Prophylactic vs. Therapeutic Effect of Probiotics on the Inflammation Mediated by the NF-κB Pathway in Inflammatory Bowel Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Cell Line
2.3. Preparation of Bacterial Cocktails
2.4. MTT Assay for MOI Determination
2.5. Challenge with Probiotics and Gram-Negative Bacteria
2.6. Total RNA Isolation and cDNA Synthesis
2.7. Quantitative Real-Time PCR (qRT-PCR)
2.8. Cytokine Assays
2.9. Statistical Analysis
3. Results
3.1. Receptors and NF-κB Pathway Modulation
3.2. Pro-Inflammatory Cytokine Modulation by Probiotic Cocktails
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaplan, G.G. The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 720–727. [Google Scholar] [CrossRef]
- Molodecky, N.A.; Soon, I.S.; Rabi, D.M.; Ghali, W.A.; Ferris, M.; Chernoff, G.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Barkema, H.W.; et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012, 142, 46–54.e42; quiz e30. [Google Scholar] [CrossRef] [Green Version]
- Fabián, O.; Kamaradová, K. Morphology of inflammatory bowel diseases (IBD). Ceskoslovenska Patol. 2022, 58, 27–37. [Google Scholar]
- Vemuri, R.; Gundamaraju, R.; Eri, R. Role of Lactic Acid Probiotic Bacteria in IBD. Curr. Pharm. Des. 2017, 23, 2352–2355. [Google Scholar] [CrossRef]
- Group, F.W.W. WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food; WHO: London, ON, Canada, 2002; Volume 30. [Google Scholar]
- Ljungh, A.; Wadström, T. Lactic acid bacteria as probiotics. Curr. Issues Intest. Microbiol. 2006, 7, 73–89. [Google Scholar]
- Kumar, M.; Hemalatha, R.; Nagpal, R.; Singh, B.; Parasannanavar, D.; Verma, V.; Kumar, A.; Marotta, F.; Catanzaro, R.; Cuffari, B.; et al. Probiotic Approaches for Targeting Inflammatory Bowel Disease: An Update on Advances and Opportunities in Managing the Disease. Int. J. Probiotics Prebiotics 2016, 11, 99–116. [Google Scholar] [PubMed]
