Insight into the Inter-Organ Crosstalk and Prognostic Role of Liver-Derived MicroRNAs in Metabolic Disease Progression
Abstract
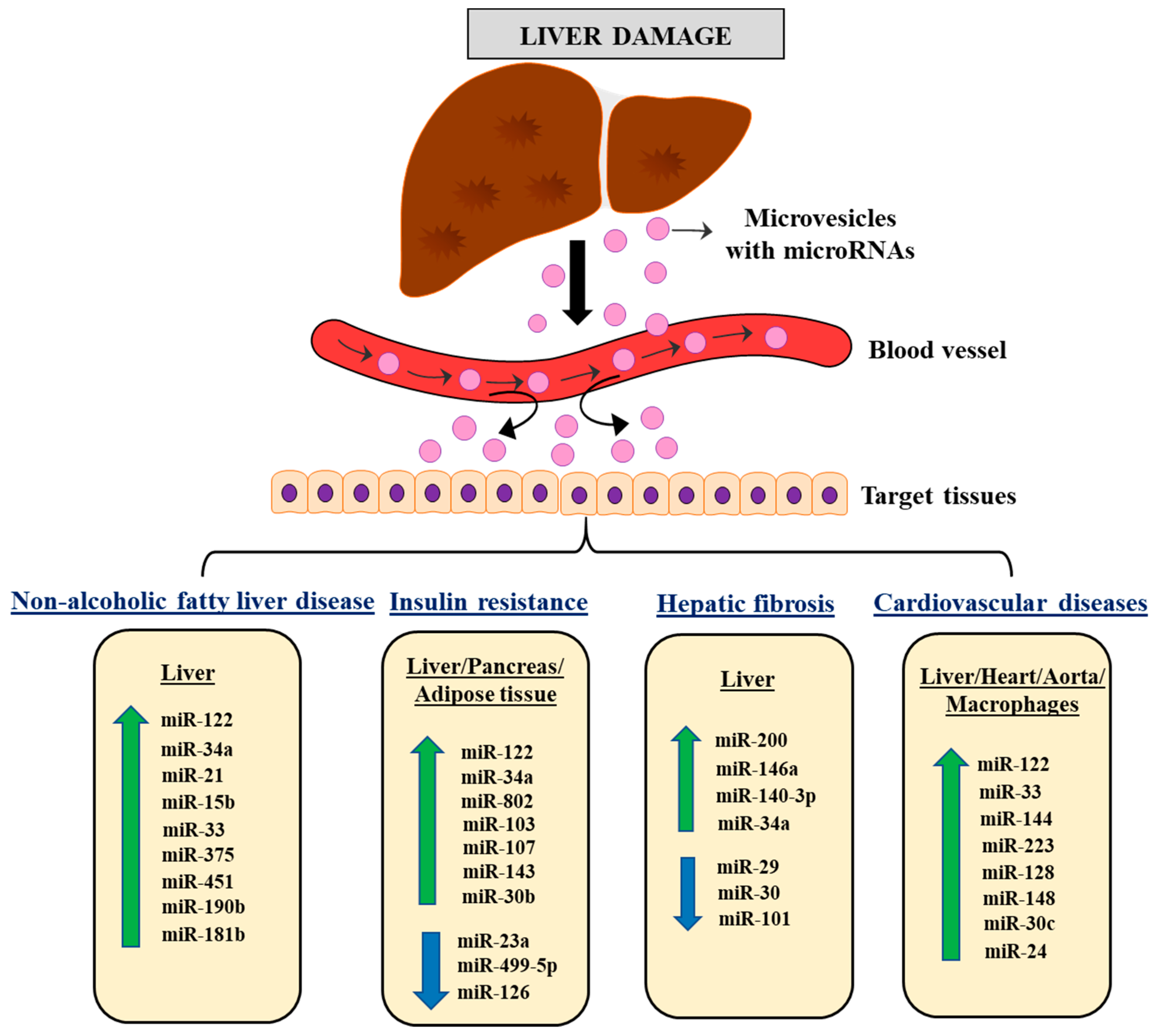
:1. Introduction
2. Biogenesis and Release of miRNA from the Liver
3. The Role of Liver-Derived miRNAs in Metabolic Regulation and Associated Pathologies
3.1. Role of Liver-Derived miRNAs in NAFLD
| miRNAs | Site of Action | Status in Disease | Targets | Physiological Effect | References |
|---|---|---|---|---|---|
| miR-122 | Serum | Increased | FAS ACC SCD-1 SREBPs HNF6 | Lipid metabolism and hepatocyte differentiation | [38,39,41,42] |
| miR-34a | Serum | Increased | P53 SIRT1 NAD-dependent deacetylase PPAR-α Liver x receptor HNF4α | Oxidative stress, lipid metabolism, hepatocyte apoptosis, and fatty acid oxidation | [43,44,45,46,47,48] |
| miR-21 | Serum Liver | Increased Increased | SREBP1 HMGCR FABP7 LPR6 FOXA2 FOXO1 HNF4α STAT3 INSIG2 PPAR-α | Lipid metabolism | [37,39,49,50,51,52,53,54,55] |
| miR-15b | Serum L02 cells | Increased Increased | LOXL1 | HSC activation, glucose metabolism, and lipid metabolism | [56,58] |
| miR-33 | Serum Liver | Increased Increased | SREBPs ABCA1 TGFβ | Lipid metabolism and insulin sensitivity | [59,61,62,63,64] |
| miR-375 | Serum | Increased | AdipoR2 | Glucose metabolism, insulin sensitivity, and inflammation | [38,67] |
| miR-451 | Serum | Increased | THRSP | Lipid metabolism | [39,68] |
| miR-190b | Liver | Increased | IGF-1 ADAMTS9 | Lipid metabolism | [69] |
| miR-181b | Serum Liver | Increased Increased | SIRT1 | Lipid metabolism | [70] |
3.2. Role of Liver-Derived miRNAs in Insulin Resistance
| miRNAs | Site of Action | Status in Disease | Targets | Physiological Effects | References |
|---|---|---|---|---|---|
| miR-122 | Plasma Serum Liver SC Pancreatic β cells WAT | Increased Increased | ACLY MTTP SREBP-1 IGF-1R | Lipid metabolism, insulin sensitivity, and insulin resistance | [78,79,80,81,82,83] |
| miR-34a | Plasma Liver Pancreatic islet | Increased Increased | HNF4α USP10 | Glucose metabolism, lipid metabolism, and insulin resistance | [48,80,84,85] |
| miR-802 | Serum Liver Pancreatic islet WAT BAT | Increased Increased | HNF1β HSP60 NeuroD1 Fzd5 | Glucose metabolism, insulin resistance, insulin metabolism, and oxidative stress | [87,89,90,91] |
| miR-126 | Plasma Serum Liver INS-1 β cells | Decreased Decreased | IRS-1 IRS-2 CCL2 | Insulin resistance, glucose metabolism, and lipid metabolism | [93,94,95,96,97] |
| miR-143 | Serum Liver Pancreas | Increased Increased | ORP8 MAPK11 | Glucose metabolism and insulin resistance | [99,100,101] |
| miR-30b | Serum Liver MIN6 cells | Increased Increased | SERCA2b PPARGC1A NeuroD1 | Endoplasmic reticulum stress, insulin sensitivity, and lipid metabolism | [102,103,104,105,106] |
| miR-23a | Serum Liver Pancreatic islets β-Cells | Decreased Decreased | NEK7 SREBP-1 FAS DP5 PUMA | Pyroptosis, lipid metabolism, inflammatory process, and insulin resistance | [107,108,109,110] |
| miR-499-5p | Plasma/serum Liver | Decreased Decreased | PTEN PI3K/AKT/ GSK | Insulin resistance | [75,112] |
| miR-103 | Serum Liver Adipose tissue | Increased Increased | Caveolin-1 | Insulin sensitivity | [113,114] |
| miR-107 | Serum Liver Adipose tissue | Increased Increased | Caveolin-1 | Insulin sensitivity | [113,115] |
3.3. Role of Liver-Derived miRNAs in Hepatic Fibrosis
| miRNAs | Site of Action | Status in Disease | Targets | Physiological Effect | References |
|---|---|---|---|---|---|
| miR-200 | Serum L02 cells | Increased Increased | SIRT1 MMP-2 PI3K/AKT FOG2 MAPK | Extracellular matrix protein accumulation | [119,120,121] |
| miR-30 | Liver HSC cells | Decreased Decreased | Beclin1 SNAI1 | HSC activation | [123,124,125,128] |
| miR-146a | Liver HSC cells Serum | Decreased Decreased Increased | TRAF6 IRAK1 TGF-β1 WNT1 WNT5a | Immunological response, inflammation, and HSC activation | [129,130,131,132] |
| miR-140-3p | Liver HSC cells Serum | Increased | PTEN | HSC activation | [136,137] |
| miR-34a | Liver HSC cells | Increased Increased | ASCL1 TGF-β SIRT1/p53 | Lipid metabolism | [138,139,140,141] |
| miR-29 | Serum Liver HSC cells | Decreased Decreased Decreased | TGF-β NF-κB PIK3/AKT | Extracellular matrix protein accumulation and HSC activation | [142,143] |
| miR-101 | Liver HSC cells | Decreased Decreased | TβRI KLF6 | HSC activation | [144,145] |
3.4. Role of Liver-Derived miRNAs in Cardiovascular Diseases
| miRNAs | Site of Action | Status in Disease | Targets | Physiological Effects | References |
|---|---|---|---|---|---|
| miR-122 | Plasma Liver Cardiac rissue | Increased | HMGCS1 HMGCR MTTP HAND2 | Lipid and cholesterol synthesis and apoptosis | [150,151,152,153,154,155,156] |
| miR-33 | Plasma Hepatocyte Cell lines Liver Aorta | Increased | ABCA1 ABCG1 SIRT6 CPT1A AMPK IRS2 | Cholesterol transport and lipid metabolism | [63,157,158,162] |
| miR-144 | Plasma Macrophages Liver Aorta | Increased | ABCA1 | Lipid metabolism and antioxidant response | [163,164,167,168,169] |
| miR-223 | Plasma Liver Heart | Increased | HMGCoA MSMO1 SCARB1 ABCA1 | Cholesterol metabolism, inflammatory process, and hypertrophy | [171,172,173,174] |
| miR-30c | Plasma Heart Cell lines | Increased Increased | PAI-1 p53 MTA1 MTTP | Fibrosis, hypertrophy, apoptosis, and lipid metabolism | [176,177,178,179,180,181] |
| miR-128 | Plasma Heart Cardiac cells | Increased | Axin1 PI3K/Akt/mTORC1 SIRT1/p53 ABCA1 | Oxidative stress and lipid metabolism | [182,183,184,185] |
| miR-148 | Plasma Serum Liver Hepatic cells Aorta | Increased | LDLR SREBP1 ABCA1 CPT1A AMPKa1 SIK1 | Lipid metabolism, lipid transport, inflammatory process, and cholesterol efflux | [186,187,188,189] |
| miR-143 | Plasma Cardiac cells | Increased | Elk-1 | Cell differentiation | [190,191] |
| miR-24 | Liver Human Hepatocytes | Increased | Insig1 | Lipid metabolism | [192] |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seto, W.K.; Mandell, M.S. Chronic liver disease: Global perspectives and future challenges to delivering quality health care. PLoS ONE 2021, 16, e0243607. [Google Scholar] [CrossRef]
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef]
- Mohammed, M.S.; Sendra, S.; Lloret, J.; Bosch, I. Systems and WBANs for Controlling Obesity. J. Healthc. Eng. 2018, 2018, 1564748. [Google Scholar] [CrossRef] [PubMed]
- Perdoncin, M.; Konrad, A.; Wyner, J.R.; Lohana, S.; Pillai, S.S.; Pereira, D.G.; Lakhani, H.V.; Sodhi, K. A Review of miRNAs as Biomarkers and Effect of Dietary Modulation in Obesity Associated Cognitive Decline and Neurodegenerative Disorders. Front. Mol. Neurosci. 2021, 14, 756499. [Google Scholar] [CrossRef] [PubMed]
