Abstract
A number of studies are underway to gain a better understanding of the role of immunity in the pathogenesis of hepatocellular carcinoma and to identify subgroups of individuals who may benefit the most from systemic therapy according to the etiology of their tumor. Human leukocyte antigens play a key role in antigen presentation to T cells. This is fundamental to the host’s defense against pathogens and tumor cells. In addition, HLA-specific interactions with innate lymphoid cell receptors, such those present on natural killer cells and innate lymphoid cell type 2, have been shown to be important activators of immune function in the context of several liver diseases. More recent studies have highlighted the key role of members of the non-classical HLA-Ib and the transcript adjacent to the HLA-F locus, FAT10, in hepatocarcinoma. The present review analyzes the major contribution of these molecules to hepatic viral infection and hepatocellular prognosis. Particular attention has been paid to the association of natural killer and Vδ2 T-cell activation, mediated by specific HLA class Ib molecules, with risk assessment and novel treatment strategies to improve immunotherapy in HCC.
1. Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancer, accounting for 70% to 90% of all diagnoses, and expected to reach 22 million cases in the next two decades [1]. The main etiologic factors for HCC are hepatitis B (HBV) and C (HCV) viruses, steatohepatitis and metabolic syndromes, alcohol abuse and aflatoxin, while cirrhosis is considered as a major indicator for its surveillance [1].
HCC is still difficult to cure with a 5-year survival rate of less than 15%. In patients with advanced HCC, systemic therapies, including targeted therapy, immunotherapy, or a combination of both, are the mainstay of treatment [2]. However, immunotherapy used successfully in most refractory malignancies has not been as effective as expected in HCC, probably because of its particularly complex immune microenvironment. Indeed, antigen-rich blood from the gut passes through the liver, and this requires particularly efficient immune surveillance against pathogens and tolerance towards non-dangerous antigens [3,4]. The response rate for immunotherapy alone in a multi-center phase III study of nivolumab (an anti-programmed cell death protein 1, PD-1, drug) vs. the standard of care sorafenib (an anti-angiogenetic drug) confirmed efficacy only in 15% of patients [5]. The scientific community is now waiting for a better result from the combination of immune drugs (anti-PD-1 or anti-PD-L1 or anti-cytotoxic T lymphocyte-associated antigen 4 [CTLA4]) with multi-tyrosine kinases (anti-vascular endothelial growth factor receptor (VEGFR1-3), fibroblast growth factor (FGF) receptors (FGFR1-4), platelet-derived growth factor (PDGF) receptor α, KIT and RET. In the meantime, research on immune markers is ongoing to better understand the role of immunity in the pathogenesis of HCC, and to recognize which subgroups of individuals will most benefit from a systemic combined therapy according to liver cancer etiology [6].
To date, it is accepted that during HCC progression, the proportion of tumor-associated macrophages (TAM) displaying an M2 phenotype and expressing the immunosuppressive cytokines interleukin (IL)-10 and transforming growth factor-β (TGF-β) is increased, while cytotoxic natural killer (NK) and CD8 T cells are both decreased in number and inhibited through activation of immune checkpoints signaling (i.e., PD-1, CTLA4, TIGIT and LAG3) [7,8,9,10,11].
2. Classical HLA Class I Molecules (HLA-Ia)
The major histocompatibility complex (MHC), also known as the human leukocyte antigens (HLA), plays a key role in antigen presentation to T cells, leading to the basic formation of host defense mechanisms against pathogens, tumor cells and transplantation outcomes. HLA complex consists of more than 200 genes on chromosome 6, and can be categorized into three groups: class I, class II and class III [12].
Immunogenic peptides binding to HLA class I (e.g., HLA-A, B, C) are presented by nucleated cells and are recognized by cytotoxic CD8+ T cells; those binding to HLA class II (e.g., HLA-DR, DP, DQ) on the surface of the antigen-presenting cells [e.g., dendritic cells, macrophages, or B cells] and tumor cells display antigen peptide to CD4+ T cells, thus providing help to the effector cells such as innate immune cells, B cells and CD8 T cells [12]. Specific HLA alleles are the main risk of acute liver failure caused by certain drugs, by drug-metabolites and drug-modified peptides that can be recognized as foreign moieties trough the presentation of HLA-antigen complex. Examples are nevirapine in patients carrying the HLA-DRB1*0101 allele, flucloxacillin in HLA-B*5701 and minocycline in HLA-B*3502 carriers [13,14,15,16,17]. Furthermore, specific interactions of classical HLA with innate lymphoid cells (ILC) receptors, such as those expressed by natural killer cell (NK) and ILC2, have been shown to be important activators of liver-disease-associated immune function [18]. Among them, several studies have associated class I HLA (mainly HLA-A-Bw4 and HLA-C) with NK cell KIR receptors and the response to interferon treatment to induce HCV and HBV viral clearance, as well as the risk of developing HCC [19,20,21,22,23,24,25].
3. Non-Classical HLA Class I Molecules (HLA-Ib)
However, most recent studies have highlighted the key role played by members of the non-classical HLA class Ib (HLA-E, HLA-G, HLA-F, MHC class I related chain A (MICA/B) and Hla-F locus adjacent transcript (FAT10)) in HCC (Table 1).

Table 1.
NK-like and NK cell receptors binding classical and non-classical HLA class I.
It has been shown that antigen-presenting cells, as a consequence of infection or in response to antigen stimulation, produce several cytokines including IL-12 and IL-23, which are crucial for the secretion of IFN-γ mainly by T-cells, including CD4+ helper type 1 (TH1) cells, CD8+ cytotoxic cells and NK cells [51]. In addition, IFN-γ further increased the IL-12 production and secretion, thus creating a positive feedback loop between IL-12 and IFN-γ signaling [52].
IFN-γ, a member of type II interferon, plays a key role in host defense and immune surveillance against microorganisms and tumor cells, primarily by acting as a potent activator of neutrophil and macrophage activities, promoting B-cell immunoglobulin isotype switching, promoting activation of CD8+ T-cells and differentiation of CD4+ T-cells towards a TH1 phenotype and, finally, up-regulates HLA class I expression and antigen presentation in tumor cells. However, IFN-γ has also recently been shown to have an immunosuppressive effect in tumors as it promotes the PD-L1/PD-L2, indoleamine 2, 3-dioxygenase 1 (IDO 1) [53,54], and the expression of HLA-Ia and HLA-Ib (i.e., HLA-E, HLA-G and MICA) that suppress T-cells function [55,56] (Figure 1). Therefore, similarly to other conditions present in different types of cancers [57,58], it has been proposed that cytokine secreted by NK cells, by modulating the immune response networks and counteracting viral infection, may be involved in liver fibrosis and poor HCC prognosis through the induction of a tolerant microenvironment [43,43,57,57,58,59,60].
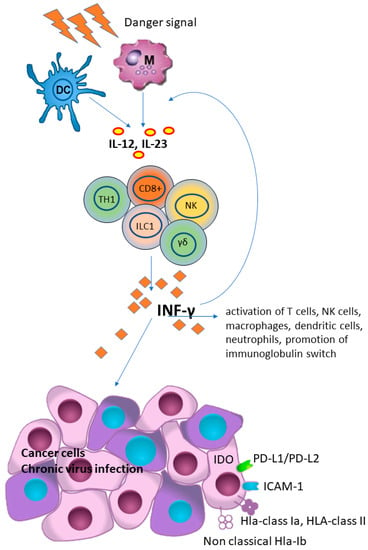
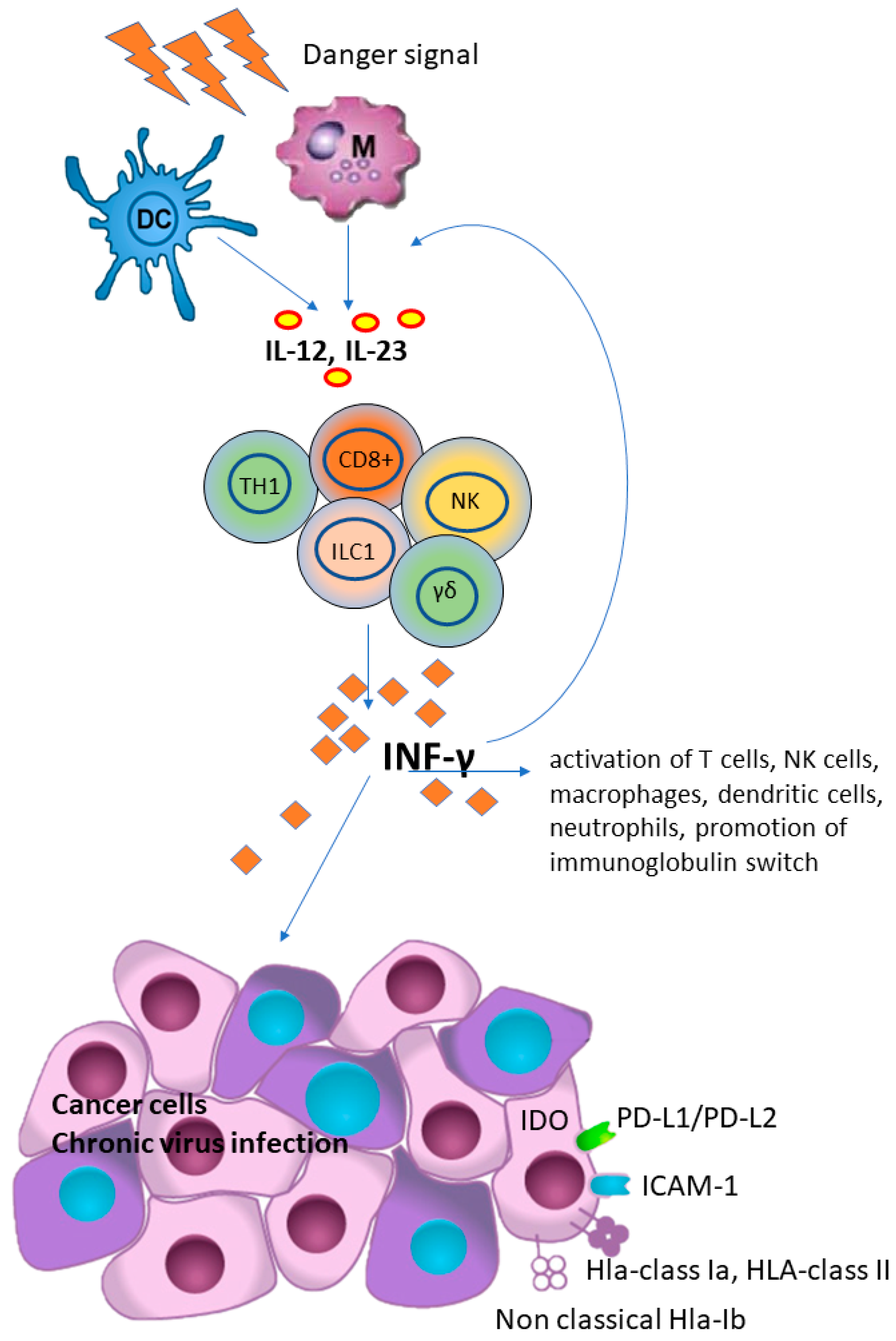
Figure 1.
Interferon-gamma (IFN-γ) plays a key role in host defense and immune surveillance against microorganisms and tumor cells, primarily by acting as a potent activator of neutrophil and macrophage activities, promoting B-cell immunoglobulin isotype switching, promoting activation of CD8+ T-cells and differentiation of CD4+ T-cells towards a TH1 phenotype and, finally, up-regulating HLA class I expression and antigen presentation in tumor cells. However, IFN-γ has also been shown to have an immunosuppressive effect in tumors, as it promotes the PD-L1/PD-L2, indoleamine 2, 3-dioxygenase 1 (IDO 1), ICAM-1 and the expression of HLA and HLA-Ib (i.e., HLA-E, HLA-G and MICA) that suppress T-cells function. (DC), dendritic cell; (M), macrophage; (IL) interleukins 12 and 23; (CD8+), cytotoxic T cells; (NK), natural killer cells; (γδ), γδ T cells; (TH1), Th1 polarized CD4+ T helper cells; (ILC1), group 1 innate lymphoid cells; (HLA), human lymphoid antigen.
Unlike classical HLA-Ia, the HLA-Ib molecules have a limited number of polymorphisms and low expression on the cell surface [12,61,62,63]. They bind receptors expressed on T, NK and NK-like T lymphocytes. In analogy to their function in trophoblasts, where they protect the fetus from maternal immune attack under special conditions, such as in the tumor microenvironment, their aberrant expression may promote the evasion of tumor cells from immune surveillance [63,64].
HLA-E has only 2 alleles, namely HLA-E*01:01 and HLA-E*01:03, which differ in one amino acid at position 107, conferring significant differences in terms of charge, conformation, relative peptide binding affinity to selected peptides and, consequently, HLA-E expression on the cell surface [65,66,67]. Studies have also shown that the HLA-E molecule derived from the HLA-E*01:03 allele is more highly expressed on the cell surface and is more stable than the HLA-E*01:01 allele. This difference may result in a more effective inhibitory function of the HLA-E*01:03 allele in response to a wide range of ligands [68]. However, HLA-E can present peptides derived from the leader of some HLA-Ia molecules (as reported in the table [67]), but also other peptides such as the heat shock protein (hsp60) [69]. Furthermore, when HLA-E is bound to peptides derived from HLA leader molecules or some viral peptides, stimulation of the CD94/NKG2A receptor inhibits lysis and cells are protected from NK cell cytotoxicity, whereas presentation of the hsp60, and probably other peptides, results in loss of ligand recognition by the inhibitory NK receptor CD94/NKG2A and enables the lytic response of NK cells against stressed cells expressing the hsp60 antigen [70]. Thus, NK inhibition by the CD94/NKG2A/HLA-E system is peptide-specific. In addition, the HLA-E molecule regulates NK cells and cytotoxic T lymphocytes, with different effects on NK cells depending on NKG2 type expression (inhibition of NKG2A, activation of NKG2B, and NKG2C), activation receptors bind HLA-E with lower affinity than the inhibitory NKG2A receptor [68,69,70] (Figure 2). Interestingly, HLA-E, the only ligand for the inhibitory CD94/NKG2A receptor, whereas NKG2-activating receptors have multiple ligands, is frequently upregulated in many tumors [71,72,73,74,75,76]. HLA-E was found to be expressed on the surface of human hepatoma cells, resulting in negative regulation of NK cell activity through CD94/NKG2A interaction [77]. A higher level of HLA-E expression has been observed in HCC tissues compared to adjacent non-cancerous liver tissues and to normal liver [42,78,79]. Furthermore, NKG2A, but not NKG2C, was also upregulated on NK cells in tumor tissue from patients with HCC compared to patients without HCC, and higher NKG2A expression was associated with increased HLA-E expression [80]. These data suggest that HLA-E has a mainly inhibitory function in HCC, and that the NKG2A/HLA-E interaction may act as a resistance mechanism as a result of repeated activation of the immune system in the tumor microenvironment. NKG2A has been thus proposed as a marker for late tumor-infiltrating proliferating lymphocytes (TILs) [81,82] and increased expression was associated with exhausted NK functions [83] and reduced disease-free and survival rates [79]. Most importantly, the NKG2A/NKG2D ratio has been shown to be closely associated with HCC progression and immunosuppression [84], whereas switching to NKG2C on the NK cell surface indicates more mature NK cells [85,86]. A decreased NKG2D expression on circulating NK cells isolated form HCC patients compared to blood donors has also been observed [87]. Furthermore, the presence of inflammatory cytokines such as IL-10, IL15 and TGFβ, particularly abundant in the TME of HCC [88], has been shown to increase NKG2A expression on NK cells [88,89,90], and recently, a specific study of the gene signature once again suggested the CD94/NKG2-HLA-E system as the likely cause of the immune evasion of NK cells in HCC samples with higher levels of infiltrated NK cells characterized by the TGF-beta–Wnt interaction signature [91].