- Hemarajata, P.; Versalovic, J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterol. 2013, 6, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oelschlaeger, T.A. Mechanisms of probiotic actions—A review. Int. J. Med. Microbiol. 2010, 300, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Chu, H. Host gene-microbiome interactions: Molecular mechanisms in inflammatory bowel disease. Genome Med. 2017, 9, 69. [Google Scholar] [CrossRef] [Green Version]
- Anjum, M.; Laitila, A.; Ouwehand, A.C.; Forssten, S.D. Current Perspectives on Gastrointestinal Models to Assess Probiotic-Pathogen Interactions. Front. Microbiol. 2022, 13, 831455. [Google Scholar] [CrossRef]
- Wang, C.S.; Li, W.B.; Wang, H.Y.; Ma, Y.M.; Zhao, X.H.; Yang, H.; Qian, J.M.; Li, J.N. VSL#3 can prevent ulcerative colitis-associated carcinogenesis in mice. World J. Gastroenterol. 2018, 24, 4254–4262. [Google Scholar] [CrossRef]
- Sood, A.; Midha, V.; Makharia, G.K.; Ahuja, V.; Singal, D.; Goswami, P.; Tandon, R.K. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin. Gastroenterol. Hepatol. 2009, 7, 1202–1209.e1. [Google Scholar] [CrossRef] [PubMed]
- Tursi, A.; Brandimarte, G.; Papa, A.; Giglio, A.; Elisei, W.; Giorgetti, G.M.; Forti, G.; Morini, S.; Hassan, C.; Pistoia, M.A.; et al. Treatment of relapsing mild-to-moderate ulcerative colitis with the probiotic VSL#3 as adjunctive to a standard pharmaceutical treatment: A double-blind, randomized, placebo-controlled study. Am. J. Gastroenterol. 2010, 105, 2218–2227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caballero-Franco, C.; Keller, K.; De Simone, C.; Chadee, K. The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G315–G322. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, S.K.; El-Bedewy, M.M. Effect of probiotics on pro-inflammatory cytokines and NF-kappaB activation in ulcerative colitis. World J. Gastroenterol. 2010, 16, 4145–4151. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakubczyk, D.; Leszczyńska, K.; Górska, S. The Effectiveness of Probiotics in the Treatment of Inflammatory Bowel Disease (IBD)—A Critical Review. Nutrients 2020, 12, 1973. [Google Scholar] [CrossRef] [PubMed]
- Khajeh Alizadeh Attar, M.; Anwar, M.A.; Eskian, M.; Keshavarz-Fathi, M.; Choi, S.; Rezaei, N. Basic understanding and therapeutic approaches to target toll-like receptors in cancerous microenvironment and metastasis. Med. Res. Rev. 2018, 38, 1469–1484. [Google Scholar] [CrossRef]