- Wree, A.; Kahraman, A.; Gerken, G.; Canbay, A. Obesity affects the liver—The link between adipocytes and hepatocytes. Digestion 2011, 83, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.B.; Hu, E.D.; Xu, L.M.; Chen, L.; Wu, J.L.; Li, H.; Chen, D.Z.; Chen, Y.P. The relationship between obesity and the severity of non-alcoholic fatty liver disease: Systematic review and meta-analysis. Expert Rev. Gastroenterol. Hepatol. 2018, 12, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.C.; Sundaram, V. Obesity and Liver Decompensation. Clin. Liver Dis. 2019, 14, 12–15. [Google Scholar] [CrossRef]
- Global, B.M.I.M.C.; Di Angelantonio, E.; Bhupathiraju Sh, N.; Wormser, D.; Gao, P.; Kaptoge, S.; Berrington de Gonzalez, A.; Cairns, B.J.; Huxley, R.; Jackson Ch, L.; et al. Body-mass index and all-cause mortality: Individual-participant-data meta-analysis of 239 prospective studies in four continents. Lancet 2016, 388, 776–786. [Google Scholar] [CrossRef]
- Polyzos, S.A.; Kountouras, J.; Mantzoros, C.S. Obesity and nonalcoholic fatty liver disease: From pathophysiology to therapeutics. Metabolism 2019, 92, 82–97. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Willeit, P.; Skroblin, P.; Kiechl, S.; Fernandez-Hernando, C.; Mayr, M. Liver microRNAs: Potential mediators and biomarkers for metabolic and cardiovascular disease? Eur. Heart J. 2016, 37, 3260–3266. [Google Scholar] [CrossRef]
- Panera, N.; Gnani, D.; Crudele, A.; Ceccarelli, S.; Nobili, V.; Alisi, A. MicroRNAs as controlled systems and controllers in non-alcoholic fatty liver disease. World J. Gastroenterol. 2014, 20, 15079–15086. [Google Scholar] [CrossRef]
- Pirola, C.J.; Scian, R.; Gianotti, T.F.; Dopazo, H.; Rohr, C.; Martino, J.S.; Castano, G.O.; Sookoian, S. Epigenetic Modifications in the Biology of Nonalcoholic Fatty Liver Disease: The Role of DNA Hydroxymethylation and TET Proteins. Medicine 2015, 94, e1480. [Google Scholar] [CrossRef]
- Mc Cormack, B.A.; Gonzalez-Canto, E.; Agababyan, C.; Espinoza-Sanchez, N.A.; Tomas-Perez, S.; Llueca, A.; Mari-Alexandre, J.; Gotte, M.; Gilabert-Estelles, J. miRNAs in the Era of Personalized Medicine: From Biomarkers to Therapeutics. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef]
- Szabo, G.; Bala, S. MicroRNAs in liver disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 542–552. [Google Scholar] [CrossRef]
- Wang, X.; He, Y.; Mackowiak, B.; Gao, B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut 2021, 70, 784–795. [Google Scholar] [CrossRef]
- Schueller, F.; Roy, S.; Vucur, M.; Trautwein, C.; Luedde, T.; Roderburg, C. The Role of miRNAs in the Pathophysiology of Liver Diseases and Toxicity. Int. J. Mol. Sci. 2018, 19, 261. [Google Scholar] [CrossRef]
- Dongiovanni, P.; Meroni, M.; Longo, M.; Fargion, S.; Fracanzani, A.L. miRNA Signature in NAFLD: A Turning Point for a Non-Invasive Diagnosis. Int. J. Mol. Sci. 2018, 19, 3966. [Google Scholar] [CrossRef]
- Celikbilek, M.; Baskol, M.; Taheri, S.; Deniz, K.; Dogan, S.; Zararsiz, G.; Gursoy, S.; Guven, K.; Ozbakir, O.; Dundar, M.; et al. Circulating microRNAs in patients with non-alcoholic fatty liver disease. World J. Hepatol. 2014, 6, 613–620. [Google Scholar] [CrossRef]
- Wang, R.; Hong, J.; Cao, Y.; Shi, J.; Gu, W.; Ning, G.; Zhang, Y.; Wang, W. Elevated circulating microRNA-122 is associated with obesity and insulin resistance in young adults. Eur. J. Endocrinol. 2015, 172, 291–300. [Google Scholar] [CrossRef]
- Jiang, X.P.; Ai, W.B.; Wan, L.Y.; Zhang, Y.Q.; Wu, J.F. The roles of microRNA families in hepatic fibrosis. Cell Biosci. 2017, 7, 34. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Heo, M.J.; Yun, J.; Kim, S.G. Role of non-coding RNAs in liver disease progression to hepatocellular carcinoma. Arch. Pharmacal Res. 2019, 42, 48–62. [Google Scholar] [CrossRef]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Hayes, C.N.; Chayama, K. MicroRNAs as Biomarkers for Liver Disease and Hepatocellular Carcinoma. Int. J. Mol. Sci. 2016, 17, 280. [Google Scholar] [CrossRef] [PubMed]
- Gjorgjieva, M.; Sobolewski, C.; Dolicka, D.; Correia de Sousa, M.; Foti, M. miRNAs and NAFLD: From pathophysiology to therapy. Gut 2019, 68, 2065–2079. [Google Scholar] [CrossRef] [PubMed]
- Szabo, G.; Momen-Heravi, F. Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Prada, I.; Meldolesi, J. Binding and Fusion of Extracellular Vesicles to the Plasma Membrane of Their Cell Targets. Int. J. Mol. Sci. 2016, 17, 1296. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Capobianco, V.; Nardelli, C.; Ferrigno, M.; Iaffaldano, L.; Pilone, V.; Forestieri, P.; Zambrano, N.; Sacchetti, L. miRNA and protein expression profiles of visceral adipose tissue reveal miR-141/YWHAG and miR-520e/RAB11A as two potential miRNA/protein target pairs associated with severe obesity. J. Proteome Res. 2012, 11, 3358–3369. [Google Scholar] [CrossRef] [PubMed]
- Guay, C.; Roggli, E.; Nesca, V.; Jacovetti, C.; Regazzi, R. Diabetes mellitus, a microRNA-related disease? Transl. Res. 2011, 157, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Sanchez, G.N.; Dominguez-Perez, M.; Uribe, M.; Chavez-Tapia, N.C.; Nuno-Lambarri, N. Non-alcoholic fatty liver disease and microRNAs expression, how it affects the development and progression of the disease. Ann. Hepatol. 2021, 21, 100212. [Google Scholar] [CrossRef]
- Azparren-Angulo, M.; Royo, F.; Gonzalez, E.; Liebana, M.; Brotons, B.; Berganza, J.; Goni-de-Cerio, F.; Manicardi, N.; Abad-Jorda, L.; Gracia-Sancho, J.; et al. Extracellular vesicles in hepatology: Physiological role, involvement in pathogenesis, and therapeutic opportunities. Pharmacol. Ther. 2021, 218, 107683. [Google Scholar] [CrossRef] [PubMed]
- Becker, P.P.; Rau, M.; Schmitt, J.; Malsch, C.; Hammer, C.; Bantel, H.; Mullhaupt, B.; Geier, A. Performance of Serum microRNAs -122, -192 and -21 as Biomarkers in Patients with Non-Alcoholic Steatohepatitis. PLoS ONE 2015, 10, e0142661. [Google Scholar] [CrossRef]
- Pirola, C.J.; Fernandez Gianotti, T.; Castano, G.O.; Mallardi, P.; San Martino, J.; Mora Gonzalez Lopez Ledesma, M.; Flichman, D.; Mirshahi, F.; Sanyal, A.J.; Sookoian, S. Circulating microRNA signature in non-alcoholic fatty liver disease: From serum non-coding RNAs to liver histology and disease pathogenesis. Gut 2015, 64, 800–812. [Google Scholar] [CrossRef]