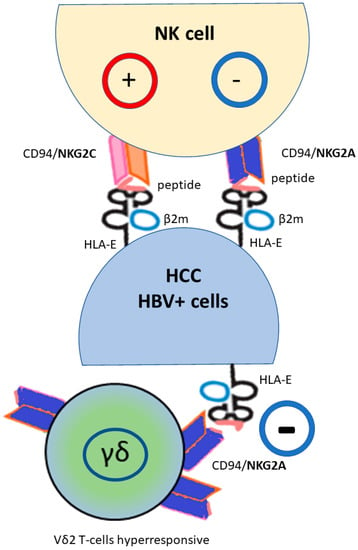
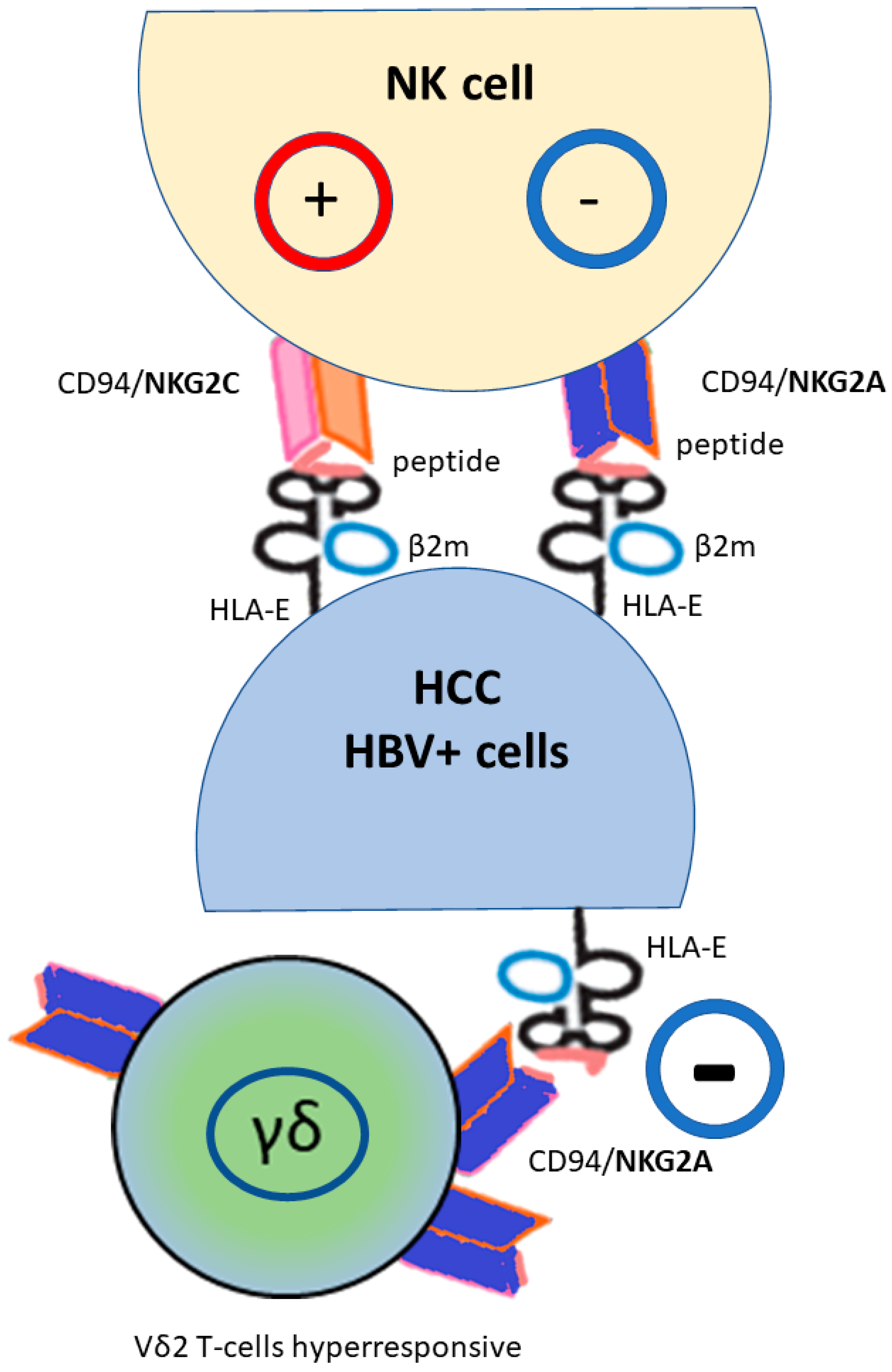
Figure 2.
HLA-E molecule regulates natural killer (NK) and cytotoxic T-lymphocyte cells via the heterodimeric receptor CD94/NKG2, with a different effect on NK cells depending on the NKG2 type (activation NKG2C, inhibition NKG2A). Moreover, HLA-E*01:01 usually presents better virus-derived peptides than the HLA-E*01:03 that is more efficient at leading HLA class Ia leader peptide. NKG2A+ found a subset of Vδ2 T-cells educated to hyperresponsiveness; their function is inhibited by HLA-E expression. (HBV+) hepatitis B virus infected cells; (HCC) hepatocellular carcinoma.
HLA-E is transcribed in most tissues but its expression on the cell surface is limited, and requires a peptide bound with a difference between the two HLA-E*01:01 and HLA-E*01:03 allele as consequence of their relative peptide affinity [66]. The peptide is mostly derived from the signal of HLA class I molecules, i.e., HLA-A, B, C and G, so the surface expression of HLA-E largely reflects the integrity of the antigen presentation pathway mediated by HLA class I molecules, or peptides derived from pathogens or cellular stress [92,93,94]. The peptides share a common motif: methionine at position 2, and leucine or isoleucine at position 9 [95], and these complexes can be detected by T cells via their antigen-specific T-cell receptor (TCR). It has been reported that the HLA-Ia leader sequence/HLA-E complex preferentially binds the inhibitory CD94/NKG2A receptor, while the HLA-G leader sequence/HLA-E complex binds the activating CD94/NKG2C receptor [96]. The viral peptide/HLA-E interaction with the CD94/NKG2 receptor was further complex. In fact, while viral infection caused CD94/NKG2A overexpression, the virus-induced reduction of HLA-Ia expression produced a minor HLA-Ia leader peptide/HLA-E complex with reduced interaction with the CD94/NKG2A receptor, resulting in virus-infected cells being able to be eliminated by CD8+ and NK cells. Moreover, in this context and as shown in susceptibility to COVID-19 infection, because HLA-E*01:01 allele is less efficient at leading HLA class Ia leader peptide than the HLA-E*01:03 allele, HLA-E*01:01 usually presents better virus-derived peptides than the HLA-E*01:03, thus affecting target cell recognition and cytolysis of virus-infected cells by CD8+ T-cells [97]. Consistent with these observations, in a study of HBV, an expanded population of NK-like CD8+ T-cells characterized by a HLA-E/CD94/NKG2C interaction was found in hepatic sinusoids, leading to activation of NK-like cells independent of TCR stimulation [98]. Because HLA-E expression increases following various cellular stresses, including cancer-related inflammation [28,99], it is probable that HLA-E/NKG2C binding is likely to strongly influence the immune response to eliminate infected cells. In addition, growing studies showed that a subset of unconventional gamma delta (γδ) T lymphocytes, harboring the Vγ9Vδ2 TCR (abbreviated as Vδ2pos) T cells, exert a potent antitumor activity in several cancers, and that the inhibitory NKG2A expression on these T cells may abolished the immune response, due to the HLA-E expression on the tumor cells in HCC [79,100] (Figure 2). Furthermore, patients with HBV infection had more exhausted NKG2A+ Vδ2+ T cells in the peripheral blood compared to uninfected patients [101]. The authors found that increased NKG2A expression is associated with more cytolytic cells, although direct stimulation by CD94/NKG2A is inhibitory. Thus, circulating NKG2A+ Vδ2+ T cells are functionally preserved in chronically HBV-infected patients, but have a suppressed function in acute infection. Interestingly, an in vitro blockade of the NKG2A receptor with a targeted antibody has been shown to restore the ability of Vδ2pos T cells even against HLA-Epos tumor cells. Thus, in general, high frequencies of NKG2A+ Vδ2 T-cells improve overall survival in HCC patients when they showed a decreased or a similar level of HLA-E compared to normal tissue. Based on these studies, and the evidence of educated and hyper-responsive NKG2A+ Vδ2 T-cells, HCC prognosis might be improved by the development of NKG2A-HLA-E checkpoint-based therapies.
HLA-G is more polymorphic than the other HLA class Ib antigens, with 117 alleles and 38 proteins [https://www.ebi.ac.uk/ipd/imgt/hla/about/statistics/, accessed on 5 June 2023]. It was first studied on the surface of extra villous cytotrophoblast cells in the placenta as a key factor that induces fetal immune tolerance during pregnancy [102]. HLA-G is an inhibitory molecule that binds immunoglobulin-like transcript 2 (ILT2), ILT4, killer inhibitory receptor 2DL4 (KIR2DL4), CD8 T-cells and CD160 receptor on NK and CD8+ T-cells [103,104,105,106,107]. LT2 binds to both classical HLA-Ia and non-classical HLA-Ib molecules. However, its binding affinity for HLA-G is three to four times higher than for the classical HLA molecule [107]. The interaction of HLA-G with ILT2 and ILT4 leads to inhibition of toxicity in monocytes/macrophages, maturation and activation of T cells in dendritic cells, phagocytosis in neutrophils, secretion of IFN-γ in NK, expression of chemokines and induction of apoptosis in T cells. Together, these effects serve to reduce the inflammatory response [39]. However, it is proposed that HLA-G expression on tumor B cells, as in Hodgkin lymphoma, may be potentially beneficial in controlling tumor cell proliferation, as in vitro HLA-G binding to ILT2 on B cell surface has been shown to cause inhibition of B cell proliferation, differentiation and antibody secretion [41]. In addition to its physiological role in maternal-fetal tolerance, HLA-G is commonly expressed by solid tumors and has been associated with poorer prognosis in several cancers [108,109], including HCC [110,111,112]. HLA-G expression has been reported in approximately 50% of primary HCC lesions, reaching 71% in advanced stage III disease. In addition, plasma-soluble HLA-G was found to be significantly higher in HCC patients than in normal controls [37]. In several studies, a neo-expression of HLA-G has also been observed in connection with a viral infection of the liver [35,113,114].
The level of secretion of soluble HLA-G (sHLA-G) was associated with haplotypes mapping in the 3′UTR of the HLA-G gene [115] (Figure 3). Expression of sHLA-G has also been associated with HBV/HCV infection and poor prognosis, via increased viral load and fibrosis [35,78,112,116]. Indeed, it has been found that increased ILT2 expression engaging HLA-G contributes to the dysfunction of CD56dimCD16+NK cells [117] and that a tumor-expressing HLA-G promotes the accumulation and suppressive activities of immune cells such as Treg and mast cells, as well as converting NK cells, T-cells and macrophages toward a pro-tumor phenotype [118]. Moreover, sHLA-G, which is associated with the UTR1 HLA-G haplotype, was found tolerogenic, and thus reduces the inflammation also associated with a better clinical course in type 1 autoimmune hepatitis [119], as well as in liver transplantation through the immunosuppressive properties of HLA-G (reviewed in [35]). Of note, a recent study demonstrated NKG2A/CD94 to be an additional receptor for HLA-G that moreover was able to distinguish amino acid differences in the HLA-G heavy chain. In fact, G*01:04 allele was more protective against NK cell-mediated lysis than the other 2 allelic variants (G*01:04 and G*01.01) [120]. Overall, these results show sHLA-G and HLA-G protein as unfavorable markers in HCC prognosis, and they were potential targets for the development of new therapeutic strategies [33,34,36,37,111,121,122].