- George, J.; Kubarenko, A.V.; Rautanen, A.; Mills, T.C.; Colak, E.; Kempf, T.; Hill, A.V.; Nieters, A.; Weber, A.N. MyD88 adaptor-like D96N is a naturally occurring loss-of-function variant of TIRAP. J. Immunol. 2010, 184, 3025–3032. [Google Scholar] [CrossRef] [Green Version]
- Koedel, U.; Merbt, U.M.; Schmidt, C.; Angele, B.; Popp, B.; Wagner, H.; Pfister, H.W.; Kirschning, C.J. Acute brain injury triggers MyD88-dependent, TLR2/4-independent inflammatory responses. Am. J. Pathol. 2007, 171, 200–213. [Google Scholar] [CrossRef] [Green Version]
- Gárate, I.; Garcia-Bueno, B.; Madrigal, J.L.; Caso, J.R.; Alou, L.; Gomez-Lus, M.L.; Micó, J.A.; Leza, J.C. Stress-induced neuroinflammation: Role of the Toll-like receptor-4 pathway. Biol. Psychiatry 2013, 73, 32–43. [Google Scholar] [CrossRef]
- Kawai, T.; Takeuchi, O.; Fujita, T.; Inoue, J.; Mühlradt, P.F.; Sato, S.; Hoshino, K.; Akira, S. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J. Immunol. 2001, 167, 5887–5894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef] [Green Version]
- Feerick, C.L.; McKernan, D.P. Understanding the regulation of pattern recognition receptors in inflammatory diseases—A ‘Nod’ in the right direction. Immunology 2017, 150, 237–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negroni, A.; Pierdomenico, M.; Cucchiara, S.; Stronati, L. NOD2 and inflammation: Current insights. J. Inflamm. Res. 2018, 11, 49–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llewellyn, A.; Foey, A. Probiotic Modulation of Innate Cell Pathogen Sensing and Signaling Events. Nutrients 2017, 9, 1156. [Google Scholar] [CrossRef] [Green Version]
- Rohani, M.; Noohi, N.; Talebi, M.; Katouli, M.; Pourshafie, M.R. Highly Heterogeneous Probiotic Lactobacillus Species in Healthy Iranians with Low Functional Activities. PLoS ONE 2015, 10, e0144467. [Google Scholar] [CrossRef]
- Eshaghi, M.; Bibalan, M.H.; Rohani, M.; Esghaei, M.; Douraghi, M.; Talebi, M.; Pourshafie, M.R. Bifidobacterium obtained from mother’s milk and their infant stool; A comparative genotyping and antibacterial analysis. Microb. Pathog. 2017, 111, 94–98. [Google Scholar] [CrossRef]