- Yamada, H.; Suzuki, K.; Ichino, N.; Ando, Y.; Sawada, A.; Osakabe, K.; Sugimoto, K.; Ohashi, K.; Teradaira, R.; Inoue, T.; et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin. Chim. Acta 2013, 424, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Iravani, F.; Hosseini, N.; Mojarrad, M. Role of MicroRNAs in Pathophysiology of Non-alcoholic Fatty Liver Disease and Non-alcoholic Steatohepatitis. Middle East J. Dig. Dis. 2018, 10, 213–219. [Google Scholar] [CrossRef]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Laudadio, I.; Manfroid, I.; Achouri, Y.; Schmidt, D.; Wilson, M.D.; Cordi, S.; Thorrez, L.; Knoops, L.; Jacquemin, P.; Schuit, F.; et al. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology 2012, 142, 119–129. [Google Scholar] [CrossRef]
- Cermelli, S.; Ruggieri, A.; Marrero, J.A.; Ioannou, G.N.; Beretta, L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS ONE 2011, 6, e23937. [Google Scholar] [CrossRef]
- Liu, X.L.; Pan, Q.; Zhang, R.N.; Shen, F.; Yan, S.Y.; Sun, C.; Xu, Z.J.; Chen, Y.W.; Fan, J.G. Disease-specific miR-34a as diagnostic marker of non-alcoholic steatohepatitis in a Chinese population. World J. Gastroenterol. 2016, 22, 9844–9852. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, M.; Cao, Y.; Ma, L.; Shen, Y.; Velikanova, A.A.; Li, X.; Sun, C.; Zhao, Y. miR-34a regulates lipid metabolism by targeting SIRT1 in non-alcoholic fatty liver disease with iron overload. Arch. Biochem. Biophys. 2020, 695, 108642. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.E.; Ferreira, D.M.; Afonso, M.B.; Borralho, P.M.; Machado, M.V.; Cortez-Pinto, H.; Rodrigues, C.M. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J. Hepatol. 2013, 58, 119–125. [Google Scholar] [CrossRef]
- Ding, J.; Li, M.; Wan, X.; Jin, X.; Chen, S.; Yu, C.; Li, Y. Effect of miR-34a in regulating steatosis by targeting PPARalpha expression in nonalcoholic fatty liver disease. Sci. Rep. 2015, 5, 13729. [Google Scholar] [CrossRef]
- Xu, Y.; Zalzala, M.; Xu, J.; Li, Y.; Yin, L.; Zhang, Y. A metabolic stress-inducible miR-34a-HNF4alpha pathway regulates lipid and lipoprotein metabolism. Nat. Commun. 2015, 6, 7466. [Google Scholar] [CrossRef]
- Loyer, X.; Paradis, V.; Henique, C.; Vion, A.C.; Colnot, N.; Guerin, C.L.; Devue, C.; On, S.; Scetbun, J.; Romain, M.; et al. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARalpha expression. Gut 2016, 65, 1882–1894. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Y.; Wu, X.; Jiang, L.; Yang, S.; Ding, Z.; Fang, Z.; Hua, H.; Kirby, M.S.; Shou, J. A circulating microRNA signature as noninvasive diagnostic and prognostic biomarkers for nonalcoholic steatohepatitis. BMC Genom. 2018, 19, 188. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ng, R.; Chen, X.; Steer, C.J.; Song, G. MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut 2016, 65, 1850–1860. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Huang, F.; Liu, X.; Xiao, X.; Yang, M.; Hu, G.; Liu, H.; Liao, L. miR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int. J. Mol. Med. 2015, 35, 847–853. [Google Scholar] [CrossRef]
- Ahn, J.; Lee, H.; Jung, C.H.; Ha, T. Lycopene inhibits hepatic steatosis via microRNA-21-induced downregulation of fatty acid-binding protein 7 in mice fed a high-fat diet. Mol. Nutr. Food Res. 2012, 56, 1665–1674. [Google Scholar] [CrossRef]
- Wang, X.M.; Wang, X.Y.; Huang, Y.M.; Chen, X.; Lu, M.H.; Shi, L.; Li, C.P. Role and mechanisms of action of microRNA-21 as regards the regulation of the WNT/beta-catenin signaling pathway in the pathogenesis of non-alcoholic fatty liver disease. Int. J. Mol. Med. 2019, 44, 2201–2212. [Google Scholar] [CrossRef] [PubMed]
- Calo, N.; Ramadori, P.; Sobolewski, C.; Romero, Y.; Maeder, C.; Fournier, M.; Rantakari, P.; Zhang, F.P.; Poutanen, M.; Dufour, J.F.; et al. Stress-activated miR-21/miR-21* in hepatocytes promotes lipid and glucose metabolic disorders associated with high-fat diet consumption. Gut 2016, 65, 1871–1881. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cheng, X.; Lu, Z.; Wang, J.; Chen, H.; Fan, W.; Gao, X.; Lu, D. Upregulation of miR-15b in NAFLD models and in the serum of patients with fatty liver disease. Diabetes Res. Clin. Pract. 2013, 99, 327–334. [Google Scholar] [CrossRef]
- Pescador, N.; Perez-Barba, M.; Ibarra, J.M.; Corbaton, A.; Martinez-Larrad, M.T.; Serrano-Rios, M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS ONE 2013, 8, e77251. [Google Scholar] [CrossRef]
- Ma, L.; Liu, J.; Xiao, E.; Ning, H.; Li, K.; Shang, J.; Kang, Y. MiR-15b and miR-16 suppress TGF-beta1-induced proliferation and fibrogenesis by regulating LOXL1 in hepatic stellate cells. Life Sci. 2021, 270, 119144. [Google Scholar] [CrossRef]
- Pan, J.H.; Cha, H.; Tang, J.; Lee, S.; Lee, S.H.; Le, B.; Redding, M.C.; Kim, S.; Batish, M.; Kong, B.C.; et al. The role of microRNA-33 as a key regulator in hepatic lipogenesis signaling and a potential serological biomarker for NAFLD with excessive dietary fructose consumption in C57BL/6N mice. Food Funct. 2021, 12, 656–667. [Google Scholar] [CrossRef]
- Vega-Badillo, J.; Gutierrez-Vidal, R.; Hernandez-Perez, H.A.; Villamil-Ramirez, H.; Leon-Mimila, P.; Sanchez-Munoz, F.; Moran-Ramos, S.; Larrieta-Carrasco, E.; Fernandez-Silva, I.; Mendez-Sanchez, N.; et al. Hepatic miR-33a/miR-144 and their target gene ABCA1 are associated with steatohepatitis in morbidly obese subjects. Liver Int. 2016, 36, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Nishino, T.; Baba, O.; Kuwabara, Y.; Nakao, T.; Nishiga, M.; Usami, S.; Izuhara, M.; Sowa, N.; Yahagi, N.; et al. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat. Commun. 2013, 4, 2883. [Google Scholar] [CrossRef]
- Price, N.L.; Rotllan, N.; Zhang, X.; Canfran-Duque, A.; Nottoli, T.; Suarez, Y.; Fernandez-Hernando, C. Specific Disruption of Abca1 Targeting Largely Mimics the Effects of miR-33 Knockout on Macrophage Cholesterol Efflux and Atherosclerotic Plaque Development. Circ. Res. 2019, 124, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Davalos, A.; Goedeke, L.; Smibert, P.; Ramirez, C.M.; Warrier, N.P.; Andreo, U.; Cirera-Salinas, D.; Rayner, K.; Suresh, U.; Pastor-Pareja, J.C.; et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 9232–9237. [Google Scholar] [CrossRef]
- Li, Z.J.; Ou-Yang, P.H.; Han, X.P. Profibrotic effect of miR-33a with Akt activation in hepatic stellate cells. Cell Signal. 2014, 26, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Rotllan, N.; Canfran-Duque, A.; Sun, J.; Toczek, J.; Moshnikova, A.; Malik, S.; Price, N.L.; Araldi, E.; Zhong, W.; et al. Targeted Suppression of miRNA-33 Using pHLIP Improves Atherosclerosis Regression. Circ. Res. 2022, 131, 77–90. [Google Scholar] [CrossRef]
