(6):1552. https://doi.org/10.3390/biomedicines11061552
Chicago/Turabian StyleKiełb, Paweł, Kamil Kowalczyk, Adam Gurwin, Łukasz Nowak, Wojciech Krajewski, Roman Sosnowski, Tomasz Szydełko, and Bartosz Małkiewicz. 2023. "Novel Histopathological Biomarkers in Prostate Cancer: Implications and Perspectives" Biomedicines 11, no. 6: 1552. https://doi.org/10.3390/biomedicines11061552