- Hasannejad Bibalan, M.; Eshaghi, M.; Rohani, M.; Pourshafie, M.R.; Talebi, M. Determination of Bacteriocin Genes and Antibacterial Activity of Lactobacillus Strains Isolated from Fecal of Healthy Individuals. Int. J. Mol. Cell. Med. 2017, 6, 50–55. [Google Scholar]
- Singla, P.; Dalal, P.; Kaur, M.; Arya, G.; Nimesh, S.; Singh, R.; Salunke, D.B. Bile Acid Oligomers and Their Combination with Antibiotics to Combat Bacterial Infections. J. Med. Chem. 2018, 61, 10265–10275. [Google Scholar] [CrossRef]
- Ghanavati, R.; Asadollahi, P.; Shapourabadi, M.B.; Razavi, S.; Talebi, M.; Rohani, M. Inhibitory effects of Lactobacilli cocktail on HT-29 colon carcinoma cells growth and modulation of the Notch and Wnt/β-catenin signaling pathways. Microb. Pathog. 2020, 139, 103829. [Google Scholar] [CrossRef]
- Chen, H.; Xia, Y.; Zhu, S.; Yang, J.; Yao, J.; Di, J.; Liang, Y.; Gao, R.; Wu, W.; Yang, Y.; et al. Lactobacillus plantarum LPOnlly alters the gut flora and attenuates colitis by inducing microbiome alteration in interleukin10 knockout mice. Mol. Med. Rep. 2017, 16, 5979–5985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Najafi, S.; Sotoodehnejadnematalahi, F.; Amiri, M.M.; Pourshafie, M.R.; Rohani, M. Decreased mucosal adhesion of Lactobacillus species in patients with inflammatory bowel disease. Caspian. J. Intern. Med. 2022, 13, 713–720. [Google Scholar] [CrossRef]
- Dejban, P.; Nikravangolsefid, N.; Chamanara, M.; Dehpour, A.; Rashidian, A. The role of medicinal products in the treatment of inflammatory bowel diseases (IBD) through inhibition of TLR4/NF-kappaB pathway. Phytother. Res. 2021, 35, 835–845. [Google Scholar] [CrossRef]
- Kim, H.; Zhao, Q.; Zheng, H.; Li, X.; Zhang, T.; Ma, X. A novel crosstalk between TLR4- and NOD2-mediated signaling in the regulation of intestinal inflammation. Sci. Rep. 2015, 5, 12018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.; Wang, D.; Yang, Z.; Wang, T. Pharmacological Effects of Polyphenol Phytochemicals on the Intestinal Inflammation via Targeting TLR4/NF-κB Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 6939. [Google Scholar] [CrossRef]