- Rotllan, N.; Ramirez, C.M.; Aryal, B.; Esau, C.C.; Fernandez-Hernando, C. Therapeutic silencing of microRNA-33 inhibits the progression of atherosclerosis in Ldlr−/− mice—Brief report. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1973–1977. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Zhou, C.; Yang, X.; Li, L. Down-regulation of microRNA-375 regulates adipokines and inhibits inflammatory cytokines by targeting AdipoR2 in non-alcoholic fatty liver disease. Clin. Exp. Pharmacol. Physiol. 2018, 45, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Zeng, N.; Huang, R.; Li, N.; Jiang, H.; Li, R.; Wang, F.; Chen, W.; Xia, M.; Wang, Q. MiR-451a attenuates free fatty acids-mediated hepatocyte steatosis by targeting the thyroid hormone responsive spot 14 gene. Mol. Cell. Endocrinol. 2018, 474, 260–271. [Google Scholar] [CrossRef]
- Xu, M.; Zheng, X.M.; Jiang, F.; Qiu, W.Q. MicroRNA-190b regulates lipid metabolism and insulin sensitivity by targeting IGF-1 and ADAMTS9 in non-alcoholic fatty liver disease. J. Cell Biochem. 2018, 119, 5864–5874. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, K.; Yu, W.; Wang, H.; Liu, L.; Wu, Q.; Li, S.; Guo, J. MiR-181b regulates steatosis in nonalcoholic fatty liver disease via targeting SIRT1. Biochem. Biophys. Res. Commun. 2017, 493, 227–232. [Google Scholar] [CrossRef]
- Huang, R.; Duan, X.; Liu, X.; Cao, H.; Wang, Y.; Fan, J.; Wang, B. Upregulation of miR-181a impairs lipid metabolism by targeting PPARalpha expression in nonalcoholic fatty liver disease. Biochem. Biophys. Res. Commun. 2019, 508, 1252–1258. [Google Scholar] [CrossRef]
- DiStefano, J.K. Beyond the Protein-Coding Sequence: Noncoding RNAs in the Pathogenesis of Type 2 Diabetes. Rev. Diabet. Stud. 2015, 12, 260–276. [Google Scholar] [CrossRef]
- Shantikumar, S.; Caporali, A.; Emanueli, C. Role of microRNAs in diabetes and its cardiovascular complications. Cardiovasc. Res. 2012, 93, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Skyler, J.S.; Bakris, G.L.; Bonifacio, E.; Darsow, T.; Eckel, R.H.; Groop, L.; Groop, P.H.; Handelsman, Y.; Insel, R.A.; Mathieu, C.; et al. Differentiation of Diabetes by Pathophysiology, Natural History, and Prognosis. Diabetes 2017, 66, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, N.; Pan, H.P.; Wang, Z.; Cao, Z.Y. MiR-499-5p Contributes to Hepatic Insulin Resistance by Suppressing PTEN. Cell. Physiol. Biochem. 2015, 36, 2357–2365. [Google Scholar] [CrossRef]
- Ezaz, G.; Trivedi, H.D.; Connelly, M.A.; Filozof, C.; Howard, K.; Mark, L.P.; Kim, M.; Herman, M.A.; Nasser, I.; Afdhal, N.H.; et al. Differential Associations of Circulating MicroRNAs With Pathogenic Factors in NAFLD. Hepatol. Commun. 2020, 4, 670–680. [Google Scholar] [CrossRef]
- Lopez-Pastor, A.R.; Infante-Menendez, J.; Escribano, O.; Gomez-Hernandez, A. miRNA Dysregulation in the Development of Non-Alcoholic Fatty Liver Disease and the Related Disorders Type 2 Diabetes Mellitus and Cardiovascular Disease. Front. Med. 2020, 7, 527059. [Google Scholar] [CrossRef]
- Willeit, P.; Skroblin, P.; Moschen, A.R.; Yin, X.; Kaudewitz, D.; Zampetaki, A.; Barwari, T.; Whitehead, M.; Ramirez, C.M.; Goedeke, L.; et al. Circulating MicroRNA-122 Is Associated with the Risk of New-Onset Metabolic Syndrome and Type 2 Diabetes. Diabetes 2017, 66, 347–357. [Google Scholar] [CrossRef]
- Dong, L.; Hou, X.; Liu, F.; Tao, H.; Zhang, Y.; Zhao, H.; Song, G. Regulation of insulin resistance by targeting the insulin-like growth factor 1 receptor with microRNA-122-5p in hepatic cells. Cell Biol. Int. 2019, 43, 553–564. [Google Scholar] [CrossRef]
- Jones, A.; Danielson, K.M.; Benton, M.C.; Ziegler, O.; Shah, R.; Stubbs, R.S.; Das, S.; Macartney-Coxson, D. miRNA Signatures of Insulin Resistance in Obesity. Obesity 2017, 25, 1734–1744. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dong, Z.; Lou, L.; Yang, L.; Qiu, J. MiR-122 Participates in Oxidative Stress and Apoptosis in STZ-Induced Pancreatic beta Cells by Regulating PI3K/AKT Signaling Pathway. Int. J. Endocrinol. 2021, 2021, 5525112. [Google Scholar] [CrossRef] [PubMed]
- Chai, C.; Cox, B.; Yaish, D.; Gross, D.; Rosenberg, N.; Amblard, F.; Shemuelian, Z.; Gefen, M.; Korach, A.; Tirosh, O.; et al. Agonist of RORA Attenuates Nonalcoholic Fatty Liver Progression in Mice via Up-regulation of MicroRNA 122. Gastroenterology 2020, 159, 999–1014e1019. [Google Scholar] [CrossRef] [PubMed]
- Mohany, K.M.; Al Rugaie, O.; Al-Wutayd, O.; Al-Nafeesah, A. Investigation of the levels of circulating miR-29a, miR-122, sestrin 2 and inflammatory markers in obese children with/without type 2 diabetes: A case control study. BMC Endocr. Disord. 2021, 21, 152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, H.; Li, D.; Sun, H.; Li, M.; Hu, H. Long noncoding RNA Mirt2 upregulates USP10 expression to suppress hepatic steatosis by sponging miR-34a-5p. Gene 2019, 700, 139–148. [Google Scholar] [CrossRef]
- Tugay, K.; Guay, C.; Marques, A.C.; Allagnat, F.; Locke, J.M.; Harries, L.W.; Rutter, G.A.; Regazzi, R. Role of microRNAs in the age-associated decline of pancreatic beta cell function in rat islets. Diabetologia 2016, 59, 161–169. [Google Scholar] [CrossRef]
- Suksangrat, T.; Phannasil, P.; Jitrapakdee, S. miRNA Regulation of Glucose and Lipid Metabolism in Relation to Diabetes and Non-alcoholic Fatty Liver Disease. Rev. Biomark. Stud. Metab. Metab. Relat. Disord. 2019, 1134, 129–148. [Google Scholar] [CrossRef]
- Kornfeld, J.W.; Baitzel, C.; Konner, A.C.; Nicholls, H.T.; Vogt, M.C.; Herrmanns, K.; Scheja, L.; Haumaitre, C.; Wolf, A.M.; Knippschild, U.; et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature 2013, 494, 111–115. [Google Scholar] [CrossRef]
- Zhen, Y.F.; Zhang, Y.J.; Zhao, H.; Ma, H.J.; Song, G.Y. MicroRNA-802 regulates hepatic insulin sensitivity and glucose metabolism. Int. J. Clin. Exp. Pathol. 2018, 11, 2440–2449. [Google Scholar]
- Wen, Z.; Li, J.; Fu, Y.; Zheng, Y.; Ma, M.; Wang, C. Hypertrophic Adipocyte-Derived Exosomal miR-802-5p Contributes to Insulin Resistance in Cardiac Myocytes through Targeting HSP60. Obesity 2020, 28, 1932–1940. [Google Scholar] [CrossRef]
- Zhang, F.; Ma, D.; Zhao, W.; Wang, D.; Liu, T.; Liu, Y.; Yang, Y.; Liu, Y.; Mu, J.; Li, B.; et al. Obesity-induced overexpression of miR-802 impairs insulin transcription and secretion. Nat. Commun. 2020, 11, 1822. [Google Scholar] [CrossRef]