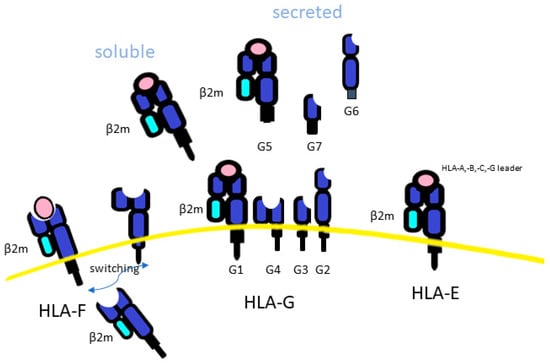
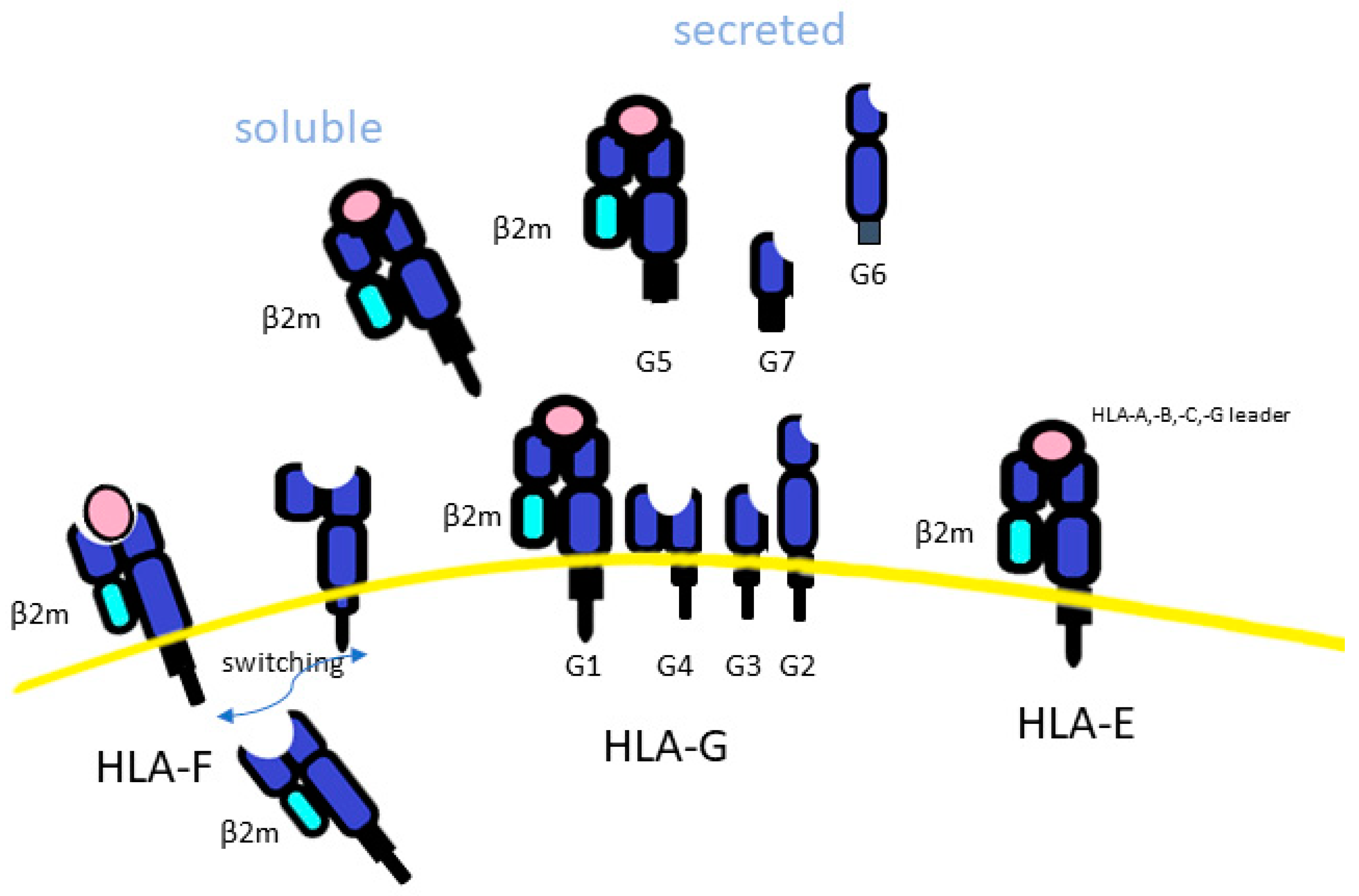
Figure 3.
The difference among HLA class Ib. HLA-F, is an empty molecule, found intracellularly. HLA-F led to the classical trimeric closed HLA conformer form (beta2-microglobulin light chain (β2m), peptide and HLA-Ia) or an open conformation (because it is without a peptide). HLA-G may be secreted as a soluble form (sHLA-G), and this is associated with haplotypes mapping in the 3′UTR of the HLA-G genes.
HLA-F, the smallest of the HLA class I molecules, had only 31 alleles, which can encode 6 distinct HLA-F proteins. HLA-F is an empty intracellular molecule, found mainly in the endoplasmic reticulum and Golgi apparatus (Figure 3). It acts as a ligand for transporters associated with antigen processing (TAP) and plays a role in the loading of endogenous peptides onto HLA class I molecules to CD8+ T-cell presentation and trans-interaction with NK receptors [123]. This led to the classical trimeric closed HLA conformer form (beta2-microglobulin light chain (β2m), peptide and HLA-Ia). However, HLA-F has also been reported to mediate HLA class I exogenous antigen cross-presentation [124]. In this case, HLA-Ia, which is in an open conformation (because it is without a peptide), can present its α1 domain free to interact with HLA-F [125,126]. Then, the HLA-I/HLA-F heterodimer is transported to the cell surface to present extracellular/foreign antigens after endocytosis to T-cells. This process has been observed to occur mostly during an inflammatory response, characterized by high levels of IFN-β, IFN-γ, and TNF-α, where overexpression of HLA-F and HLA-I in open conformation were shown in lymphocytes, monocytes and tumor cells [126,127,128,129,130,131,132,133]. In addition to HLA-peptide complex presentation to TCR, when classical HLA class I molecules were downregulated, thus reducing peptides presentation for immune recognition, HLA-F in open conformer may be exported on the cell surface and result in a marker of activated lymphocytes [134,135], conferring an inhibition of T and NK cell function [127]. It was then demonstrated that HLA-F in open conformation had a high affinity for the NK activator KIR3DS1 and a low affinity for the inhibitory receptors KIR3DL1 and KIR3DL2 [126,136,137], while HLA-F, expressed on the cell surface associated with β2 microglobulin, exhibited, as a consequence of peptide presentation, a reduced interaction between HLA-F and KIR3DS1. Moreover, NK cells from homozygous KIR3DS1 carriers showed a higher antiviral capacity compared to those carrying homologous inhibitory KIRDL3 homozygosity [138]. Overall, HLA-F in open conformer by activation of NK cell function has been associated with the resolution of some infections, including HCV and HIV infection, while hemoglobin peptides had been proposed to have a role in HIV progression, due to a reduction of HLA-F/KIR3DS1 interaction [138,139,140,141]. Previous studies, including one of ours, when the HLA-F function of NKs was still unknown, highlighted the potential role of balance between KIR3DL1 and KIR3DS1 on NK function and outcomes of HCV, HBV and HCC [19,24,25,142,143]. Although the clinical relevance of HLA-F expression in HCC patients is still unclear, HLA-F has attracted attention as an important immunosuppressive molecule, the expression of which was found to correlate with the degree of lymphatic or venous invasion in HCC [144].
More recently, a ubiquitin-like protein that targets proteins for degradation by being part of the ubiquitin-proteasome system (UPS), and thus affecting the peptide/HLA-I presentation, has been reported adjacent to the HLA-F gene region. This protein was called human leukocyte antigen F locus adjacent transcript 10 (FAT10), and has been associated with a susceptibility to inflammation-driven diseases, like nonalcoholic fatty liver disease (NAFLD), non-alcoholic steatohepatitis (NASH), and HCC [145]. Inflammatory stimuli, fibrogenesis and cytokines induced the expression of FAT10 [50,145,146,147]. FAT10 has been involved in the Mallory–Denk Bodies (MDBs), aggresomal molecules formed from undigested ubiquitinated short-lived proteins resulting from a reduced proteasome’s degradation rate, observed in hepatitis, alcoholic steatohepatitis, NAH and HCC [146,148,149,150,151]. Moreover, the upregulated expression of FAT10, related with IFNγ and TNF-α in hepatocytes, was found to accelerate the proliferation and progression of HCC-derived cells in vitro [152,153]. Conversely, FAT10 limits type I INF production in order to avoid exceeding inflammation and tissue destruction [154]. Based on all these results, FAT10 was proposed as a new biomarker in cancer, including the HCC [155,156].
4. NK and NK-like T-Cells
Cytotoxic CD8+ T-cells, natural killer (NK) cells and NK-like T-cells are effector lymphocytes involved in tumor immunosurveillance, and their abundance was lower in HCC tissues than in the adjacent normal tissues [80].
CD8+ T cell response is related to the tumor cell ability to recognize classic HLA-Ia antigen complex presentation to specific CD8+ T-cell receptors (TCR). Of note, tyrosine kinase inhibitors, used in HCC treatment, were also found to stimulate HLA-Ia expression in HCC, showing that downregulation of HLA is reversible [157,158]. However, the liver shows disrupted activation of most CD8+ T cells leading, in case of high antigen load, to antigen persistence accompanied by T-cell exhaustion and hepatitis development [159,160]. Tumor endothelial marker 1 (TEM1), epithelial cell adhesion molecule (EpCAM) melanoma-associated gene-A1 (MAGE-A1), New York esophageal squamous cell carcinoma 1 (NY-ESO-1), AFP and glypican-3 (GPC-3) have been identified as specifically associated with HCC and leading to a T-cell response in the blood in about 50% of patients [161]. These antigens are now used for the development of several CAR-T cells, an adoptive T-cell transfer therapeutic strategy, to avoid the involvement of TCR [e.g., Clinical Trials.gov Identifier: NCT03198546 and NCT02395250]. However, mainly due to the specific immunological tolerance of the liver as the organ that filters the blood effluent from the intestine [3], the efficacy of CAR-T cells in HCC showed a low efficacy if compared to hematologic and lymphatic cancer, and needs to be improved [162,163,164]. In addition, serious toxicities such as immune effector-cell-associated neurotoxicity syndrome (ICANS) or even life-threatening immune-related toxicities, such as cytokine release syndrome (CRS), are not uncommon with the use of the CAR-T approach, requiring multidisciplinary management with the need to develop algorithms that can identify patients at high risk of toxicity [165,166].
An alternative to CAR-T may be represented by innate immune cells like NK, gamma delta T-cells (γδT) and myeloid cells [167,168,169].
NK cells are highly enriched in the human liver, accounting for about 30% of the intrahepatic lymphocytes, and are highly associated with the outcomes of patients with HCC. The infiltration of NK cells into the liver increases further after surgery, due to the inflammation that occurs post-surgery, and this finding further supports importance of NK cells in the liver pathogenesis [170]. Unlike the effector function of CD8+ T cells, NK cells depend on a combination of activating and inhibitory receptors that bind primarily to HLA molecules in their peptide-binding region [Table 1]. The differences in receptors, both as genetic variation (allele) and in number, placed on individual NK cells lead to different repertories among individuals. Tolerance to one’s own molecules depends on the recognition of HLA antigens present on the target cells by the inhibitory receptors, especially KIR2DL, present on NK cells. Signals received through several inhibitory receptors during NK cell development promotes cell function, termed NK cell education [171]. Meanwhile, NK cells can sense the absence of HLA expression on cell surfaces, and some activating receptors such as killer-cell immunoglobulin-like receptors (KIR) and NKG2C receptors, which have variable expression over the time, can also detect HLA molecules as stress signals, causing the activation of NK effector functions. Moreover, it has been proved that activation of an NK receptor from differentially educated NK cells, for example by carrying more inhibitory receptors such as NKG2A or KIR3DL1 in addition to KIR2DL, showed enhanced NK cell activity [83]. Therefore, modulation of HLA expression and changes in the peptide-HLA immunocomplex repertoire may influence the functional activity of NK cells, resulting in a complex relationship between tumor microenvironment, difference in NK cell formation and expression of activating NK receptors [62]. In addition, the expression of gene encoding for immune checkpoint proteins, such as KLRD1/HLA-E, PD-L1/CD274/PD-CD1 and CTLA4/CD86, may in turn influence NK cell activation such as cytolysis, antibody-dependent cellular cytotoxicity (ADCC) and cytokine release, their survival, and the abundance and inhibition of NK cells in patients with HCC. In the end, as for the increase in activated CD8+ T cells, increased number of NK cells was found to prolong both relapse-free and overall survival of HCC patients [62].
The main strategies employed by viruses to evade NK cells are the reduced expression of ligands for activating receptors or the increased levels of inhibitory ligands. After viral infection, a variety of stress-induced molecules are expressed on the surface of infected cells, which can be recognized by activating receptors on NK cell receptors; the expression of HLA-C, a molecule able to interacts with the inhibitory KIR2DL receptor, is among the molecules most used by a virus to supply a reduction of NK functions. As different combinations of HLA ligands and KIR receptors in HCC have been shown to produce a different responses to treatment with anti-PD-1 immunotherapies in patients with virus-related HCC compared to non-alcoholic steatohepatitis (NASH) or controls, data had supported this concept [57].
More recently, a distinct population of NK-like hepatic CD8+ T-cells, characterized by a CD56 hi CD161−CD8+ phenotype and activated by a NKG2C/HLA-E complex independent from TCR, had been reported expanded in the liver sinusoids of the liver, and showed a strong immune response [98].
5. Non-Classical NKG2A HLA-E as a Novel Potential Checkpoint Targeted Therapy for HCC
Researchers have recently turned their attention to immunotherapy, as chemotherapy and radiotherapy have shown little survival benefit for HCC patients. Previous studies have confirmed that levels of tumor-infiltrating CD8+ T leukocytes are associated with better prognosis for both overall survival and disease-free survival in HCC patients, but most are inhibited by receptor-ligand interactions [172]. The most-studied targets of immune checkpoint inhibitors (ICIs) are the programmed cell death receptor (PD-1) on T cells and its ligands PD-L1 and PD-L2 on tumor cells. However, only 15–20% of patients have shown benefit from anti-PD-L1 monoclonal antibodies, which inhibit interactions with PD-1 and restore the role of T cells in the TME [173,174]. In addition, due to their toxicity, biologic drugs should be used with caution, especially in liver transplant patients [175].
Indeed, combination therapies, including PD-1/PD-L1 ICIs, have shown promise and several trials are ongoing in advanced or unresectable HCC. Among these, a combination of anti-PD1 with SP94-PB-sorafenib-Cy5.5 nanoparticles, a tyrosine kinase inhibitor (TKI), plus near-infrared therapy has shown suppression of distant metastases [176] and, in patients with early HCC recurrence after radical resection, TKI plus PD-1 therapy had shown survival benefit [177]. Data from IMbrave150, a phase III first-line trial in unresectable HCC, showed that after a median follow-up of 15.6 months, atezolizumab (anti-PD-L1) in combination with bevacizumab (anti-angiogenetic VEGF drug) reduced the risk of OS by 34%, with a median OS of 19.2 months, compared to 13.4 months for the TKI sorafenib. The next study, the HYMALAYA trial, approved in 2022, combined two different ICIs, the durvalumab [PD-1/CD80(B7.1), PD-L1 ICI] and tremelimumab (CTLA-4 ICI), in comparison to the sorafenib. The results of the study showed an OS of 16.4 months versus 13.8 months (p = 0.0035) after a median follow-up of 33.2 months, with a notable durable response. For advanced HCC, immunotherapy has now become the standard of care. A comprehensive characterization of the molecular patterns associated with response and resistance in patients with advanced HCC treated with anti-PD1 has recently been reported, and therefore has the potential to maximize the efficiency of anti-PD1 application [178]. However, phase III trials are needed to assess the ultimate value of this signature.