- Neurath, M.F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 2014, 14, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Kordjazy, N.; Haj-Mirzaian, A.; Haj-Mirzaian, A.; Rohani, M.M.; Gelfand, E.W.; Rezaei, N.; Abdolghaffari, A.H. Role of toll-like receptors in inflammatory bowel disease. Pharmacol. Res. 2018, 129, 204–215. [Google Scholar] [CrossRef]
- Hill, D.A.; Artis, D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu. Rev. Immunol. 2010, 28, 623–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bron, P.A.; Kleerebezem, M.; Brummer, R.J.; Cani, P.D.; Mercenier, A.; MacDonald, T.T.; Garcia-Rodenas, C.L.; Wells, J.M. Can probiotics modulate human disease by impacting intestinal barrier function? Br. J. Nutr. 2017, 117, 93–107. [Google Scholar] [CrossRef] [Green Version]
- Van Niel, C.W. Probiotics: Not just for treatment anymore. Pediatrics 2005, 115, 174–177. [Google Scholar] [CrossRef]
- Chapman, C.M.; Gibson, G.R.; Rowland, I. In vitro evaluation of single- and multi-strain probiotics: Inter-species inhibition between probiotic strains, and inhibition of pathogens. Anaerobe 2012, 18, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Ramírez, L.M.; Pérez-Solano, R.A.; Castañón-Alonso, S.L.; Moreno Guerrero, S.S.; Ramírez Pacheco, A.; García Garibay, M.; Eslava, C. Probiotic Lactobacillus Strains Stimulate the Inflammatory Response and Activate Human Macrophages. J. Immunol. Res. 2017, 2017, 4607491. [Google Scholar] [CrossRef] [Green Version]
- Duary, R.K.; Batish, V.K.; Grover, S. Immunomodulatory activity of two potential probiotic strains in LPS-stimulated HT-29 cells. Genes Nutr. 2014, 9, 398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkar, A.; Mandal, S. Bifidobacteria-Insight into clinical outcomes and mechanisms of its probiotic action. Microbiol. Res. 2016, 192, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Li, S.C.; Hsu, W.F.; Chang, J.S.; Shih, C.K. Combination of Lactobacillus acidophilus and Bifidobacterium animalis subsp. lactis Shows a Stronger Anti-Inflammatory Effect than Individual Strains in HT-29 Cells. Nutrients 2019, 11, 969. [Google Scholar] [CrossRef] [Green Version]