- Higuchi, C.; Nakatsuka, A.; Eguchi, J.; Teshigawara, S.; Kanzaki, M.; Katayama, A.; Yamaguchi, S.; Takahashi, N.; Murakami, K.; Ogawa, D.; et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism 2015, 64, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xing, H.; Liu, J.; Yang, L.; Ma, H.; Ma, H. MicroRNA-802 increases hepatic oxidative stress and induces insulin resistance in high-fat fed mice. Mol. Med. Rep. 2019, 20, 1230–1240. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Wang, M.M.; Zhang, M.; Zhang, S.P.; Wang, C.H.; Yuan, W.J.; Sun, T.; He, L.J.; Hu, Q.K. MiR-126 Suppresses the Glucose-Stimulated Proliferation via IRS-2 in INS-1 beta Cells. PLoS ONE 2016, 11, e0149954. [Google Scholar] [CrossRef] [PubMed]
- Arner, E.; Mejhert, N.; Kulyte, A.; Balwierz, P.J.; Pachkov, M.; Cormont, M.; Lorente-Cebrian, S.; Ehrlund, A.; Laurencikiene, J.; Heden, P.; et al. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes 2012, 61, 1986–1993. [Google Scholar] [CrossRef]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef]
- Zhang, T.; Lv, C.; Li, L.; Chen, S.; Liu, S.; Wang, C.; Su, B. Plasma miR-126 is a potential biomarker for early prediction of type 2 diabetes mellitus in susceptible individuals. Biomed Res. Int. 2013, 2013, 761617. [Google Scholar] [CrossRef]
- Parrizas, M.; Novials, A. Circulating microRNAs as biomarkers for metabolic disease. Best Pr. Res. Clin. Endocrinol. Metab. 2016, 30, 591–601. [Google Scholar] [CrossRef]
- Liang, T.; Liu, C.; Ye, Z. Deep sequencing of small RNA repertoires in mice reveals metabolic disorders-associated hepatic miRNAs. PLoS ONE 2013, 8, e80774. [Google Scholar] [CrossRef]
- Jordan, S.D.; Kruger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Bronneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Bottger, T.; et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 2011, 13, 434–446. [Google Scholar] [CrossRef]
- Chen, X.; Luo, J.; Yang, L.; Guo, Y.; Fan, Y.; Liu, J.; Sun, J.; Zhang, Y.; Jiang, Q.; Chen, T.; et al. miR-143-Mediated Responses to Betaine Supplement Repress Lipogenesis and Hepatic Gluconeogenesis by Targeting MAT1a and MAPK11. J. Agric. Food Chem. 2022, 70, 7981–7992. [Google Scholar] [CrossRef]
- Xihua, L.; Shengjie, T.; Weiwei, G.; Matro, E.; Tingting, T.; Lin, L.; Fang, W.; Jiaqiang, Z.; Fenping, Z.; Hong, L. Circulating miR-143-3p inhibition protects against insulin resistance in Metabolic Syndrome via targeting of the insulin-like growth factor 2 receptor. Transl. Res. 2019, 205, 33–43. [Google Scholar] [CrossRef]
- Dai, L.L.; Li, S.D.; Ma, Y.C.; Tang, J.R.; Lv, J.Y.; Zhang, Y.Q.; Miao, Y.L.; Ma, Y.Q.; Li, C.M.; Chu, Y.Y.; et al. MicroRNA-30b regulates insulin sensitivity by targeting SERCA2b in non-alcoholic fatty liver disease. Liver Int. 2019, 39, 1504–1513. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, Z.; Tu, Y.; Shen, H.; Dai, Z.; Lin, J.; Zhou, Z. miR-101a and miR-30b contribute to inflammatory cytokine-mediated beta-cell dysfunction. Lab. Investig. 2015, 95, 1387–1397. [Google Scholar] [CrossRef]
- Zhang, Q.; Ma, X.F.; Dong, M.Z.; Tan, J.; Zhang, J.; Zhuang, L.K.; Liu, S.S.; Xin, Y.N. MiR-30b-5p regulates the lipid metabolism by targeting PPARGC1A in Huh-7 cell line. Lipids Health Dis. 2020, 19, 76. [Google Scholar] [CrossRef]
- Stepien, E.L.; Durak-Kozica, M.; Kaminska, A.; Targosz-Korecka, M.; Libera, M.; Tylko, G.; Opalinska, A.; Kapusta, M.; Solnica, B.; Georgescu, A.; et al. Circulating ectosomes: Determination of angiogenic microRNAs in type 2 diabetes. Theranostics 2018, 8, 3874–3890. [Google Scholar] [CrossRef]
- Latorre, J.; Moreno-Navarrete, J.M.; Mercader, J.M.; Sabater, M.; Rovira, O.; Girones, J.; Ricart, W.; Fernandez-Real, J.M.; Ortega, F.J. Decreased lipid metabolism but increased FA biosynthesis are coupled with changes in liver microRNAs in obese subjects with NAFLD. Int. J. Obes. 2017, 41, 620–630. [Google Scholar] [CrossRef]
- Chang, H.; Chang, H.; Cheng, T.; Lee, G.D.; Chen, X.; Qi, K. Micro-ribonucleic acid-23a-3p prevents the onset of type 2 diabetes mellitus by suppressing the activation of nucleotide-binding oligomerization-like receptor family pyrin domain containing 3 inflammatory bodies-caused pyroptosis through negatively regulating NIMA-related kinase 7. J. Diabetes Investig. 2021, 12, 334–345. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, H.; Si, H.; Li, X.; Ding, X.; Sheng, Q.; Chen, P.; Zhang, H. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014, 51, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, X.; Ren, H.; Huang, X.; Shen, T.; Tang, W.; Dou, L.; Li, J. miR-23a/b-3p promotes hepatic lipid accumulation by regulating Srebp-1c and Fas. J. Mol. Endocrinol. 2021, 68, 35–49. [Google Scholar] [CrossRef]
- Grieco, F.A.; Sebastiani, G.; Juan-Mateu, J.; Villate, O.; Marroqui, L.; Ladriere, L.; Tugay, K.; Regazzi, R.; Bugliani, M.; Marchetti, P.; et al. MicroRNAs miR-23a-3p, miR-23b-3p, and miR-149-5p Regulate the Expression of Proapoptotic BH3-Only Proteins DP5 and PUMA in Human Pancreatic beta-Cells. Diabetes 2017, 66, 100–112. [Google Scholar] [CrossRef] [PubMed]
- de Candia, P.; Spinetti, G.; Specchia, C.; Sangalli, E.; La Sala, L.; Uccellatore, A.; Lupini, S.; Genovese, S.; Matarese, G.; Ceriello, A. A unique plasma microRNA profile defines type 2 diabetes progression. PLoS ONE 2017, 12, e0188980. [Google Scholar] [CrossRef]
- Fluitt, M.B.; Kumari, N.; Nunlee-Bland, G.; Nekhai, S.; Gambhir, K.K. miRNA-15a, miRNA-15b, and miRNA-499 are Reduced in Erythrocytes of Pre-Diabetic African-American Adults. Jacobs J. Diabetes Endocrinol. 2016, 2, 14. [Google Scholar]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Li, Y.; Shang, Y.F.; Wang, H.L.; Yao, M.X. miRNA-103: Molecular link between insulin resistance and nonalcoholic fatty liver disease. World J. Gastroenterol. 2015, 21, 511–516. [Google Scholar] [CrossRef]
- Simoniene, D.; Stukas, D.; Dauksa, A.; Velickiene, D. Clinical Role of Serum miR107 in Type 2 Diabetes and Related Risk Factors. Biomolecules 2022, 12. [Google Scholar] [CrossRef]
- Lambrecht, J.; Mannaerts, I.; van Grunsven, L.A. The role of miRNAs in stress-responsive hepatic stellate cells during liver fibrosis. Front. Physiol. 2015, 6, 209. [Google Scholar] [CrossRef]
- Toosi, A.E. Liver Fibrosis: Causes and Methods of Assessment, A Review. Rom. J. Intern. Med. 2015, 53, 304–314. [Google Scholar] [CrossRef]