In addition to CD8+ T-cell inhibition, lower levels of NK cell infiltration have been found in HCC and more often in recurrent tumors in comparison to adjacent normal tissues [91]. Specific expression levels of NK cell immune checkpoint gene signatures highlighted a high correlation with the CD94/NKG2-HLA-E system as the probable cause of immune evasion in HCC with higher infiltrated NK cells, which are characterized by TGF-beta and Wnt interaction signature, in the HCC tissue. In addition, CD94 and KIR2DL4 (activation receptor for HLA-G) were highly expressed in patients responsive to targeted therapy with sorafenib, a kinase inhibitor.
Although liver transplantation and hepatectomy remain successful therapeutic strategies for patients with recurrent HCC, cytokine-induced killing (CIK) cell-based immunotherapy using ex vivo expanded T lymphocytes expressing the T-cell markers CD3+ or CD3−CD56+ have gained popularity as a promising new adjuvant therapy approach, targeting both exhausted T cells and NK cells to reduce HCC recurrence rates [179,180].
Several clinical trials have shown that CIK cell-based immunotherapy increases RFS in HCC patients who have undergone surgical resection (reviewed on [181]). The 18-month recurrence rate after adjuvant CIK therapy was 15.6%, compared to 40.0% in the control group receiving ablation therapy alone [182]. The rationale is that the irreversible cellular injury caused by ablation (RFA or cryoablation) allows an inflammatory TME with release of tumor neoantigens. However, the benefit of neoantigens in priming the immune response against tumor cells is hindered by the intrinsic inhibition of tumor cells. Therefore, the combination of ablation and CIK may synergize with the benefit of ablation.
Recently, the inhibitory NKG2A/HLA-E receptor has received attention as a new type of immune checkpoint blocker, and has also been proposed to disrupt the escape of circulating tumor cells from solid tumors [183]. In addition, it has been shown that HLA-G is also a ligand for the CD94/NKG2A receptor, but with a different affinity for the different HLA-G variants [120]. NKG2A blockade as a standalone therapy has appeared poorly effective in mouse tumor models; however, in the presence of activated T cells, for example, induced by PD-1/PD-L1 blockade, it exerts strongly enhanced efficacy. Clinical trials showed the safety of the humanized NKG2A-blocking antibody, monalizumab, and the first results of the phase II study had proved encouraging durable response rates in gynecologic cancers and head and neck carcinoma, including regressions of target lesions in recurrent disease. An increased effect for monalizumab has been shown by an enhanced ADCC function of NK cell and CTL cell effector, indicating that NKG2A-blocking directly impacts on NK cell effector functions [184,185].
Granulin-epithelin precursor (GEP), a hepatic oncofetal protein, was found to up-regulate NKG2A and HLA-E in the HCC microenvironment to enhance HCC cell invasion and resistance to human NK cell cytotoxicity, while downregulated MICA (MHC class I chain-related molecule A), the ligand for the stimulatory receptor NKG2D [186], a blockade of GEP by monoclonal antibody was demonstrated able to sensitized HCC cells to NK cytotoxicity. It was also shown that, as liver cancer progresses, the beneficial effect of stimulatory NKG2D immune responses can be also inhibited by increased expression of inhibitory receptors influenced by tumor-related metabolic disruption (tumor glycolysis and oxidative metabolism) and by tissue/cell mechanical properties, such as pathological conditions that drastically alter tissue stiffness, such as HCC [186,187,188].
In recent years, adoptive cell therapy has become one of the pillars in the treatment of cancer. One of the most important branches of this therapeutic approach, chimeric antigen receptor (CAR-T) therapy, has shown remarkable efficacy in the treatment of hematological cancers, solid tumor derived cell lines and in patients with glioblastoma but with an attention to severe toxicity [189,190].
Based on the observation that TGF-β increases the expression of the inhibitor NKG2A in HCC, a CAR NK cell engineered to convert the TGF-β-induced suppressive signal into an activating signal was developed [88,90]. Interestingly, these cells were better attracted to TGF-β-expressing tumor cells, inhibited the differentiation of human naïve CD4+ T cells into Treg cells, and could inhibit tumor growth in vivo in a xenograft model of HCC. Several clinical trials (NCT02906397, NCT02947165, NCT02423343, NCT02178358NKGD2) of monoclonal antibodies or CAR-T cells for TG-B blockade in HCC have been completed or are currently ongoing.
NKG2D-based CAR-T and CAR-NK cells have also been developed. In preclinical studies, they proved potent cytotoxicity against NKG2DL+ cell lines in vitro, therapeutic activity against NKG2DL+ cell xenografts in vivo and were shown to be safe. Two phase I trials were ongoing in patients with relapsed/refractory NKG2DL+ solid tumors, with a focus on NKG2DL+ HCC. [NCT05131763; NCT04550663].
The development of new therapeutic approaches for HCC through a targeted blockade of the NKG2A/HLA-E interaction of Vδ2pos T cells is of particular interest today. NKG2A has been shown to play a role in the development of NK cells. Indeed, NKG2A/HLA-E interaction makes NK cells educated and hyperreactive, increasing their effector functions in response to stimulatory activation. Conversely, NK cells that fail to interact because they lack NKG2A are poorly educated and unresponsive. However, educated NK cells are inhibited by the aberrant expression of HLA-E on HCC cells, resulting in an inhibitory function on Vδ2pos T cells, which are also unable to perform ADCC and escape immunological surveillance by the tumor. In preclinical studies, blocking the NKG2A receptor with a targeted antibody has been shown to restore the capacity of Vδ2pos T cells even against HLA-E+ tumor cells. It is thought that a sizable number of NKG2A+ Vδ2 T cells may improve overall survival in HCC when the level of HLA-E is reduced, or similar to, normal tissue. Based on these studies and the evidence that educated NKG2A+ Vδ2 T cells are more responsive, the prognosis of HCC may be improved by the development of therapies based on NKG2A-HLA-E immune checkpoint inhibition [191,192]. Most studies are ongoing, but currently only in preclinical models and in different cancer types, e.g., [193,194,195,196]. However, this new immune approach, NKG2A/HLA-E ICI, appears promising for patients with HCC, and the biological evidence obtained so far in vitro is encouraging.
6. Conclusions
This review reports on the increasing understanding of the key role of non-classical HLA-1b molecules in HCC pathogenesis and prognosis. Based on the current data, clinical applications of HLA-Ib in HCC risk assessment have been proposed.
Today’s findings identify NKG2A/HLA-E as an immune-checkpoint inhibitor and it seems the most promising target for new therapeutic approaches involving the HLA-Ib antigens to improve NK and, in particular, Vδ2 T cell function. Promising preclinical results have been obtained by combining NKG2A inhibitors with existing ICI therapies such as PD-1/PD-L1, multi-kinase inhibitors or anti-angiogenics. A NKG2D-based CAR-T cell therapy has been developed for the treatment of relapsed or refractory NKG2DL+ solid tumors [197] and a phase I clinical trial focus on HCC is ongoing [NCT04550663; NCT05131763].
The information on HLA-G and, in particular, HLA-F and FAT10, is too recent to be translated into therapeutic approaches at present. However, it would not be surprising if this were to happen in the near future, given the important information that has recently been reported on tumors and infections involving these molecules.
However, due to the limited number of studies available on the association between more recent HLA-Ib discoveries, compared to classical HLA class I and clinical outcome in patients with HCC, further research is needed to be more conclusive on these issues.
Taken together, the evidence described demonstrates that NK cells are dysfunctional in HCC, that their revitalization appears to be feasible, and these findings are driving the development of novel ICIs aimed at harnessing NK cell-mediated effector functions.
Author Contributions
Conceptualization, V.D.R.; writing—original draft preparation, V.D.R.; writing—review and editing, M.L.T., V.R., M.P. and A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Italian Ministry of Health (Ricerca Corrente) [no grant number provided].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data was in the text.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Samant, H.; Amiri, H.S.; Zibari, G.B. Addressing the worldwide hepatocellular carcinoma: Epidemiology, prevention and management. J. Gastrointest. Oncol. 2021, 12, S361–S373. [Google Scholar] [CrossRef] [PubMed]
- Gordan, J.D.; Kennedy, E.B.; Abou-Alfa, G.K.; Beg, M.S.; Brower, S.T.; Gade, T.P.; Goff, L.; Gupta, S.; Guy, J.; Harris, W.P.; et al. Systemic Therapy for Advanced Hepatocellular Carcinoma: ASCO Guideline. J. Clin. Oncol. 2020, 38, 4317–4345. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, A.; De Re, V.; Steffan, A.; Ravaioli, M.; Miolo, G.; Leone, P.; Racanelli, V.; Uzzau, A.; Baccarani, U.; Cescon, M. Carcinogenesis and Metastasis in Liver: Cell Physiological Basis. Cancers 2019, 11, 1731. [Google Scholar] [CrossRef] [PubMed]
- Kubes, P.; Jenne, C. Immune Responses in the Liver. Annu. Rev. Immunol. 2018, 36, 247–277. [Google Scholar] [CrossRef] [PubMed]
- Yau, T.; Park, J.-W.; Finn, R.S.; Cheng, A.-L.; Mathurin, P.; Edeline, J.; Kudo, M.; Harding, J.J.; Merle, P.; Rosmorduc, O.; et al. Nivolumab versus sorafenib in advanced hepatocellular carcinoma (CheckMate 459): A randomised, multicentre, open-label, phase 3 trial. Lancet Oncol. 2021, 23, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.S.; Villeret, F.; Decaens, T.; Merle, P.; Nahon, P. Immunotherapy in hepatocellular carcinoma: How does underlying liver disease influence therapeutic strategy and outcomes? Liver Int. 2023, 43, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; He, Y.; Luo, N.; Patel, S.J.; Han, Y.; Gao, R.; Modak, M.; Carotta, S.; Haslinger, C.; Kind, D.; et al. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell 2019, 179, 829–845.e20. [Google Scholar] [CrossRef]
- Tian, Z.; Hou, X.; Liu, W.; Han, Z.; Wei, L. Macrophages and hepatocellular carcinoma. Cell Biosci. 2019, 9, 79. [Google Scholar] [CrossRef]
- Ding, T.; Xu, J.; Wang, F.; Shi, M.; Zhang, Y.; Li, S.-P.; Zheng, L. High tumor-infiltrating macrophage density predicts poor prognosis in patients with primary hepatocellular carcinoma after resection. Hum. Pathol. 2009, 40, 381–389. [Google Scholar] [CrossRef]
- Moeini, A.; Torrecilla, S.; Tovar, V.; Montironi, C.; Andreu-Oller, C.; Peix, J.; Higuera, M.; Pfister, D.; Ramadori, P.; Pinyol, R.; et al. An Immune Gene Expression Signature Associated With Development of Human Hepatocellular Carcinoma Identifies Mice That Respond to Chemopreventive Agents. Gastroenterology 2019, 157, 1383–1397.e11. [Google Scholar] [CrossRef]
- Lim, C.J.; Lee, Y.H.; Pan, L.; Lai, L.; Chua, C.; Wasser, M.; Lim, T.K.H.; Yeong, J.; Toh, H.C.; Lee, S.Y.; et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus-related hepatocellular carcinoma. Gut 2019, 68, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Pishesha, N.; Harmand, T.J.; Ploegh, H.L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 2022, 22, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Urban, T.J.; Nicoletti, P.; Chalasani, N.; Serrano, J.; Stolz, A.; Daly, A.K.; Aithal, G.P.; Dillon, J.; Navarro, V.; Odin, J.; et al. Minocycline hepatotoxicity: Clinical characterization and identification of HLA-B∗35:02 as a risk factor. J. Hepatol. 2017, 67, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, M.; Hagihara, K.; Abe, K.; Ando, O.; Hirayama, N. Interaction of Nevirapine with the Peptide Binding Groove of HLA-DRB1*01:01 and Its Effect on the Conformation of HLA-Peptide Complex. Int. J. Mol. Sci. 2018, 19, 1660. [Google Scholar] [CrossRef] [PubMed]