- Candela, M.; Perna, F.; Carnevali, P.; Vitali, B.; Ciati, R.; Gionchetti, P.; Rizzello, F.; Campieri, M.; Brigidi, P. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: Adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 2008, 125, 286–292. [Google Scholar] [CrossRef]
- Sheikhi, A.; Shakerian, M.; Giti, H.; Baghaeifar, M.; Jafarzadeh, A.; Ghaed, V.; Heibor, M.R.; Baharifar, N.; Dadafarin, Z.; Bashirpour, G. Probiotic Yogurt Culture Bifidobacterium Animalis Subsp. Lactis BB-12 and Lactobacillus Acidophilus LA-5 Modulate the Cytokine Secretion by Peripheral Blood Mononuclear Cells from Patients with Ulcerative Colitis. Drug Res. 2016, 66, 300–305. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.L.; Romanuk, T.N. A meta-analysis of probiotic efficacy for gastrointestinal diseases. PLoS ONE 2012, 7, e34938. [Google Scholar] [CrossRef] [Green Version]
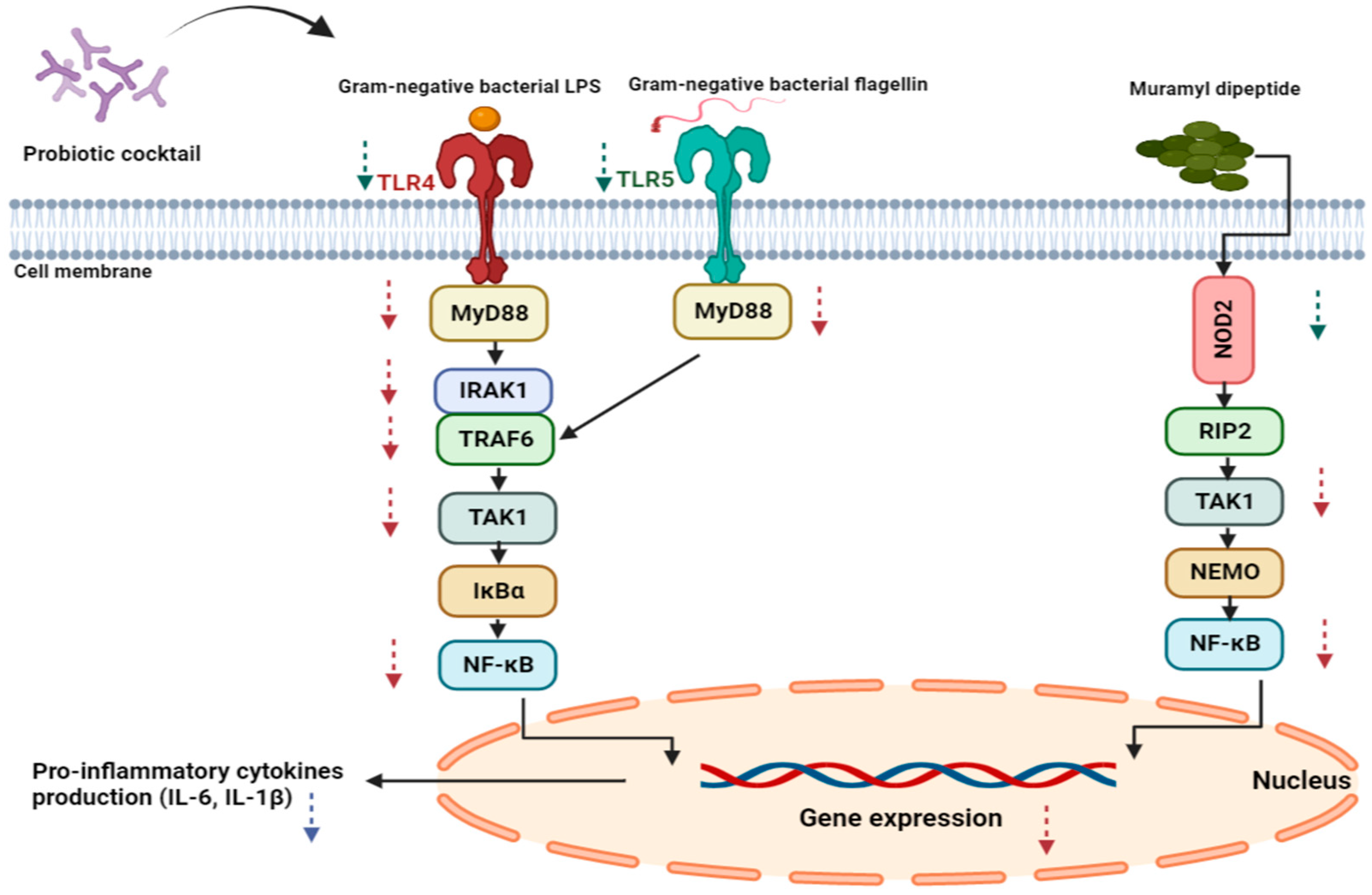

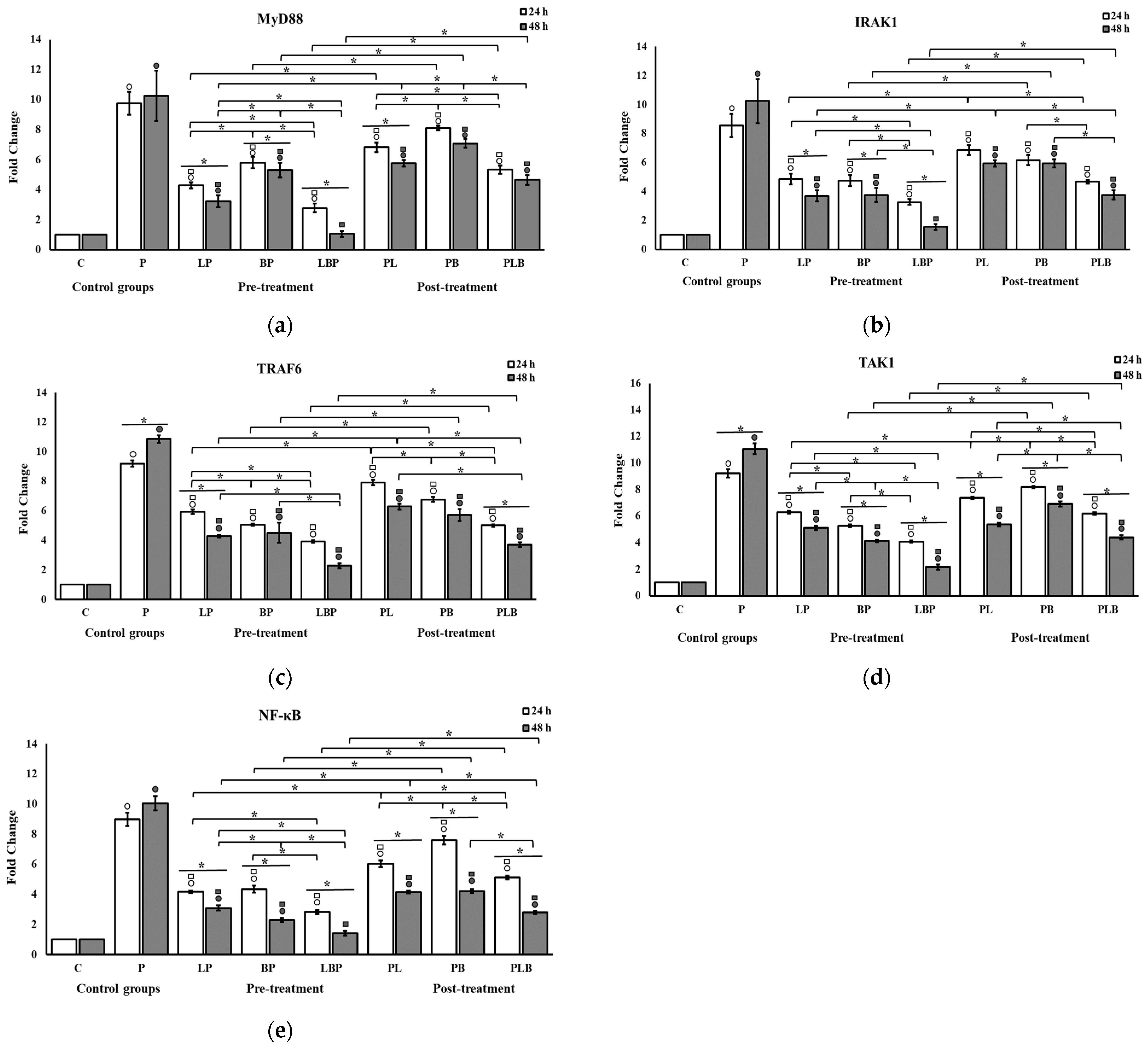
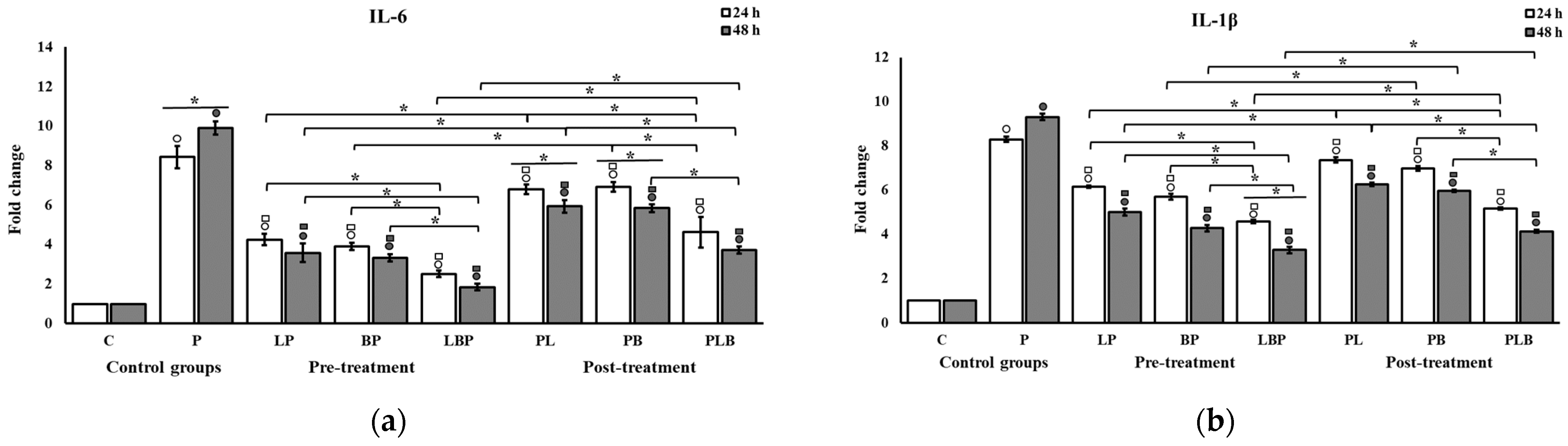

| Treatment Setups | Description |
|---|---|
| Pre-treatment | LP: HT-29 cells challenged with Gram-negative bacteria after Lactobacillus cocktail treatment |
| BP: HT-29 cells challenged with Gram-negative bacteria after Bifidobacterium cocktail treatment | |
| LBP: HT-29 cells challenged with Gram-negative bacteria after Lactobacillus and Bifidobacterium cocktail treatment | |
| Post-treatment | PL: HT-29 cells treated with Lactobacillus cocktail after Gram-negative bacteria challenge |
| PB: HT-29 cells treated with Bifidobacterium cocktail after Gram-negative bacteria challenge | |
| PLB: HT-29 cells treated with Lactobacillus and Bifidobacterium cocktail after Gram-negative bacteria challenge |
| Name | Forward Primer (5′ > 3′) | Reverse Primer (5′ > 3′) | Product Size (bp) | Tm (°C) | Primer Bank ID |
|---|---|---|---|---|---|
| GAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG | 197 | 61 | 378404907c1 |