- He, Y.; Huang, C.; Zhang, S.P.; Sun, X.; Long, X.R.; Li, J. The potential of microRNAs in liver fibrosis. Cell. Signal. 2012, 24, 2268–2272. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, Z.; Guan, L.; Ao, R. GRHL2 induces liver fibrosis and intestinal mucosal barrier dysfunction in non-alcoholic fatty liver disease via microRNA-200 and the MAPK pathway. J. Cell. Mol. Med. 2020, 24, 6107–6119. [Google Scholar] [CrossRef]
- Ma, T.; Cai, X.; Wang, Z.; Huang, L.; Wang, C.; Jiang, S.; Hua, Y.; Liu, Q. miR-200c Accelerates Hepatic Stellate Cell-Induced Liver Fibrosis via Targeting the FOG2/PI3K Pathway. Biomed Res. Int. 2017, 2017, 2670658. [Google Scholar] [CrossRef]
- Murakami, Y.; Toyoda, H.; Tanaka, M.; Kuroda, M.; Harada, Y.; Matsuda, F.; Tajima, A.; Kosaka, N.; Ochiya, T.; Shimotohno, K. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS ONE 2011, 6, e16081. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Wang, S.; Sun, P.; Qie, J. Integrated MicroRNA Expression Profile Reveals Dysregulated miR-20a-5p and miR-200a-3p in Liver Fibrosis. Biomed Res. Int. 2021, 2021, 9583932. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yu, Y.; Li, S.; Liu, Y.; Zhou, S.; Cao, S.; Yin, J.; Li, G. MicroRNA-30a ameliorates hepatic fibrosis by inhibiting Beclin1-mediated autophagy. J. Cell. Mol. Med. 2017, 21, 3679–3692. [Google Scholar] [CrossRef]
- Roy, S.; Benz, F.; Vargas Cardenas, D.; Vucur, M.; Gautheron, J.; Schneider, A.; Hellerbrand, C.; Pottier, N.; Alder, J.; Tacke, F.; et al. miR-30c and miR-193 are a part of the TGF-beta-dependent regulatory network controlling extracellular matrix genes in liver fibrosis. J. Dig. Dis. 2015, 16, 513–524. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, W.; Yu, F.; Dong, P.; Chen, B.; Zhou, M.T. MicroRNA-30a Suppresses the Activation of Hepatic Stellate Cells by Inhibiting Epithelial-to-Mesenchymal Transition. Cell. Physiol. Biochem. 2018, 46, 82–92. [Google Scholar] [CrossRef]
- Hernandez-Gea, V.; Ghiassi-Nejad, Z.; Rozenfeld, R.; Gordon, R.; Fiel, M.I.; Yue, Z.; Czaja, M.J.; Friedman, S.L. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology 2012, 142, 938–946. [Google Scholar] [CrossRef]
- Xiao, Y.; Qu, C.; Ge, W.; Wang, B.; Wu, J.; Xu, L.; Chen, Y. Depletion of thymosin beta4 promotes the proliferation, migration, and activation of human hepatic stellate cells. Cell. Physiol. Biochem. 2014, 34, 356–367. [Google Scholar] [CrossRef]
- Geng, W.; Li, C.; Zhan, Y.; Zhang, R.; Zheng, J. Thymoquinone alleviates liver fibrosis via miR-30a-mediated epithelial-mesenchymal transition. J. Cell Physiol. 2020, 236, 3629–3640. [Google Scholar] [CrossRef]
- Zou, Y.; Cai, Y.; Lu, D.; Zhou, Y.; Yao, Q.; Zhang, S. MicroRNA-146a-5p attenuates liver fibrosis by suppressing profibrogenic effects of TGFbeta1 and lipopolysaccharide. Cell. Signal. 2017, 39, 1–8. [Google Scholar] [CrossRef]
- He, Y.; Huang, C.; Sun, X.; Long, X.R.; Lv, X.W.; Li, J. MicroRNA-146a modulates TGF-beta1-induced hepatic stellate cell proliferation by targeting SMAD4. Cell Signal. 2012, 24, 1923–1930. [Google Scholar] [CrossRef]
- Du, J.; Niu, X.; Wang, Y.; Kong, L.; Wang, R.; Zhang, Y.; Zhao, S.; Nan, Y. MiR-146a-5p suppresses activation and proliferation of hepatic stellate cells in nonalcoholic fibrosing steatohepatitis through directly targeting Wnt1 and Wnt5a. Sci. Rep. 2015, 5, 16163. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, Q.; Ying, Y.; Lu, C.; Li, W.; Huang, C.; Xu, W.; Li, Q.; Qi, X.; Zhang, X.; et al. Using Next-generation Sequencing to Identify Novel Exosomal miRNAs as Biomarkers for Significant Hepatic Fibrosis. Discov. Med. 2021, 31, 147–159. [Google Scholar]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Yuan, B.Y.; Chen, Y.H.; Wu, Z.F.; Zhuang, Y.; Chen, G.W.; Zhang, L.; Zhang, H.G.; Cheng, J.C.; Lin, Q.; Zeng, Z.C. MicroRNA-146a-5p Attenuates Fibrosis-related Molecules in Irradiated and TGF-beta1-Treated Human Hepatic Stellate Cells by Regulating PTPRA-SRC Signaling. Radiat. Res. 2019, 192, 621–629. [Google Scholar] [CrossRef]
- Wu, S.M.; Li, T.H.; Yun, H.; Ai, H.W.; Zhang, K.H. miR-140-3p Knockdown Suppresses Cell Proliferation and Fibrogenesis in Hepatic Stellate Cells via PTEN-Mediated AKT/mTOR Signaling. Yonsei Med. J. 2019, 60, 561–569. [Google Scholar] [CrossRef]
- Guo, C.J.; Pan, Q.; Cheng, T.; Jiang, B.; Chen, G.Y.; Li, D.G. Changes in microRNAs associated with hepatic stellate cell activation status identify signaling pathways. FEBS J. 2009, 276, 5163–5176. [Google Scholar] [CrossRef]
- El-Ahwany, E.; Nagy, F.; Zoheiry, M.; Shemis, M.; Nosseir, M.; Taleb, H.A.; El Ghannam, M.; Atta, R.; Zada, S. Circulating miRNAs as Predictor Markers for Activation of Hepatic Stellate Cells and Progression of HCV-Induced Liver Fibrosis. Electron. Physician 2016, 8, 1804–1810. [Google Scholar] [CrossRef]
- Yan, G.; Li, B.; Xin, X.; Xu, M.; Ji, G.; Yu, H. MicroRNA-34a Promotes Hepatic Stellate Cell Activation via Targeting ACSL1. Med. Sci. Monit. 2015, 21, 3008–3015. [Google Scholar] [CrossRef]
- Li, B.; Liu, J.; Xin, X.; Zhang, L.; Zhou, J.; Xia, C.; Zhu, W.; Yu, H. MiR-34c promotes hepatic stellate cell activation and Liver Fibrogenesis by suppressing ACSL1 expression. Int. J. Med. Sci. 2021, 18, 615–625. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, H.; Yao, L.; Zhao, P.; Wu, X. MiR-34a promotes fibrosis of hepatic stellate cells via the TGF-beta pathway. Ann. Transl. Med. 2021, 9, 1520. [Google Scholar] [CrossRef]
- Tian, X.F.; Ji, F.J.; Zang, H.L.; Cao, H. Activation of the miR-34a/SIRT1/p53 Signaling Pathway Contributes to the Progress of Liver Fibrosis via Inducing Apoptosis in Hepatocytes but Not in HSCs. PLoS ONE 2016, 11, e0158657. [Google Scholar] [CrossRef] [PubMed]
- Roderburg, C.; Urban, G.W.; Bettermann, K.; Vucur, M.; Zimmermann, H.; Schmidt, S.; Janssen, J.; Koppe, C.; Knolle, P.; Castoldi, M.; et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 2011, 53, 209–218. [Google Scholar] [CrossRef]
- Wang, J.; Chu, E.S.; Chen, H.Y.; Man, K.; Go, M.Y.; Huang, X.R.; Lan, H.Y.; Sung, J.J.; Yu, J. microRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell activation and inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget 2015, 6, 7325–7338. [Google Scholar] [CrossRef] [PubMed]
- Tu, X.; Zhang, H.; Zhang, J.; Zhao, S.; Zheng, X.; Zhang, Z.; Zhu, J.; Chen, J.; Dong, L.; Zang, Y.; et al. MicroRNA-101 suppresses liver fibrosis by targeting the TGFbeta signalling pathway. J. Pathol. 2014, 234, 46–59. [Google Scholar] [CrossRef]