- Monshi, M.M.; Faulkner, L.; Gibson, A.; Jenkins, R.E.; Farrell, J.; Earnshaw, C.J.; Alfirevic, A.; Cederbrant, K.; Daly, A.K.; French, N.; et al. Human leukocyte antigen (HLA)-B*57:01-restricted activation of drug-specific T cells provides the immunological basis for flucloxacillin-induced liver injury. Hepatology 2013, 57, 727–739. [Google Scholar] [CrossRef]
- Jee, A.; Sernoskie, S.C.; Uetrecht, J. Idiosyncratic Drug-Induced Liver Injury: Mechanistic and Clinical Challenges. Int. J. Mol. Sci. 2021, 22, 2954. [Google Scholar] [CrossRef]
- Daly, A.K.; Day, C.P. Genetic association studies in drug-induced liver injury. Drug Metab. Rev. 2012, 44, 116–126. [Google Scholar] [CrossRef]
- Ebbo, M.; Crinier, A.; Vély, F.; Vivier, E. Innate lymphoid cells: Major players in inflammatory diseases. Nat. Rev. Immunol. 2017, 17, 665–678. [Google Scholar] [CrossRef]
- Umemura, T.; Joshita, S.; Saito, H.; Wakabayashi, S.-I.; Kobayashi, H.; Yamashita, Y.; Sugiura, A.; Yamazaki, T.; Ota, M. Investigation of the Effect of KIR–HLA Pairs on Hepatocellular Carcinoma in Hepatitis C Virus Cirrhotic Patients. Cancers 2021, 13, 3267. [Google Scholar] [CrossRef]
- Joshita, S.; Ota, M.; Kobayashi, H.; Wakabayashi, S.-I.; Yamashita, Y.; Sugiura, A.; Yamazaki, T.; Tanaka, E.; Umemura, T. Association analysis of KIR/HLA genotype with liver cirrhosis, hepatocellular carcinoma, and NUC freedom in chronic hepatitis B patients. Sci. Rep. 2021, 11, 21424. [Google Scholar] [CrossRef]
- Khakoo, S.I.; Thio, C.L.; Martin, M.P.; Brooks, C.R.; Gao, X.; Astemborski, J.; Cheng, J.; Goedert, J.J.; Vlahov, D.; Hilgartner, M.; et al. HLA and NK Cell Inhibitory Receptor Genes in Resolving Hepatitis C Virus Infection. Science 2004, 305, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Hydes, T.J.; Moesker, B.; Traherne, J.A.; Ashraf, S.; Alexander, G.J.; Dimitrov, B.D.; Woelk, C.H.; Trowsdale, J.; Khakoo, S.I. The interaction of genetic determinants in the outcome of HCV infection: Evidence for discrete immunological pathways. Tissue Antigens 2015, 86, 267–275. [Google Scholar] [CrossRef] [PubMed]
- López-Vázquez, A.; Rodrigo, L.; Martínez-Borra, J.; Pérez, R.; Rodríguez, M.; Fdez-Morera, J.L.; Fuentes, D.; Rodríguez-Rodero, S.; González, S.; López-Larrea, C. Protective Effect of the HLA-Bw4I80 Epitope and the Killer Cell Immunoglobulin-Like Receptor 3DS1 Gene against the Development of Hepatocellular Carcinoma in Patients with Hepatitis C Virus Infection. J. Infect. Dis. 2005, 192, 162–165. [Google Scholar] [CrossRef] [PubMed]
- De Re, V.; Caggiari, L.; De Zorzi, M.; Repetto, O.; Zignego, A.L.; Izzo, F.; Tornesello, M.L.; Buonaguro, F.M.; Mangia, A.; Sansonno, D.; et al. Genetic Diversity of the KIR/HLA System and Susceptibility to Hepatitis C Virus-Related Diseases. PLoS ONE 2015, 10, e0117420. [Google Scholar] [CrossRef] [PubMed]
- Di Bona, D.; Aiello, A.; Colomba, C.; Bilancia, M.; Accardi, G.; Rubino, R.; Giannitrapani, L.; Tuttolomondo, A.; Cascio, A.; Caiaffa, M.F.; et al. KIR2DL3 and the KIR ligand groups HLA-A-Bw4 and HLA-C2 predict the outcome of hepatitis B virus infection. J. Viral Hepat. 2017, 24, 768–775. [Google Scholar] [CrossRef]
- Fan, Q.R.; Long, E.O.; Wiley, D.C. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1–HLA-Cw4 complex. Nat. Immunol. 2001, 2, 452–460. [Google Scholar] [CrossRef]
- Husain, Z.; Levitan, E.; Larsen, C.; Mirza, N.M.; Younes, S.; Yunis, E.J.; Alper, C.A.; Dubey, D.P. HLA-Cw7 zygosity affects the size of a subset of CD158b+ natural killer cells. J. Clin. Immunol. 2002, 22, 28–36. [Google Scholar] [CrossRef]
- Boudreau, J.E.; Hsu, K.C. Natural killer cell education in human health and disease. Curr. Opin. Immunol. 2018, 50, 102–111. [Google Scholar] [CrossRef]
- Hansasuta, P.; Dong, T.; Thananchai, H.; Weekes, M.; Willberg, C.; Aldemir, H.; Rowland-Jones, S.; Braud, V.M. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur. J. Immunol. 2004, 34, 1673–1679. [Google Scholar] [CrossRef]
- Bowness, P.; Ridley, A.; Shaw, J.; Chan, A.T.; Wong-Baeza, I.; Fleming, M.; Cummings, F.; McMichael, A.; Kollnberger, S. Th17 Cells Expressing KIR3DL2+ and Responsive to HLA-B27 Homodimers Are Increased in Ankylosing Spondylitis. J. Immunol. 2011, 186, 2672–2680. [Google Scholar] [CrossRef]
- Mouchet, N.; Vu, N.; Turlin, B.; Rioux-Leclercq, N.; Jouneau, S.; Samson, M.; Amiot, L. HLA-G Is Widely Expressed by Mast Cells in Regions of Organ Fibrosis in the Liver, Lung and Kidney. Int. J. Mol. Sci. 2021, 22, 12490. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Li, N.; Zhu, Q.; Li, Z.; Zhang, G.; Chen, J.; Lv, Y.; Wang, Y.; Liu, Z.; Hao, C. Association of serum soluble human leukocyte antigen-G levels with chronic hepatitis B virus infection. Clin. Exp. Med. 2014, 14, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, S.; Jia, S.; Zhu, Z.; Gao, X.; Dong, D.; Gao, Y.; Rizzo, R.; Bergamini, G.; Bortolotti, D.; et al. Association of HLA-G3′ UTR 14-bp Insertion/Deletion Polymorphism with Hepatocellular Carcinoma Susceptibility in a Chinese Population. DNA Cell Biol. 2011, 30, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Amiot, L.; Vu, N.; Samson, M. Biology of the immunomodulatory molecule HLA-G in human liver diseases. J. Hepatol. 2015, 62, 1430–1437. [Google Scholar] [CrossRef]
- Amiot, L.; Vu, N.; Rauch, M.; L’helgoualc’h, A.; Chalmel, F.; Gascan, H.; Turlin, B.; Guyader, D.; Samson, M. Expression of HLA-G by mast cells is associated with hepatitis C virus-induced liver fibrosis. J. Hepatol. 2014, 60, 245–252. [Google Scholar] [CrossRef]
- Park, Y.; Lim, H.S.; Kim, Y.S.; Hong, D.J.; Kim, H.S. Soluble human leukocyte antigen-G expression in hepatitis B virus infection and hepatocellular carcinoma. Tissue Antigens 2012, 79, 97–103. [Google Scholar] [CrossRef]
- Lin, A.; Chen, H.-X.; Zhu, C.-C.; Zhang, X.; Xu, H.-H.; Zhang, J.-G.; Wang, Q.; Zhou, W.-J.; Yan, W.-H. Aberrant human leucocyte antigen-G expression and its clinical relevance in hepatocellular carcinoma. J. Cell. Mol. Med. 2010, 14, 2162–2171. [Google Scholar] [CrossRef]
- Gao, G.F.; Willcox, B.E.; Wyer, J.R.; Boulter, J.M.; O’Callaghan, C.A.; Maenaka, K.; Stuart, D.I.; Jones, E.Y.; Van Der Merwe, P.A.; Bell, J.I.; et al. Classical and Nonclassical Class I Major Histocompatibility Complex Molecules Exhibit Subtle Conformational Differences That Affect Binding to CD8αα*. J. Biol. Chem. 2000, 275, 15232–15238. [Google Scholar] [CrossRef]
- Zhuang, B.; Shang, J.; Yao, Y. HLA-G: An Important Mediator of Maternal-Fetal Immune-Tolerance. Front. Immunol. 2021, 12, 744324. [Google Scholar] [CrossRef]
- Sanders, S.K.; Giblin, P.A.; Kavathas, P.B. Cell-cell adhesion mediated by CD8 and human histocompatibility leukocyte antigen G, a nonclassical major histocompatibility complex class 1 molecule on cytotrophoblasts. J. Exp. Med. 1991, 174, 737–740. [Google Scholar] [CrossRef]
- Attia, J.V.D.; Dessens, C.E.; van de Water, R.; Houvast, R.D.; Kuppen, P.J.K.; Krijgsman, D. The Molecular and Functional Characteristics of HLA-G and the Interaction with Its Receptors: Where to Intervene for Cancer Immunotherapy? Int. J. Mol. Sci. 2020, 21, 8678. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Shen, Y.; Xia, M.; Xu, L.; Pan, N.; Yin, Y.; Miao, F.; Shen, C.; Xie, W.; Zhang, J. Expression of the nonclassical HLA class I and MICA/B molecules in human hepatocellular carcinoma. Neoplasma 2011, 58, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Luo, W.; Zhu, Q.; Huang, H.; Peng, H.; Liu, R.; Xie, M.; Li, S.; Li, M.; Hu, X.; et al. Tumor-Derived Soluble MICA Obstructs the NKG2D Pathway to Restrain NK Cytotoxicity. Aging Dis. 2020, 11, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Cariani, E.; Pilli, M.; Zerbini, A.; Rota, C.; Olivani, A.; Zanelli, P.; Zanetti, A.; Trenti, T.; Ferrari, C.; Missale, G. HLA and Killer Immunoglobulin-like Receptor Genes as Outcome Predictors of Hepatitis C Virus–Related Hepatocellular Carcinoma. Clin. Cancer Res. 2013, 19, 5465–5473. [Google Scholar] [CrossRef] [PubMed]
- Sim, M.J.W.; Rajagopalan, S.; Altmann, D.M.; Boyton, R.J.; Sun, P.D.; Long, E.O. Human NK cell receptor KIR2DS4 detects a conserved bacterial epitope presented by HLA-C. Proc. Natl. Acad. Sci. USA 2019, 116, 12964–12973. [Google Scholar] [CrossRef]
- Graef, T.; Moesta, A.K.; Norman, P.J.; Abi-Rached, L.; Vago, L.; Aguilar, A.M.O.; Gleimer, M.; Hammond, J.A.; Guethlein, L.A.; Bushnell, D.A.; et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J. Exp. Med. 2009, 206, 2557–2572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Pan, L.; Chen, L.; Feng, X.; Zhou, L.; Zheng, S. Non-classical MHC-Ι genes in chronic hepatitis B and hepatocellular carcinoma. Immunogenetics 2012, 64, 251–258. [Google Scholar] [CrossRef]
- Brandt, C.S.; Baratin, M.; Yi, E.C.; Kennedy, J.; Gao, Z.; Fox, B.; Haldeman, B.; Ostrander, C.D.; Kaifu, T.; Chabannon, C.; et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 2009, 206, 1495–1503. [Google Scholar] [CrossRef]
- Cheung, P.F.Y.; Yip, C.W.; Wong, N.C.L.; Fong, D.Y.T.; Ng, L.W.C.; Wan, A.M.Y.; Wong, C.K.; Cheung, T.T.; Ng, I.O.L.; Poon, R.T.P.; et al. Granulin-Epithelin Precursor Renders Hepatocellular Carcinoma Cells Resistant to Natural Killer Cytotoxicity. Cancer Immunol. Res. 2014, 2, 1209–1219. [Google Scholar] [CrossRef]
- Yan, J.; Lei, J.; Chen, L.; Deng, H.; Dong, D.; Jin, T.; Liu, X.; Yuan, R.; Qiu, Y.; Ge, J.; et al. Human Leukocyte Antigen F Locus Adjacent Transcript 10 Overexpression Disturbs WISP1 Protein and mRNA Expression to Promote Hepatocellular Carcinoma Progression. Hepatology 2018, 68, 2268–2284. [Google Scholar] [CrossRef]
- Trinchieri, G. Interleukin-12: A cytokine produced by antigen-presenting cells with immunoregulatory functions in the generation of T-helper cells type 1 and cytotoxic lymphocytes. Blood 1994, 84, 4008–4027. [Google Scholar] [CrossRef] [PubMed]
- Schulz, E.G.; Mariani, L.; Radbruch, A.; Höfer, T. Sequential Polarization and Imprinting of Type 1 T Helper Lymphocytes by Interferon-γ and Interleukin-12. Immunity 2009, 30, 673–683. [Google Scholar] [CrossRef]
- Chen, Y.-C.; He, X.-L.; Qi, L.; Shi, W.; Yuan, L.-W.; Huang, M.-Y.; Xu, Y.-L.; Chen, X.; Gu, L.; Zhang, L.-L.; et al. Myricetin inhibits interferon-γ-induced PD-L1 and IDO1 expression in lung cancer cells. Biochem. Pharmacol. 2022, 197, 114940. [Google Scholar] [CrossRef]
- Garcia-Diaz, A.; Shin, D.S.; Moreno, B.H.; Saco, J.; Escuin-Ordinas, H.; Rodriguez, G.A.; Zaretsky, J.M.; Sun, L.; Hugo, W.; Wang, X.; et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017, 19, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Jaw, J.J.; Stutzman, N.C.; Zou, Z.; Sun, P.D. Natural killer cell-produced IFN-γ and TNF-α induce target cell cytolysis through up-regulation of ICAM-1. J. Leukoc. Biol. 2011, 91, 299–309. [Google Scholar] [CrossRef]
- Amadei, B.; Urbani, S.; Cazaly, A.; Fisicaro, P.; Zerbini, A.; Ahmed, P.; Missale, G.; Ferrari, C.; Khakoo, S.I. Activation of Natural Killer Cells During Acute Infection With Hepatitis C Virus. Gastroenterology 2010, 138, 1536–1545. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, L.; Li, W.; Chen, B.; Liu, Y.; Wang, H.; Zhao, S.; Ye, L.; He, Y.; Zhou, C. Killer immunoglobulin-like receptors/human leukocyte antigen class-I, a crucial immune pathway in cancer. Ann. Transl. Med. 2020, 8, 244. [Google Scholar] [CrossRef] [PubMed]