| TLR4 | AGACCTGTCCCTGAACCCTAT | CGATGGACTTCTAAACCAGCCA | 147 | 61 | 373432602c1 |
| TLR5 | TCCCTGAACTCACGAGTCTTT | TGGTTGTCAAGTCCGTAAAATGC | 109 | 61 | 281427130c3 |
| NOD2 | TGGTTGGTTCAGCCTCTCACGATGA | CAGGACACTCTCGAAGCCTT | 157 | 61 | 11545911c1 |
| MyD88 | GGCGGCTGCTCTCAACATGCGA | CTGTCTGTGTCCGCACGTTCAAGA | 61 | 61 | 289546652c1 |
| IRAK1 | TGAGGAACACGGTGTATGCTG | GTTT GTTTGGGTGACGAAACCTGGA | 119 | 61 | 68800242c2 |
| TRAF6 | TTTGCTCTTATGGATTGTCCCC | CATTGATGCAGCACAGTTGTC | 120 | 61 | 332000008c2 |
| NF-κB | GAAGCACGAATGACAGAGGC | GCTTGGCGGATTAGCTCTTTT | 137 | 56 | 259155300c2 |
| TAK1 | ATTGTAGAGCTTCGGCAGTTATC | CTGTAAACACCAACTCATTGC | 186 | 66 | 21735563c2 |
| IL-1β | ATGATGGCTTATTACAGTGGCAA | GTCGGAGATTCGTAGCTGGA | 132 | 61 | 27894305c1 |
| IL-6 | ACTCACCTCTTCAGAACGAATTG | CCATCTTTGGAAGGTTCAGGTTG | 149 | 61 | 224831235c1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Najafi, S.; Sotoodehnejadnematalahi, F.; Amiri, M.M.; Pourshafie, M.R.; Rohani, M. Prophylactic vs. Therapeutic Effect of Probiotics on the Inflammation Mediated by the NF-κB Pathway in Inflammatory Bowel Conditions. Biomedicines 2023, 11, 1675. https://doi.org/10.3390/biomedicines11061675
Najafi S, Sotoodehnejadnematalahi F, Amiri MM, Pourshafie MR, Rohani M. Prophylactic vs. Therapeutic Effect of Probiotics on the Inflammation Mediated by the NF-κB Pathway in Inflammatory Bowel Conditions. Biomedicines. 2023; 11(6):1675. https://doi.org/10.3390/biomedicines11061675
Chicago/Turabian StyleNajafi, Saeideh, Fattah Sotoodehnejadnematalahi, Mohammad Mehdi Amiri, Mohammad Reza Pourshafie, and Mahdi Rohani. 2023. "Prophylactic vs. Therapeutic Effect of Probiotics on the Inflammation Mediated by the NF-κB Pathway in Inflammatory Bowel Conditions" Biomedicines 11, no. 6: 1675. https://doi.org/10.3390/biomedicines11061675
APA StyleNajafi, S., Sotoodehnejadnematalahi, F., Amiri, M. M., Pourshafie, M. R., & Rohani, M. (2023). Prophylactic vs. Therapeutic Effect of Probiotics on the Inflammation Mediated by the NF-κB Pathway in Inflammatory Bowel Conditions. Biomedicines, 11(6), 1675. https://doi.org/10.3390/biomedicines11061675