- Yu, F.; Lu, Z.; Cai, J.; Huang, K.; Chen, B.; Li, G.; Dong, P.; Zheng, J. MALAT1 functions as a competing endogenous RNA to mediate Rac1 expression by sequestering miR-101b in liver fibrosis. Cell Cycle 2015, 14, 3885–3896. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Chen, X.; Yu, C.; Deng, Y.; Zhang, Y.; Chen, S.; Chen, X.; Chen, K.; Yang, Y.; Ling, W. Gut Microbially Produced Indole-3-Propionic Acid Inhibits Atherosclerosis by Promoting Reverse Cholesterol Transport and Its Deficiency Is Causally Related to Atherosclerotic Cardiovascular Disease. Circ. Res. 2022, 131, 404–420. [Google Scholar] [CrossRef]
- Jusic, A.; Thomas, P.B.; Wettinger, S.B.; Dogan, S.; Farrugia, R.; Gaetano, C.; Tuna, B.G.; Pinet, F.; Robinson, E.L.; Tual-Chalot, S.; et al. Noncoding RNAs in age-related cardiovascular diseases. Ageing Res. Rev. 2022, 77, 101610. [Google Scholar] [CrossRef]
- Hassen, G.; Singh, A.; Belete, G.; Jain, N.; De la Hoz, I.; Camacho-Leon, G.P.; Dargie, N.K.; Carrera, K.G.; Alemu, T.; Jhaveri, S.; et al. Nonalcoholic Fatty Liver Disease: An Emerging Modern-Day Risk Factor for Cardiovascular Disease. Cureus 2022, 14, e25495. [Google Scholar] [CrossRef]
- Huang, D.Q.; Downes, M.; Evans, R.M.; Witztum, J.L.; Glass, C.K.; Loomba, R. Shared Mechanisms between Cardiovascular Disease and NAFLD. Semin. Liver Dis. 2022, 42, 455–464. [Google Scholar] [CrossRef]
- Rotllan, N.; Fernandez-Hernando, C. MicroRNA Regulation of Cholesterol Metabolism. Cholesterol 2012, 2012, 847849. [Google Scholar] [CrossRef]
- Krutzfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Chai, C.; Rivkin, M.; Berkovits, L.; Simerzin, A.; Zorde-Khvalevsky, E.; Rosenberg, N.; Klein, S.; Yaish, D.; Durst, R.; Shpitzen, S.; et al. Metabolic Circuit Involving Free Fatty Acids, microRNA 122, and Triglyceride Synthesis in Liver and Muscle Tissues. Gastroenterology 2017, 153, 1404–1415. [Google Scholar] [CrossRef]
- Vickers, K.C.; Rye, K.A.; Tabet, F. MicroRNAs in the onset and development of cardiovascular disease. Clin. Sci. 2014, 126, 183–194. [Google Scholar] [CrossRef]
- Gao, W.; He, H.W.; Wang, Z.M.; Zhao, H.; Lian, X.Q.; Wang, Y.S.; Zhu, J.; Yan, J.J.; Zhang, D.G.; Yang, Z.J.; et al. Plasma levels of lipometabolism-related miR-122 and miR-370 are increased in patients with hyperlipidemia and associated with coronary artery disease. Lipids Health Dis. 2012, 11, 55. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Wang, L.; Qiao, S.; Lu, X.; Wu, Y.; Xu, B.; Li, H.; Gu, D. Plasma miR-122 and miR-3149 Potentially Novel Biomarkers for Acute Coronary Syndrome. PLoS ONE 2015, 10, e0125430. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, Z.; Yin, Q.; Fu, C.; Barszczyk, A.; Zhang, X.; Wang, J.; Yang, D. Cardiac-specific overexpression of miR-122 induces mitochondria-dependent cardiomyocyte apoptosis and promotes heart failure by inhibiting Hand2. J. Cell. Mol. Med. 2021, 25, 5326–5334. [Google Scholar] [CrossRef]
- Najafi-Shoushtari, S.H.; Kristo, F.; Li, Y.; Shioda, T.; Cohen, D.E.; Gerszten, R.E.; Naar, A.M. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010, 328, 1566–1569. [Google Scholar] [CrossRef]
- Horie, T.; Baba, O.; Kuwabara, Y.; Chujo, Y.; Watanabe, S.; Kinoshita, M.; Horiguchi, M.; Nakamura, T.; Chonabayashi, K.; Hishizawa, M.; et al. MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE−/− mice. J. Am. Heart Assoc. 2012, 1, e003376. [Google Scholar] [CrossRef]
- Price, N.L.; Rotllan, N.; Canfran-Duque, A.; Zhang, X.; Pati, P.; Arias, N.; Moen, J.; Mayr, M.; Ford, D.A.; Baldan, A.; et al. Genetic Dissection of the Impact of miR-33a and miR-33b during the Progression of Atherosclerosis. Cell Rep. 2017, 21, 1317–1330. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Suarez, Y.; Davalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernandez-Hernando, C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, D.; Thrush, A.B.; Nguyen, M.A.; Richards, L.; Geoffrion, M.; Singaravelu, R.; Ramphos, E.; Shangari, P.; Ouimet, M.; Pezacki, J.P.; et al. Macrophage Mitochondrial Energy Status Regulates Cholesterol Efflux and Is Enhanced by Anti-miR33 in Atherosclerosis. Circ. Res. 2015, 117, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Reddy, L.L.; Shah, S.A.V.; Ponde, C.K.; Rajani, R.M.; Ashavaid, T.F. Circulating miRNA-33: A potential biomarker in patients with coronary artery disease. Biomarkers 2019, 24, 36–42. [Google Scholar] [CrossRef] [PubMed]
- de Aguiar Vallim, T.Q.; Tarling, E.J.; Kim, T.; Civelek, M.; Baldan, A.; Esau, C.; Edwards, P.A. MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ. Res. 2013, 112, 1602–1612. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, C.M.; Rotllan, N.; Vlassov, A.V.; Davalos, A.; Li, M.; Goedeke, L.; Aranda, J.F.; Cirera-Salinas, D.; Araldi, E.; Salerno, A.; et al. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ. Res. 2013, 112, 1592–1601. [Google Scholar] [CrossRef]
- Azzimato, V.; Jager, J.; Chen, P.; Morgantini, C.; Levi, L.; Barreby, E.; Sulen, A.; Oses, C.; Willerbrords, J.; Xu, C.; et al. Liver macrophages inhibit the endogenous antioxidant response in obesity-associated insulin resistance. Sci. Transl. Med. 2020, 12, eaaw9709. [Google Scholar] [CrossRef]
- Azzimato, V.; Chen, P.; Barreby, E.; Morgantini, C.; Levi, L.; Vankova, A.; Jager, J.; Sulen, A.; Diotallevi, M.; Shen, J.X.; et al. Hepatic miR-144 Drives Fumarase Activity Preventing NRF2 Activation During Obesity. Gastroenterology 2021, 161, 1982–1997.e1911. [Google Scholar] [CrossRef]
- Cheng, J.; Cheng, A.; Clifford, B.L.; Wu, X.; Hedin, U.; Maegdefessel, L.; Pamir, N.; Sallam, T.; Tarling, E.J.; de Aguiar Vallim, T.Q. MicroRNA-144 Silencing Protects Against Atherosclerosis in Male, but Not Female Mice. Arter. Thromb. Vasc. Biol. 2020, 40, 412–425. [Google Scholar] [CrossRef]
- Hu, Y.W.; Hu, Y.R.; Zhao, J.Y.; Li, S.F.; Ma, X.; Wu, S.G.; Lu, J.B.; Qiu, Y.R.; Sha, Y.H.; Wang, Y.C.; et al. An agomir of miR-144-3p accelerates plaque formation through impairing reverse cholesterol transport and promoting pro-inflammatory cytokine production. PLoS ONE 2014, 9, e94997. [Google Scholar] [CrossRef]
- Chen, B.; Luo, L.; Wei, X.; Gong, D.; Jin, L. Altered Plasma miR-144 as a Novel Biomarker for Coronary Artery Disease. Ann. Clin. Lab. Sci. 2018, 48, 440–445. [Google Scholar]
- Zhang, M.W.; Shen, Y.J.; Shi, J.; Yu, J.G. MiR-223-3p in Cardiovascular Diseases: A Biomarker and Potential Therapeutic Target. Front. Cardiovasc. Med. 2020, 7, 610561. [Google Scholar] [CrossRef]
- Vickers, K.C.; Landstreet, S.R.; Levin, M.G.; Shoucri, B.M.; Toth, C.L.; Taylor, R.C.; Palmisano, B.T.; Tabet, F.; Cui, H.L.; Rye, K.A.; et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, 14518–14523. [Google Scholar] [CrossRef]