- Hiraoka, N.; Ino, Y.; Hori, S.; Yamazaki-Itoh, R.; Naito, C.; Shimasaki, M.; Esaki, M.; Nara, S.; Kishi, Y.; Shimada, K.; et al. Expression of classical human leukocyte antigen class I antigens, HLA-E and HLA-G, is adversely prognostic in pancreatic cancer patients. Cancer Sci. 2020, 111, 3057–3070. [Google Scholar] [CrossRef]
- Wei, Y.; Bingyu, W.; Lei, Y.; Xingxing, Y. The antifibrotic role of natural killer cells in liver fibrosis. Exp. Biol. Med. 2022, 247, 1235–1243. [Google Scholar] [CrossRef]
- Wijaya, R.S.; Read, S.A.; Schibeci, S.; Eslam, M.; Azardaryany, M.K.; El-Khobar, K.; van der Poorten, D.; Lin, R.; Yuen, L.; Lam, V.; et al. KLRG1+ natural killer cells exert a novel antifibrotic function in chronic hepatitis B. J. Hepatol. 2019, 71, 252–264. [Google Scholar] [CrossRef]
- Rodgers, J.R.; Cook, R.G. MHC class Ib molecules bridge innate and acquired immunity. Nat. Rev. Immunol. 2005, 5, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Rascle, P.; Woolley, G.; Jost, S.; Manickam, C.; Reeves, R.K. NK cell education: Physiological and pathological influences. Front. Immunol. 2023, 14, 1087155. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, L.L.; Hviid, T.V.F. HLA Class Ib-receptor interactions during embryo implantation and early pregnancy. Hum. Reprod. Updat. 2022, 28, 435–454. [Google Scholar] [CrossRef]
- Rouas-Freiss, N.; Gonçalves, R.M.-B.; Menier, C.; Dausset, J.; Carosella, E.D. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc. Natl. Acad. Sci. USA 1997, 94, 11520–11525. [Google Scholar] [CrossRef]
- Lu, L.; Ikizawa, K.; Hu, D.; Werneck, M.B.; Wucherpfennig, K.W.; Cantor, H. Regulation of Activated CD4+ T Cells by NK Cells via the Qa-1–NKG2A Inhibitory Pathway. Immunity 2007, 26, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Soloski, M.J.; Decloux, A.; Aldrich, C.J.; Forman, J. Structural and Functional Characteristics of the Class Ib Molecule, Qa-1. Immunol. Rev. 1995, 147, 67–89. [Google Scholar] [CrossRef]
- Borst, L.; van der Burg, S.H.; van Hall, T. The NKG2A–HLA-E Axis as a Novel Checkpoint in the Tumor Microenvironment. Clin. Cancer Res. 2020, 26, 5549–5556. [Google Scholar] [CrossRef]
- Hosseini, E.; Schwarer, A.P.; Ghasemzadeh, M. Do human leukocyte antigen E polymorphisms influence graft-versus-leukemia after allogeneic hematopoietic stem cell transplantation? Exp. Hematol. 2015, 43, 149–157. [Google Scholar] [CrossRef]
- Michaëlsson, J.; De Matos, C.T.; Achour, A.; Lanier, L.L.; Kärre, K.; Söderström, K. A Signal Peptide Derived from hsp60 Binds HLA-E and Interferes with CD94/NKG2A Recognition. J. Exp. Med. 2002, 196, 1403–1414. [Google Scholar] [CrossRef]
- Fisher, J.G.; Doyle, A.D.P.; Graham, L.V.; Khakoo, S.I.; Blunt, M.D. Disruption of the NKG2A:HLA-E Immune Checkpoint Axis to Enhance NK Cell Activation against Cancer. Vaccines 2022, 10, 1993. [Google Scholar] [CrossRef]
- Nguyen, S.; Beziat, V.; Dhedin, N.; Kuentz, M.; Vernant, J.P.; Debre, P.; Vieillard, V. HLA-E upregulation on IFN-γ-activated AML blasts impairs CD94/NKG2A-dependent NK cytolysis after haplo-mismatched hematopoietic SCT. Bone Marrow Transplant. 2008, 43, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.G.; Crispim, J.C.O.; Miranda, F.A.; Hassumi, M.K.; de Mello, J.M.Y.; Simões, R.T.; Souto, F.; Soares, E.G.; Donadi, E.A.; Soares, C.P. Expression of the Nonclassical HLA-G and HLA-E Molecules in Laryngeal Lesions as Biomarkers of Tumor Invasiveness. Histol. Histopathol. 2011, 26, 1487–1497. [Google Scholar] [CrossRef] [PubMed]
- Levy, E.M.; Bianchini, M.; Von Euw, E.M.; Barrio, M.M.; Bravo, A.I.; Furman, D.; Domenichini, E.; Macagno, C.; Pinsky, V.; Zucchini, C.; et al. Human leukocyte antigen-E protein is overexpressed in primary human colorectal cancer. Int. J. Oncol. 2008, 32, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Andersson, E.; Poschke, I.; Villabona, L.; Carlson, J.W.; Lundqvist, A.; Kiessling, R.; Seliger, B.; Masucci, G.V. Non-classical HLA-class I expression in serous ovarian carcinoma: Correlation with the HLA-genotype, tumor infiltrating immune cells and prognosis. Oncoimmunology 2016, 5, e1052213. [Google Scholar] [CrossRef] [PubMed]
- Gooden, M.; Lampen, M.; Jordanova, E.S.; Leffers, N.; Trimbos, J.B.; van der Burg, S.H.; Nijman, H.; van Hall, T. HLA-E expression by gynecological cancers restrains tumor-infiltrating CD8+ T lymphocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 10656–10661. [Google Scholar] [CrossRef] [PubMed]
- de Kruijf, E.M.; Sajet, A.; van Nes, J.G.H.; Natanov, R.; Putter, H.; Smit, V.T.H.B.M.; Liefers, G.J.; van den Elsen, P.J.; van de Velde, C.J.H.; Kuppen, P.J.K. HLA-E and HLA-G Expression in Classical HLA Class I-Negative Tumors Is of Prognostic Value for Clinical Outcome of Early Breast Cancer Patients. J. Immunol. 2010, 185, 7452–7459. [Google Scholar] [CrossRef]
- Jinushi, M.; Takehara, T.; Tatsumi, T.; Kanto, T.; Miyagi, T.; Suzuki, T.; Kanazawa, Y.; Hiramatsu, N.; Hayashi, N. Negative Regulation of NK Cell Activities by Inhibitory Receptor CD94/NKG2A Leads to Altered NK Cell-Induced Modulation of Dendritic Cell Functions in Chronic Hepatitis C Virus Infection. J. Immunol. 2004, 173, 6072–6081. [Google Scholar] [CrossRef]
- Wang, X.-K.; Liao, X.-W.; Yang, C.-K.; Yu, T.-D.; Liu, Z.-Q.; Gong, Y.-Z.; Huang, K.-T.; Zeng, X.-M.; Han, C.-Y.; Zhu, G.-Z.; et al. Diagnostic and prognostic biomarkers of Human Leukocyte Antigen complex for hepatitis B virus-related hepatocellular carcinoma. J. Cancer 2019, 10, 5173–5190. [Google Scholar] [CrossRef]
- Sun, C.; Xu, J.; Huang, Q.; Huang, M.; Wen, H.; Zhang, C.; Wang, J.; Song, J.; Zheng, M.; Sun, H.; et al. High NKG2A expression contributes to NK cell exhaustion and predicts a poor prognosis of patients with liver cancer. Oncoimmunology 2017, 6, e1264562. [Google Scholar] [CrossRef]
- Jacquelot, N.; Seillet, C.; Souza-Fonseca-Guimaraes, F.; Sacher, A.G.; Belz, G.T.; Ohashi, P.S. Natural Killer Cells and Type 1 Innate Lymphoid Cells in Hepatocellular Carcinoma: Current Knowledge and Future Perspectives. Int. J. Mol. Sci. 2021, 22, 9044. [Google Scholar] [CrossRef]
- Borst, L.; Sluijter, M.; Sturm, G.; Charoentong, P.; Santegoets, S.J.; van Gulijk, M.; van Elsas, M.J.; Groeneveldt, C.; van Montfoort, N.; Finotello, F.; et al. NKG2A Is a Late Immune Checkpoint on CD8 T Cells and Marks Repeated Stimulation and Cell Division. Int. J. Cancer 2022, 150, 688–704. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Llano, M.; Carretero, M.; Ishitani, A.; Navarro, F.; López-Botet, M.; Geraghty, D.E. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA 1998, 95, 5199–5204. [Google Scholar] [CrossRef] [PubMed]
- Highton, A.J.; Diercks, B.-P.; Möckl, F.; Martrus, G.; Sauter, J.; Schmidt, A.H.; Bunders, M.J.; Körner, C.; Guse, A.H.; Altfeld, M. High Metabolic Function and Resilience of NKG2A-Educated NK Cells. Front. Immunol. 2020, 11, 559576. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Sun, L.; Liu, X.; Wang, X.; Yan, H.; Pu, Q.; Xie, Y.; Jiang, Y.; Du, J.; Yang, Z. The imbalance between NKG2A and NKG2D expression is involved in NK cell immunosuppression and tumor progression of patients with hepatitis B virus-related hepatocellular carcinoma. Hepatol. Res. 2023, 53, 417–431. [Google Scholar] [CrossRef]
- Tesi, B.; Schlums, H.; Cichocki, F.; Bryceson, Y.T. Epigenetic Regulation of Adaptive NK Cell Diversification. Trends Immunol. 2016, 37, 451–461. [Google Scholar] [CrossRef]
- Rölle, A.; Meyer, M.; Calderazzo, S.; Jäger, D.; Momburg, F. Distinct HLA-E Peptide Complexes Modify Antibody-Driven Effector Functions of Adaptive NK Cells. Cell Rep. 2018, 24, 1967–1976.e4. [Google Scholar] [CrossRef]
- Mantovani, S.; Oliviero, B.; Lombardi, A.; Varchetta, S.; Mele, D.; Sangiovanni, A.; Rossi, G.; Donadon, M.; Torzilli, G.; Soldani, C.; et al. Deficient Natural Killer Cell NKp30-Mediated Function and Altered NCR3 Splice Variants in Hepatocellular Carcinoma. Hepatology 2019, 69, 1165–1179. [Google Scholar] [CrossRef]
- Gonzalez-Sanchez, E.; Vaquero, J.; Férnandez-Barrena, M.G.; Lasarte, J.J.; Avila, M.A.; Sarobe, P.; Reig, M.; Calvo, M.; Fabregat, I. The TGF-β Pathway: A Pharmacological Target in Hepatocellular Carcinoma? Cancers 2021, 13, 3248. [Google Scholar] [CrossRef]
- Lassen, M.G.; Lukens, J.R.; Dolina, J.S.; Brown, M.G.; Hahn, Y.S. Intrahepatic IL-10 Maintains NKG2A+Ly49− Liver NK Cells in a Functionally Hyporesponsive State. J. Immunol. 2010, 184, 2693–2701. [Google Scholar] [CrossRef]
- Gunturi, A.; Berg, R.E.; Crossley, E.; Murray, S.; Forman, J. The role of TCR stimulation and TGF-Beta? in controlling the expression of CD94/NKG2A receptors on CD8 T?cells. Eur. J. Immunol. 2005, 35, 766–775. [Google Scholar] [CrossRef]
- Wu, M.; Mei, F.; Liu, W.; Jiang, J. Comprehensive characterization of tumor infiltrating natural killer cells and clinical significance in hepatocellular carcinoma based on gene expression profiles. Biomed. Pharmacother. 2020, 121, 109637. [Google Scholar] [CrossRef] [PubMed]
- Grant, E.J.; Nguyen, A.T.; Lobos, C.A.; Szeto, C.; Chatzileontiadou, D.S.M.; Gras, S. The Unconventional Role of HLA-E: The Road Less Traveled. Mol. Immunol. 2020, 120, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Goodlett, D.R.; Ishitani, A.; Marquardt, H.; Geraghty, D.E. HLA-E Surface Expression Depends on Binding of TAP-Dependent Peptides Derived from Certain HLA Class I Signal Sequences. J. Immunol. 1998, 160, 4951–4960. [Google Scholar] [CrossRef] [PubMed]
- Pump, W.C.; Kraemer, T.; Huyton, T.; Hò, G.-G.T.; Blasczyk, R.; Bade-Doeding, C. Between Innate and Adaptive Immune Responses: NKG2A, NKG2C, and CD8+ T Cell Recognition of HLA-E Restricted Self-Peptides Acquired in the Absence of HLA-Ia. Int. J. Mol. Sci. 2019, 20, 1454. [Google Scholar] [CrossRef] [PubMed]
- Braud, V.; Jones, E.Y.; McMichael, A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur. J. Immunol. 1997, 27, 1164–1169. [Google Scholar] [CrossRef]
- Jucaud, V.; Ravindranath, M.H.; Terasaki, P.I. Immunobiology of HLA Class-Ib Molecules in Transplantation. SOJ Immunol. 2015, 3, 1–15. [Google Scholar] [CrossRef]
- Hosseini, E.; Minagar, A.; Ghasemzadeh, M.; Arabkhazaeli, A.; Ghasemzadeh, A. HLA-E*01:01 + HLA-E*01:01 genotype confers less susceptibility to COVID-19, while HLA-E*01:03 + HLA-E*01:03 genotype is associated with more severe disease. Hum. Immunol. 2023, 84, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.-Y.; Rha, M.-S.; Choi, S.J.; Lee, H.S.; Han, J.W.; Nam, H.; Kim, D.-U.; Lee, J.G.; Kim, M.S.; Park, J.Y.; et al. Identification of a distinct NK-like hepatic T-cell population activated by NKG2C in a TCR-independent manner. J. Hepatol. 2022, 77, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Marín, R.; Ruiz-Cabello, F.; Pedrinaci, S.; Méndez, R.; Jiménez, P.; Geraghty, D.E.; Garrido, F. Analysis of HLA-E expression in human tumors. Immunogenetics 2003, 54, 767–775. [Google Scholar] [CrossRef]
- Cazzetta, V.; Bruni, E.; Terzoli, S.; Carenza, C.; Franzese, S.; Piazza, R.; Marzano, P.; Donadon, M.; Torzilli, G.; Cimino, M.; et al. NKG2A expression identifies a subset of human Vδ2 T cells exerting the highest antitumor effector functions. Cell Rep. 2021, 37, 109871. [Google Scholar] [CrossRef]
- Chang, K.-M.; Traum, D.; Park, J.-J.; Ho, S.; Ojiro, K.; Wong, D.K.; Wahed, A.S.; Terrault, N.A.; Khalili, M.; Sterling, R.K.; et al. Distinct phenotype and function of circulating Vδ1+ and Vδ2+ γδT-cells in acute and chronic hepatitis B. PLoS Pathog. 2019, 15, e1007715. [Google Scholar] [CrossRef] [PubMed]
- Kovats, S.; Main, E.K.; Librach, C.; Stubblebine, M.; Fisher, S.J.; DeMars, R. A Class I Antigen, HLA-G, Expressed in Human Trophoblasts. Science 1990, 248, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Fons, P.; Chabot, S.; Cartwright, J.E.; Lenfant, F.; L’Faqihi, F.; Giustiniani, J.; Herault, J.-P.; Gueguen, G.; Bono, F.; Savi, P.; et al. Soluble HLA-G1 inhibits angiogenesis through an apoptotic pathway and by direct binding to CD160 receptor expressed by endothelial cells. Blood 2006, 108, 2608–2615. [Google Scholar] [CrossRef] [PubMed]
- Schwich, E.; Hò, G.-G.T.; LeMaoult, J.; Bade-Döding, C.; Carosella, E.D.; Horn, P.A.; Rebmann, V. Soluble HLA-G and HLA-G Bearing Extracellular Vesicles Affect ILT-2 Positive and ILT-2 Negative CD8 T Cells Complementary. Front. Immunol. 2020, 11, 2046. [Google Scholar] [CrossRef]
- Dumont, C.; Jacquier, A.; Verine, J.; Noel, F.; Goujon, A.; Wu, C.-L.; Hung, T.-M.; Desgrandchamps, F.; Culine, S.; Carosella, E.D.; et al. CD8+PD-1–ILT2+ T Cells Are an Intratumoral Cytotoxic Population Selectively Inhibited by the Immune-Checkpoint HLA-G. Cancer Immunol. Res. 2019, 7, 1619–1632. [Google Scholar] [CrossRef]
- Rajagopalan, S.; Long, E.O. KIR2DL4 (CD158d): An activation receptor for HLA-G. Front. Immunol. 2012, 3, 258. [Google Scholar] [CrossRef]
- Shiroishi, M.; Tsumoto, K.; Amano, K.; Shirakihara, Y.; Colonna, M.; Braud, V.M.; Allan, D.S.J.; Makadzange, A.; Rowland-Jones, S.; Willcox, B.; et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl. Acad. Sci. USA 2003, 100, 8856–8861. [Google Scholar] [CrossRef]
- Adolf, I.C.; Almars, A.; Dharsee, N.; Mselle, T.; Akan, G.; Nguma, I.J.; Nateri, A.S.; Atalar, F. HLA-G and single nucleotide polymorphism (SNP) associations with cancer in African populations: Implications in personal medicine. Genes Dis. 2021, 9, 1220–1233. [Google Scholar] [CrossRef]
- Krijgsman, D.; Roelands, J.; Hendrickx, W.; Bedognetti, D.; Kuppen, P.J.K. HLA-G: A New Immune Checkpoint in Cancer? Int. J. Mol. Sci. 2020, 21, 4528. [Google Scholar] [CrossRef]
- Bian, X.; Si, Y.; Zhang, M.; Wei, R.; Yang, X.; Ren, H.; Zheng, G.; Wang, C.; Zhang, Y. Down-expression of miR-152 lead to impaired anti-tumor effect of NK via upregulation of HLA-G. Tumor Biol. 2015, 37, 3749–3756. [Google Scholar] [CrossRef]
- Cai, M.-Y.; Xu, Y.-F.; Qiu, S.-J.; Ju, M.-J.; Gao, Q.; Li, Y.-W.; Zhang, B.-H.; Zhou, J.; Fan, J. Human Leukocyte Antigen-G Protein Expression Is an Unfavorable Prognostic Predictor of Hepatocellular Carcinoma following Curative Resection. Clin. Cancer Res. 2009, 15, 4686–4693. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ye, Z.; Meng, X.-Q.; Zheng, S.-S. Expression of HLA-G in patients with hepatocellular carcinoma. Hepatobiliary Pancreat. Dis. Int. 2011, 10, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Catamo, E.; Zupin, L.; Crovella, S.; Celsi, F.; Segat, L. Non-classical MHC-I human leukocyte antigen (HLA-G) in hepatotropic viral infections and in hepatocellular carcinoma. Hum. Immunol. 2014, 75, 1225–1231. [Google Scholar] [CrossRef] [PubMed]
- Catamo, E.; Zupin, L.; Freato, N.; Polesello, V.; Celsi, F.; Crocè, S.L.; Masutti, F.; Pozzato, G.; Segat, L.; Crovella, S. HLA-G regulatory polymorphisms are associated with susceptibility to HCV infection. HLA 2017, 89, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Martelli-Palomino, G.; Pancotto, J.A.; Muniz, Y.C.; Mendes-Junior, C.T.; Castelli, E.C.; Massaro, J.D.; Krawice-Radanne, I.; Poras, I.; Rebmann, V.; Carosella, E.D.; et al. Polymorphic Sites at the 3′ Untranslated Region of the HLA-G Gene Are Associated with Differential hla-g Soluble Levels in the Brazilian and French Population. PLoS ONE 2013, 8, e71742. [Google Scholar] [CrossRef] [PubMed]
- Souto, F.J.D.; Crispim, J.C.O.; Ferreira, S.C.; da Silva, A.S.M.; Bassi, C.L.; Soares, C.P.; Zucoloto, S.; Rouas-Freiss, N.; Moreau, P.; Martinelli, A.L.C.; et al. Liver HLA-G expression is associated with multiple clinical and histopathological forms of chronic hepatitis B virus infection. J. Viral Hepat. 2011, 18, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tong, S.; Li, S.; Wang, X.; Ren, H.; Yin, W. Increased ILT2 expression contributes to dysfunction of CD56dimCD16+NK cells in chronic hepatitis B virus infection. Antivir. Res. 2022, 205, 105385. [Google Scholar] [CrossRef]
- Liu, L.; Wang, L.; Zhao, L.; He, C.; Wang, G. The Role of HLA-G in Tumor Escape: Manipulating the Phenotype and Function of Immune Cells. Front. Oncol. 2020, 10, 597468. [Google Scholar] [CrossRef]
- Littera, R.; Perra, A.; Miglianti, M.; Piras, I.S.; Mocci, S.; Lai, S.; Melis, M.; Zolfino, T.; Balestrieri, C.; Conti, M.; et al. The double-sided of human leukocyte antigen-G molecules in type 1 autoimmune hepatitis. Front. Immunol. 2022, 13, 1007647. [Google Scholar] [CrossRef]
- Hò, G.-G.T.; Celik, A.A.; Huyton, T.; Hiemisch, W.; Blasczyk, R.; Simper, G.S.; Bade-Doeding, C. NKG2A/CD94 Is a New Immune Receptor for HLA-G and Distinguishes Amino Acid Differences in the HLA-G Heavy Chain. Int. J. Mol. Sci. 2020, 21, 4362. [Google Scholar] [CrossRef]
- Carosella, E.D.; Gregori, S.; Tronik-Le Roux, D. HLA-G/LILRBs: A Cancer Immunotherapy Challenge. Trends Cancer 2021, 7, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Morandi, F.; Airoldi, I. HLA-G and Other Immune Checkpoint Molecules as Targets for Novel Combined Immunotherapies. Int. J. Mol. Sci. 2022, 23, 2925. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, S.D.; Biro, P.A.; Holmes, C.H. HLA-F Is a Predominantly Empty, Intracellular, TAP-Associated MHC Class Ib Protein with a Restricted Expression Pattern. J. Immunol. 2000, 164, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Goodridge, J.P.; Lee, N.; Burian, A.; Pyo, C.-W.; Tykodi, S.S.; Warren, E.H.; Yee, C.; Riddell, S.R.; Geraghty, D.E. HLA-F and MHC-I Open Conformers Cooperate in a MHC-I Antigen Cross-Presentation Pathway. J. Immunol. 2013, 191, 1567–1577. [Google Scholar] [CrossRef] [PubMed]
- Goodridge, J.P.; Burian, A.; Lee, N.; Geraghty, D.E. HLA-F Complex without Peptide Binds to MHC Class I Protein in the Open Conformer Form. J. Immunol. 2010, 184, 6199–6208. [Google Scholar] [CrossRef]
- Arosa, F.A.; Esgalhado, A.J.; Reste-Ferreira, D.; Cardoso, E.M. Open MHC Class I Conformers: A Look through the Looking Glass. Int. J. Mol. Sci. 2021, 22, 9738. [Google Scholar] [CrossRef]
- Schulte, D.; Vogel, M.; Langhans, B.; Krämer, B.; Körner, C.; Nischalke, H.D.; Steinberg, V.; Michalk, M.; Berg, T.; Rockstroh, J.K.; et al. The HLA-E(R)/HLA-E(R) Genotype Affects the Natural Course of Hepatitis C Virus (HCV) Infection and Is Associated with HLA-E-Restricted Recognition of an HCV-Derived Peptide by Interferon-Gamma-Secreting Human CD8(+) T Cells. J. Infect. Dis. 2009, 200, 1397–1401. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Selvan, S.R.; Terasaki, P.I. Augmentation of anti-HLA-E antibodies with concomitant HLA-Ia reactivity in IFNγ-treated autologous melanoma cell vaccine recipients. J. Immunotoxicol. 2012, 9, 282–291. [Google Scholar] [CrossRef]
- Morinaga, T.; Iwatsuki, M.; Yamashita, K.; Yasuda-Yoshihara, N.; Yamane, T.; Matsumoto, C.; Harada, K.; Eto, K.; Kurashige, J.; Ishimoto, T.; et al. Dynamic Alteration in HLA-E Expression and Soluble HLA-E via Interaction with Natural Killer Cells in Gastric Cancer. Ann. Surg. Oncol. 2023, 30, 1240–1252. [Google Scholar] [CrossRef]
- Barrett, D.M.; Gustafson, K.S.; Wang, J.; Wang, S.Z.; Ginder, G.D. A GATA Factor Mediates Cell Type-Restricted Induction of HLA-E Gene Transcription by Gamma Interferon. Mol. Cell. Biol. 2004, 24, 6194–6204. [Google Scholar] [CrossRef]
- Jørgensen, N.; Sayed, A.; Jeppesen, H.B.; Persson, G.; Weisdorf, I.; Funck, T.; Hviid, T.V.F. Characterization of HLA-G Regulation and HLA Expression in Breast Cancer and Malignant Melanoma Cell Lines upon IFN-γ Stimulation and Inhibition of DNA Methylation. Int. J. Mol. Sci. 2020, 21, 4307. [Google Scholar] [CrossRef] [PubMed]
- Kany, S.; Vollrath, J.T.; Relja, B. Cytokines in Inflammatory Disease. Int. J. Mol. Sci. 2019, 20, 6008. [Google Scholar] [CrossRef] [PubMed]
- Rea, I.M.; Gibson, D.S.; McGilligan, V.; McNerlan, S.E.; Alexander, H.D.; Ross, O.A. Age and Age-Related Diseases: Role of Inflammation Triggers and Cytokines. Front. Immunol. 2018, 9, 586. [Google Scholar] [CrossRef]
- Boyle, L.H.; Gillingham, A.K.; Munro, S.; Trowsdale, J. Selective Export of HLA-F by Its Cytoplasmic Tail. J. Immunol. 2006, 176, 6464–6472. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Ishitani, A.; Geraghty, D.E. HLA-F is a surface marker on activated lymphocytes. Eur. J. Immunol. 2010, 40, 2308–2318. [Google Scholar] [CrossRef] [PubMed]
- Burian, A.; Wang, K.L.; Finton, K.A.K.; Lee, N.; Ishitani, A.; Strong, R.K.; Geraghty, D.E. HLA-F and MHC-I Open Conformers Bind Natural Killer Cell Ig-Like Receptor KIR3DS1. PLoS ONE 2016, 11, e0163297. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Hölzemer, A.; Martrus, G.; Chung, A.W.; Pacheco, Y.; Simoneau, C.R.; Rucevic, M.; Lamothe-Molina, P.A.; Pertel, T.; Kim, T.-E.; et al. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat. Immunol. 2016, 17, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Lunemann, S.; Schöbel, A.; Kah, J.; Fittje, P.; Hölzemer, A.; Langeneckert, A.E.; Hess, L.U.; Poch, T.; Martrus, G.; Garcia-Beltran, W.F.; et al. Interactions Between KIR3DS1 and HLA-F Activate Natural Killer Cells to Control HCV Replication in Cell Culture. Gastroenterology 2018, 155, 1366–1371.e3. [Google Scholar] [CrossRef]
- Oliva, A.; Kinter, A.L.; Vaccarezza, M.; Rubbert, A.; Catanzaro, A.; Moir, S.; Monaco, J.; Ehler, L.; Mizell, S.; Jackson, R.; et al. Natural killer cells from human immunodeficiency virus (HIV)-infected individuals are an important source of CC-chemokines and suppress HIV-1 entry and replication in vitro. J. Clin. Investig. 1998, 102, 223–231. [Google Scholar] [CrossRef]
- Rivero-Juárez, A.; Gonzalez, R.; Camacho, A.; Manzanares-Martin, B.; Caruz, A.; Martínez-Peinado, A.; Torre-Cisneros, J.; Pineda, J.A.; Peña, J.; Rivero, A. Natural Killer KIR3DS1 Is Closely Associated with HCV Viral Clearance and Sustained Virological Response in HIV/HCV Patients. PLoS ONE 2013, 8, e61992. [Google Scholar] [CrossRef]
- Hò, G.-G.T.; Hiemisch, W.; Pich, A.; Behrens, G.M.N.; Blasczyk, R.; Bade-Doeding, C. The Loss of HLA-F/KIR3DS1 Ligation Is Mediated by Hemoglobin Peptides. Int. J. Mol. Sci. 2020, 21, 8012. [Google Scholar] [CrossRef] [PubMed]
- Mele, D.; Pasi, A.; Cacciatore, R.; Mantovani, S.; Oliviero, B.; Mondelli, M.U.; Varchetta, S. Decreased interferon-γ production by NK cells from KIR haplotype B carriers in hepatitis C virus infection. Liver Int. 2019, 39, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Frias, M.; Rivero-Juárez, A.; Rodriguez-Cano, D.; Camacho, Á.; López-López, P.; Risalde, M..; Manzanares-Martín, B.; Brieva, T.; Machuca, I.; Rivero, A. HLA-B, HLA-C and KIR improve the predictive value of IFNL3 for Hepatitis C spontaneous clearance. Sci. Rep. 2018, 8, 659. [Google Scholar] [CrossRef]
- Xu, Y.; Han, H.; Zhang, F.; Lv, S.; Li, Z.; Fang, Z. Lesion human leukocyte antigen-F expression is associated with a poor prognosis in patients with hepatocellular carcinoma. Oncol. Lett. 2015, 9, 300–304. [Google Scholar] [CrossRef]
- Wimalarathne, M.M.; Wilkerson-Vidal, Q.C.; Hunt, E.C.; Love-Rutledge, S.T. The case for FAT10 as a novel target in fatty liver diseases. Front. Pharmacol. 2022, 13, 972320. [Google Scholar] [CrossRef]
- Middleton, P.; Vergis, N. Mitochondrial dysfunction and liver disease: Role, relevance, and potential for therapeutic modulation. Ther. Adv. Gastroenterol. 2021, 14, 17562848211031394. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Buerger, S.; Groettrup, M. The ubiquitin-like modifier FAT10 in antigen processing and antimicrobial defense. Mol. Immunol. 2015, 68, 129–132. [Google Scholar] [CrossRef]
- Jia, Y.; French, B.; Tillman, B.; French, S. Different roles of FAT10, FOXO1, and ADRA2A in hepatocellular carcinoma tumorigenesis in patients with alcoholic steatohepatitis (ASH) vs non-alcoholic steatohepatitis (NASH). Exp. Mol. Pathol. 2018, 105, 144–149. [Google Scholar] [CrossRef]