- Wang, K.; Long, B.; Liu, F.; Wang, J.X.; Liu, C.Y.; Zhao, B.; Zhou, L.Y.; Sun, T.; Wang, M.; Yu, T.; et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016, 37, 2602–2611. [Google Scholar] [CrossRef]
- Kin, K.; Miyagawa, S.; Fukushima, S.; Shirakawa, Y.; Torikai, K.; Shimamura, K.; Daimon, T.; Kawahara, Y.; Kuratani, T.; Sawa, Y. Tissue- and plasma-specific MicroRNA signatures for atherosclerotic abdominal aortic aneurysm. J. Am. Heart Assoc. 2012, 1, e000745. [Google Scholar] [CrossRef]
- Alexandru, N.; Constantin, A.; Nemecz, M.; Comarita, I.K.; Vilcu, A.; Procopciuc, A.; Tanko, G.; Georgescu, A. Hypertension Associated with Hyperlipidemia Induced Different MicroRNA Expression Profiles in Plasma, Platelets, and Platelet-Derived Microvesicles; Effects of Endothelial Progenitor Cell Therapy. Front. Med. 2019, 6, 280. [Google Scholar] [CrossRef]
- Wijnen, W.J.; van der Made, I.; van den Oever, S.; Hiller, M.; de Boer, B.A.; Picavet, D.I.; Chatzispyrou, I.A.; Houtkooper, R.H.; Tijsen, A.J.; Hagoort, J.; et al. Cardiomyocyte-specific miRNA-30c over-expression causes dilated cardiomyopathy. PLoS ONE 2014, 9, e96290. [Google Scholar] [CrossRef]
- Patel, N.; Tahara, S.M.; Malik, P.; Kalra, V.K. Involvement of miR-30c and miR-301a in immediate induction of plasminogen activator inhibitor-1 by placental growth factor in human pulmonary endothelial cells. Biochem. J. 2011, 434, 473–482. [Google Scholar] [CrossRef]
- Li, J.; Donath, S.; Li, Y.; Qin, D.; Prabhakar, B.S.; Li, P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010, 6, e1000795. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, X.; Xun, Q.; Yu, D.; Ling, J.; Guo, F.; Yan, Y.; Shi, J.; Hu, Y. microRNA-30c negatively regulates endometrial cancer cells by targeting metastasis-associated gene-1. Oncol. Rep. 2012, 27, 807–812. [Google Scholar] [CrossRef]
- Soh, J.; Iqbal, J.; Queiroz, J.; Fernandez-Hernando, C.; Hussain, M.M. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat. Med. 2013, 19, 892–900. [Google Scholar] [CrossRef]
- Soh, J.; Hussain, M.M. Supplementary site interactions are critical for the regulation of microsomal triglyceride transfer protein by microRNA-30c. Nutr. Metab. 2013, 10, 56. [Google Scholar] [CrossRef]
- Luo, M.; Wang, G.; Xu, C.; Zeng, M.; Lin, F.; Wu, J.; Wan, Q. Circulating miR-30c as a predictive biomarker of type 2 diabetes mellitus with coronary heart disease by regulating PAI-1/VN interactions. Life Sci. 2019, 239, 117092. [Google Scholar] [CrossRef]
- Zhan, H.; Huang, F.; Niu, Q.; Jiao, M.; Han, X.; Zhang, K.; Ma, W.; Mi, S.; Guo, S.; Zhao, Z. Downregulation of miR-128 Ameliorates Ang II-Induced Cardiac Remodeling via SIRT1/PIK3R1 Multiple Targets. Oxidative Med. Cell. Longev. 2021, 2021, 8889195. [Google Scholar] [CrossRef] [PubMed]
- Newman, L.A.; Useckaite, Z.; Johnson, J.; Sorich, M.J.; Hopkins, A.M.; Rowland, A. Selective Isolation of Liver-Derived Extracellular Vesicles Redefines Performance of miRNA Biomarkers for Non-Alcoholic Fatty Liver Disease. Biomedicines 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Li, X.C.; Tang, Y.L. Upregulation of miR-128 Mediates Heart Injury by Activating Wnt/beta-catenin Signaling Pathway in Heart Failure Mice. Organogenesis 2021, 17, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Adlakha, Y.K.; Khanna, S.; Singh, R.; Singh, V.P.; Agrawal, A.; Saini, N. Pro-apoptotic miRNA-128-2 modulates ABCA1, ABCG1 and RXRalpha expression and cholesterol homeostasis. Cell Death Dis. 2013, 4, e780. [Google Scholar] [CrossRef]
- Goedeke, L.; Rotllan, N.; Canfran-Duque, A.; Aranda, J.F.; Ramirez, C.M.; Araldi, E.; Lin, C.S.; Anderson, N.N.; Wagschal, A.; de Cabo, R.; et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat. Med. 2015, 21, 1280–1289. [Google Scholar] [CrossRef]
- Wagschal, A.; Najafi-Shoushtari, S.H.; Wang, L.; Goedeke, L.; Sinha, S.; deLemos, A.S.; Black, J.C.; Ramirez, C.M.; Li, Y.; Tewhey, R.; et al. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat. Med. 2015, 21, 1290–1297. [Google Scholar] [CrossRef]
- Doghish, A.S.; Elsisi, A.M.; Amin, A.I.; Abulsoud, A.I. Circulating miR-148a-5p and miR-21-5p as Novel Diagnostic Biomarkers in Adult Egyptian Male Patients with Metabolic Syndrome. Can. J. Diabetes 2021, 45, 614–618. [Google Scholar] [CrossRef]
- Rotllan, N.; Zhang, X.; Canfran-Duque, A.; Goedeke, L.; Grinan, R.; Ramirez, C.M.; Suarez, Y.; Fernandez-Hernando, C. Antagonism of miR-148a attenuates atherosclerosis progression in APOB(TG)Apobec(−/−)Ldlr(+/−) mice: A brief report. Biomed. Pharmacother. 2022, 153, 113419. [Google Scholar] [CrossRef]
- Mehta, R.; Otgonsuren, M.; Younoszai, Z.; Allawi, H.; Raybuck, B.; Younossi, Z. Circulating miRNA in patients with non-alcoholic fatty liver disease and coronary artery disease. BMJ Open Gastroenterol. 2016, 3, e000096. [Google Scholar] [CrossRef]
- Cordes, K.R.; Sheehy, N.T.; White, M.P.; Berry, E.C.; Morton, S.U.; Muth, A.N.; Lee, T.H.; Miano, J.M.; Ivey, K.N.; Srivastava, D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009, 460, 705–710. [Google Scholar] [CrossRef]
- Ng, R.; Wu, H.; Xiao, H.; Chen, X.; Willenbring, H.; Steer, C.J.; Song, G. Inhibition of microRNA-24 expression in liver prevents hepatic lipid accumulation and hyperlipidemia. Hepatology 2014, 60, 554–564. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goncalves, B.d.S.; Meadows, A.; Pereira, D.G.; Puri, R.; Pillai, S.S. Insight into the Inter-Organ Crosstalk and Prognostic Role of Liver-Derived MicroRNAs in Metabolic Disease Progression. Biomedicines 2023, 11, 1597. https://doi.org/10.3390/biomedicines11061597
Goncalves BdS, Meadows A, Pereira DG, Puri R, Pillai SS. Insight into the Inter-Organ Crosstalk and Prognostic Role of Liver-Derived MicroRNAs in Metabolic Disease Progression. Biomedicines. 2023; 11(6):1597. https://doi.org/10.3390/biomedicines11061597
Chicago/Turabian StyleGoncalves, Bruno de Souza, Avery Meadows, Duane G. Pereira, Raghav Puri, and Sneha S. Pillai. 2023. "Insight into the Inter-Organ Crosstalk and Prognostic Role of Liver-Derived MicroRNAs in Metabolic Disease Progression" Biomedicines 11, no. 6: 1597. https://doi.org/10.3390/biomedicines11061597
APA StyleGoncalves, B. d. S., Meadows, A., Pereira, D. G., Puri, R., & Pillai, S. S. (2023). Insight into the Inter-Organ Crosstalk and Prognostic Role of Liver-Derived MicroRNAs in Metabolic Disease Progression. Biomedicines, 11(6), 1597. https://doi.org/10.3390/biomedicines11061597