- Jia, Y.; Ji, P.; French, S.W. The Role of FAT10 in Alcoholic Hepatitis Pathogenesis. Biomedicines 2020, 8, 189. [Google Scholar] [CrossRef]
- Yuan, R.; Wang, K.; Hu, J.; Yan, C.; Li, M.; Yu, X.; Liu, X.; Lei, J.; Guo, W.; Wu, L.; et al. Ubiquitin-like Protein FAT10 Promotes the Invasion and Metastasis of Hepatocellular Carcinoma by Modifying β-Catenin Degradation. Cancer Res. 2014, 74, 5287–5300. [Google Scholar] [CrossRef]
- Liu, H.; Gong, M.; French, B.A.; Li, J.; Tillman, B.; French, S.W. Mallory–Denk Body (MDB) formation modulates ufmylation expression epigenetically in alcoholic hepatitis (AH) and non-alcoholic steatohepatitis (NASH). Exp. Mol. Pathol. 2014, 97, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.; Lee, A.S. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene 2013, 32, 805–818. [Google Scholar] [CrossRef] [PubMed]
- De Re, V.; Simula, M.P.; Cannizzaro, R.; Sansonno, D.E.; Canzonieri, V.; Gloghini, A.; Carbone, A.; Colombatti, A.; Marin, M.D.; De Zorzi, M.; et al. HCV inhibits antigen processing and presentation and induces oxidative stress response in gastric mucosa. Proteom. Clin. Appl. 2008, 2, 1290–1299. [Google Scholar] [CrossRef] [PubMed]
- Mah, M.M.; Roverato, N.; Groettrup, M. Regulation of Interferon Induction by the Ubiquitin-Like Modifier FAT10. Biomolecules 2020, 10, 951. [Google Scholar] [CrossRef]
- Arshad, M.; Abdul Hamid, N.; Chan, M.C.; Ismail, F.; Tan, G.C.; Pezzella, F.; Tan, K.-L. NUB1 and FAT10 Proteins as Potential Novel Biomarkers in Cancer: A Translational Perspective. Cells 2021, 10, 2176. [Google Scholar] [CrossRef]
- Zhang, Y.; Zuo, Z.; Liu, B.; Yang, P.; Wu, J.; Han, L.; Han, T.; Chen, T. FAT10 promotes hepatocellular carcinoma (HCC) carcinogenesis by mediating P53 degradation and acts as a prognostic indicator of HCC. J. Gastrointest. Oncol. 2021, 12, 1823–1837. [Google Scholar] [CrossRef]
- Sadagopan, A.; Michelakos, T.; Boyiadzis, G.; Ferrone, C.; Ferrone, S. Human Leukocyte Antigen Class I Antigen-Processing Machinery Upregulation by Anticancer Therapies in the Era of Checkpoint Inhibitors. JAMA Oncol. 2022, 8, 462. [Google Scholar] [CrossRef]
- Takahashi, A.; Umemura, A.; Yano, K.; Okishio, S.; Kataoka, S.; Okuda, K.; Seko, Y.; Yamaguchi, K.; Moriguchi, M.; Okanoue, T.; et al. Tyrosine Kinase Inhibitors Stimulate HLA Class I Expression by Augmenting the IFNγ/STAT1 Signaling in Hepatocellular Carcinoma Cells. Front. Oncol. 2021, 11, 707473. [Google Scholar] [CrossRef]
- Sung, P.S.; Racanelli, V.; Shin, E.-C. CD8+ T-Cell Responses in Acute Hepatitis C Virus Infection. Front. Immunol. 2014, 5, 266. [Google Scholar] [CrossRef]
- Crispe, I.N. Hepatocytes as Immunological Agents. J. Immunol. 2016, 196, 17–21. [Google Scholar] [CrossRef]
- Flecken, T.; Schmidt, N.; Hild, S.; Gostick, E.; Drognitz, O.; Zeiser, R.; Schemmer, P.; Bruns, H.; Eiermann, T.; Price, D.A.; et al. Immunodominance and functional alterations of tumor-associated antigen-specific CD8+ T-cell responses in hepatocellular carcinoma. Hepatology 2013, 59, 1415–1426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-Z.; Wang, T.; Wang, X.-F.; Zhang, Y.-Q.; Song, S.-X.; Ma, C.-Q. Improving the ability of CAR-T cells to hit solid tumors: Challenges and strategies. Pharmacol. Res. 2021, 175, 106036. [Google Scholar] [CrossRef] [PubMed]
- Khatwani, N.; Romee, R.; Pillai, A.B. Editorial: Innate immune cell therapy of cancer. Front. Immunol. 2022, 13, 4415. [Google Scholar] [CrossRef]
- Shen, J.; Yang, D.; Ding, Y. Advances in Promoting the Efficacy of Chimeric Antigen Receptor T Cells in the Treatment of Hepatocellular Carcinoma. Cancers 2022, 14, 5018. [Google Scholar] [CrossRef]
- Ragoonanan, D.; Khazal, S.J.; Abdel-Azim, H.; McCall, D.; Cuglievan, B.; Tambaro, F.P.; Ahmad, A.H.; Rowan, C.M.; Gutierrez, C.; Schadler, K.; et al. Diagnosis, grading and management of toxicities from immunotherapies in children, adolescents and young adults with cancer. Nat. Rev. Clin. Oncol. 2021, 18, 435–453. [Google Scholar] [CrossRef]
- Valiullina, A.K.; Zmievskaya, E.A.; Ganeeva, I.A.; Zhuravleva, M.N.; Garanina, E.E.; Rizvanov, A.A.; Petukhov, A.V.; Bulatov, E.R. Evaluation of CAR-T Cells’ Cytotoxicity against Modified Solid Tumor Cell Lines. Biomedicines 2023, 11, 626. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-R.; Brown, J.; Yu, Y.; Lee, D.; Zhou, K.; Dunn, Z.S.; Hon, R.; Wilson, M.; Kramer, A.; Zhu, Y.; et al. Targeting Immunosuppressive Tumor-Associated Macrophages Using Innate T Cells for Enhanced Antitumor Reactivity. Cancers 2022, 14, 2749. [Google Scholar] [CrossRef]
- Johanna, I.; Hernández-López, P.; Heijhuurs, S.; Scheper, W.; Bongiovanni, L.; de Bruin, A.; Beringer, D.X.; Oostvogels, R.; Straetemans, T.; Sebestyen, Z.; et al. Adding Help to an HLA-A*24:02 Tumor-Reactive γδTCR Increases Tumor Control. Front. Immunol. 2021, 12, 752699. [Google Scholar] [CrossRef]
- Rossi, J.-F.; Lu, Z.Y.; Massart, C.; Levon, K. Dynamic Immune/Inflammation Precision Medicine: The Good and the Bad Inflammation in Infection and Cancer. Front. Immunol. 2021, 12, 595722. [Google Scholar] [CrossRef]
- Huang, M.; Cai, H.; Han, B.; Xia, Y.; Kong, X.; Gu, J. Natural Killer Cells in Hepatic Ischemia-Reperfusion Injury. Front. Immunol. 2022, 13, 870038. [Google Scholar] [CrossRef]
- Boudreau, J.E.; Hsu, K.C. Natural Killer Cell Education and the Response to Infection and Cancer Therapy: Stay Tuned. Trends Immunol. 2018, 39, 222–239. [Google Scholar] [CrossRef]
- Ding, W.; Xu, X.; Qian, Y.; Xue, W.; Wang, Y.; Du, J.; Jin, L.; Tan, Y. Prognostic Value of Tumor-Infiltrating Lymphocytes in Hepatocellular Carcinoma: A Meta-Analysis. Medicine 2018, 97, e13301. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Xu, Q.; Li, X.; Guo, Y.; Zhang, B.; Jin, Y.; Zhu, C.; Shen, Y.; Yang, P.; Shi, Y.; et al. Heterogeneity induced GZMA-F2R communication inefficient impairs antitumor immunotherapy of PD-1 mAb through JAK2/STAT1 signal suppression in hepatocellular carcinoma. Cell Death Dis. 2022, 13, 213. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Toso, C.; Meeberg, G.; Hernandez-Alejandro, R.; Dufour, J.; Marotta, P.; Majno, P.; Kneteman, N.M. Total tumor volume and alpha-fetoprotein for selection of transplant candidates with hepatocellular carcinoma: A prospective validation. Hepatology 2015, 62, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Liang, X.; Wang, P.; Hu, Y.; Qi, Y.; Jin, Y.; Du, Y.; Fang, C.; Tian, J. A Hepatocellular Carcinoma Targeting Nanostrategy with Hypoxia-Ameliorating and Photothermal Abilities that, Combined with Immunotherapy, Inhibits Metastasis and Recurrence. ACS Nano 2020, 14, 12679–12696. [Google Scholar] [CrossRef]
- Li, Z.; Han, N.; Ren, X.; Zhang, Y.; Chu, X. Effectiveness of TKI Inhibitors Combined With PD-1 in Patients With Postoperative Early Recurrence of HCC: A Real-World Study. Front. Oncol. 2022, 12, 833884. [Google Scholar] [CrossRef]
- Haber, P.K.; Castet, F.; Torres-Martin, M.; Andreu-Oller, C.; Puigvehí, M.; Miho, M.; Radu, P.; Dufour, J.-F.; Verslype, C.; Zimpel, C.; et al. Molecular Markers of Response to Anti-PD1 Therapy in Advanced Hepatocellular Carcinoma. Gastroenterology 2023, 164, 72–88.e18. [Google Scholar] [CrossRef]
- Zhang, Y.; Schmidt-Wolf, I.G.H. Ten-year update of the international registry on cytokine-induced killer cells in cancer immunotherapy. J. Cell. Physiol. 2020, 235, 9291–9303. [Google Scholar] [CrossRef]
- Yang, C.-K.; Huang, C.-H.; Hu, C.-H.; Fang, J.-H.; Chen, T.-C.; Lin, Y.-C.; Lin, C.-Y. Immunophenotype and antitumor activity of cytokine-induced killer cells from patients with hepatocellular carcinoma. PLoS ONE 2023, 18, e0280023. [Google Scholar] [CrossRef]
- Kim, N.J.; Yoon, J.H.; Tuomi, A.C.; Lee, J.; Kim, D. In-situ tumor vaccination by percutaneous ablative therapy and its synergy with immunotherapeutics: An update on combination therapy. Front. Immunol. 2023, 14, 1118845. [Google Scholar] [CrossRef] [PubMed]
- Weng, D.-S.; Zhou, J.; Zhou, Q.-M.; Zhao, M.; Wang, Q.-J.; Huang, L.-X.; Li, Y.-Q.; Chen, S.-P.; Wu, P.-H.; Xia, J.-C. Minimally Invasive Treatment Combined With Cytokine-induced Killer Cells Therapy Lower the Short-term Recurrence Rates of Hepatocellular Carcinomas. J. Immunother. 2008, 31, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Song, J.; Zhang, H.; Liu, X.; Zuo, F.; Zhao, Y.; Zhao, Y.; Yin, X.; Guo, X.; Wu, X.; et al. Immune checkpoint HLA-E:CD94-NKG2A mediates evasion of circulating tumor cells from NK cell surveillance. Cancer Cell 2023, 41, 272–287.e9. [Google Scholar] [CrossRef]
- Tinker, A.V.; Hirte, H.W.; Provencher, D.; Butler, M.; Ritter, H.; Tu, D.; Azim, H.A.; Paralejas, P.; Grenier, N.; Hahn, S.-A.; et al. Dose-Ranging and Cohort-Expansion Study of Monalizumab (IPH2201) in Patients with Advanced Gynecologic Malignancies: A Trial of the Canadian Cancer Trials Group (CCTG): IND221. Clin. Cancer Res. 2019, 25, 6052–6060. [Google Scholar] [CrossRef]
- André, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Bléry, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743.e13. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Shi, J.; Zhang, Y.; Liu, J.; An, C.; Zhu, H.; Wu, P.; Hu, W.; Qin, R.; Yao, D.; et al. NKG2D discriminates diverse ligands through selectively mechano-regulated ligand conformational changes. EMBO J. 2022, 41, e107739. [Google Scholar] [CrossRef]
- Han, Y.; Sun, F.; Zhang, X.; Wang, T.; Jiang, J.; Cai, J.; Gao, Q.; Hezam, K.; Liu, Y.; Xie, J.; et al. CD24 targeting bi-specific antibody that simultaneously stimulates NKG2D enhances the efficacy of cancer immunotherapy. J. Cancer Res. Clin. Oncol. 2019, 145, 1179–1190. [Google Scholar] [CrossRef]
- Jeong, W.-I.; Park, O.; Gao, B. Abrogation of the Antifibrotic Effects of Natural Killer Cells/Interferon-Gamma Contributes to Alcohol Acceleration of Liver Fibrosis. Gastroenterology 2008, 134, 248–258. [Google Scholar] [CrossRef]
- Pant, A.; Lim, M. CAR-T Therapy in GBM: Current Challenges and Avenues for Improvement. Cancers 2023, 15, 1249. [Google Scholar] [CrossRef]
- Titov, A.; Zmievskaya, E.; Ganeeva, I.; Valiullina, A.; Petukhov, A.; Rakhmatullina, A.; Miftakhova, R.; Fainshtein, M.; Rizvanov, A.; Bulatov, E. Adoptive Immunotherapy beyond CAR T-Cells. Cancers 2021, 13, 743. [Google Scholar] [CrossRef]
- Haanen, J.B.; Cerundolo, V. NKG2A, a New Kid on the Immune Checkpoint Block. Cell 2018, 175, 1720–1722. [Google Scholar] [CrossRef] [PubMed]
- Cazzetta, V.; Depierreux, D.; Colucci, F.; Mikulak, J.; Mavilio, D. NKG2A Immune Checkpoint in Vδ2 T Cells: Emerging Application in Cancer Immunotherapy. Cancers 2023, 15, 1264. [Google Scholar] [CrossRef]
- Wu, S.-J.; Lin, C.-T.; Liao, C.H.; Lin, C.-M. Immunotherapeutic potential of blinatumomab-secreting γ9δ2 T Cells. Transl. Oncol. 2023, 31, 101650. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Bai, Y.; Lin, A.; Jiang, A.; Zhou, C.; Cheng, Q.; Liu, Z.; Chen, X.; Zhang, J.; Luo, P. Gamma delta T-cell-based immune checkpoint therapy: Attractive candidate for antitumor treatment. Mol. Cancer 2023, 22, 31. [Google Scholar] [CrossRef] [PubMed]
- Silva-Santos, B.; Mensurado, S.; Coffelt, S.B. γδ T cells: Pleiotropic immune effectors with therapeutic potential in cancer. Nat. Rev. Cancer 2019, 19, 392–404. [Google Scholar] [CrossRef]
- Zumwalde, N.A.; Sharma, A.; Xu, X.; Ma, S.; Schneider, C.L.; Romero-Masters, J.C.; Hudson, A.W.; Gendron-Fitzpatrick, A.; Kenney, S.C.; Gumperz, J.E. Adoptively transferred Vγ9Vδ2 T cells show potent antitumor effects in a preclinical B cell lymphomagenesis model. J. Clin. Investig. 2017, 2, e93179. [Google Scholar] [CrossRef]
- Lonez, C.; Hendlisz, A.; Shaza, L.; Aftimos, P.; Vouche, M.; Donckier, V.; Machiels, J.-P.H.; Van Den Eynde, M.; Canon, J.-L.; Carrasco, J.; et al. Celyad’s Novel CAR T-Cell Therapy for Solid Malignancies. Curr. Res. Transl. Med. 2018, 66, 53–56. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).