Integrating Proteomics and Lipidomics for Evaluating the Risk of Breast Cancer Progression: A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Characteristics of Study Population
2.2. Sample Collection
2.3. Targeted Proteomics via LC–MRM MS
2.4. Untargeted Lipidomics by LC–MS/MS
2.5. Statistical Analysis
3. Results
3.1. BC Metastasis Biomarkers in the Blood
3.2. Building of a Binary Classifiers for BC Metastasis Diagnosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S.; et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer, World Health Organization: Cancer Today/Population Fact Sheets: Russia. Available online: https://gco.iarc.fr/today/home (accessed on 18 January 2022).
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA. Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Akram, M.; Iqbal, M.; Daniyal, M.; Khan, A.U. Awareness and current knowledge of breast cancer. Biol. Res. 2017, 50, 33. [Google Scholar] [CrossRef]
- Gote, V.; Nookala, A.R.; Bolla, P.K.; Pal, D. Drug resistance in metastatic breast cancer: Tumor targeted nanomedicine to the rescue. Int. J. Mol. Sci. 2021, 22, 4673. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y. Breast Cancer Metastasis. Adv. Exp. Med. Biol. 2021, 1187, 183–204. [Google Scholar] [CrossRef]
- Morrow, M.; Waters, J.; Morris, E. MRI for breast cancer screening, diagnosis, and treatment. Lancet 2011, 378, 1804–1811. [Google Scholar] [CrossRef]
- Pesapane, F.; Suter, M.B.; Rotili, A.; Penco, S.; Nigro, O.; Cremonesi, M.; Bellomi, M.; Jereczek-Fossa, B.A.; Pinotti, G.; Cassano, E. Will traditional biopsy be substituted by radiomics and liquid biopsy for breast cancer diagnosis and characterisation? Med. Oncol. 2020, 37, 29. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, L.C.; Chen, J.Q. Sentinel lymph node biopsy compared with axillary lymph node dissection in early breast cancer: A meta-analysis. Breast Cancer Res. Treat. 2011, 129, 675–689. [Google Scholar] [CrossRef]
- Lyman, G.H.; Giuliano, A.E.; Somerfield, M.R.; Benson, A.B.; Bodurka, D.C.; Burstein, H.J.; Cochran, A.J.; Cody, H.S.; Edge, S.B.; Galper, S.; et al. American Society of Clinical Oncology guideline recommendations for sentinel lymph node biopsy in early-stage breast cancer. J. Clin. Oncol. 2005, 23, 7703–7720. [Google Scholar] [CrossRef]
- Nicolini, A.; Ferrari, P.; Duffy, M.J. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin. Cancer Biol. 2018, 52, 56–73. [Google Scholar] [CrossRef]
- Li, J.; Guan, X.; Fan, Z.; Ching, L.M.; Li, Y.; Wang, X.; Cao, W.M.; Liu, D.X. Non-invasive biomarkers for early detection of breast cancer. Cancers 2020, 12, 2767. [Google Scholar] [CrossRef] [PubMed]
- Najjar, S.; Allison, K.H. Updates on breast biomarkers. Virchows Arch. 2022, 480, 163–176. [Google Scholar] [CrossRef]
- Benacka, R.; Szabóová, D.; Gul’ašová, Z.; Hertelyová, Z.; Radonák, J. Classic and New Markers in Diagnostics and Classification of Breast Cancer. Cancers 2022, 14, 5444. [Google Scholar] [CrossRef]
- Rossi, C.; Cicalini, I.; Cufaro, M.C.; Consalvo, A.; Upadhyaya, P.; Sala, G.; Antonucci, I.; Del Boccio, P.; Stuppia, L.; De Laurenzi, V. Breast cancer in the era of integrating “Omics” approaches. Oncogenesis 2022, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.B.; Kang, U.B.; Moon, H.G.; Lee, J.; Lee, K.M.; Yi, M.; Park, Y.S.; Lee, J.W.; Yu, J.H.; Choi, S.H.; et al. Development and validation of a novel plasma protein signature for breast cancer diagnosis by using multiple reaction monitoring-based mass spectrometry. Anticancer. Res. 2015, 35, 6271–6279. [Google Scholar] [PubMed]
- Kim, Y.; Kang, U.B.; Kim, S.; Lee, H.B.; Moon, H.G.; Han, W.; Noh, D.Y. A validation study of a multiple reaction monitoring-based proteomic assay to diagnose breast cancer. J. Breast Cancer 2020, 23, 113–114. [Google Scholar] [CrossRef]
- Lee, E.S.; Kim, Y.; Shin, H.C.; Hwang, K.T.; Min, J.; Kim, M.K.; Ahn, S.K.; Jung, S.Y.; Shin, H.; Chung, M.S.; et al. Diagnostic accuracy of a three-protein signature in women with suspicious breast lesions: A multicenter prospective trial. Breast Cancer Res. 2023, 25, 20. [Google Scholar] [CrossRef]
- Sunshine, H.; Iruela-Arispe, M.L. Membrane Lipids and Cell Signaling. Curr. Opin. Lipidol. 2017, 28, 408–413. [Google Scholar] [CrossRef]
- Vasseur, S.; Guillaumond, F. Lipids in cancer: A global view of the contribution of lipid pathways to metastatic formation and treatment resistance. Oncogenesis 2022, 11, 46. [Google Scholar] [CrossRef]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-Y.; Lee, K.-M.; Kim, S.-H.; Kwon, Y.-J.; Chun, Y.-J.; Choi, H.-K. Comparative metabolic and lipidomic profiling of human breast cancer cells with different metastatic potentials. Oncotarget 2016, 7, 67111. [Google Scholar] [CrossRef] [PubMed]
- Eiriksson, F.F.; Rolfsson, O.; Ogmundsdottir, H.M.; Haraldsson, G.G.; Thorsteinsdottir, M.; Halldorsson, S. Altered plasmalogen content and fatty acid saturation following epithelial to mesenchymal transition in breast epithelial cell lines. Int. J. Biochem. Cell Biol. 2018, 103, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Chen, Z.; Feng, H.; Chen, Y.; Zhang, C.; Yu, J.; Luo, Y.; Zhao, L.; Jiang, X.; Shi, F. Sphingomyelin synthase 2 promotes an aggressive breast cancer phenotype by disrupting the homoeostasis of ceramide and sphingomyelin. Cell Death Dis. 2019, 10, 157. [Google Scholar] [CrossRef]
- Nouri, M.; Mohsenpour, M.A.; Katsiki, N.; Ghobadi, S.; Jafari, A.; Faghih, S.; Banach, M.; Mazidi, M. Effect of Serum Lipid Profile on the Risk of Breast Cancer: Systematic Review and Meta-Analysis of 1,628,871 Women. J. Clin. Med. 2022, 11, 4503. [Google Scholar] [CrossRef]
- Ghahremanfard, F.; Mirmohammadkhani, M.; Shahnazari, B.; Gholami, G.; Mehdizadeh, J. The valuable role of measuring serum lipid profile in cancer progression. Oman Med. J. 2015, 30, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zheng, X.; Zhou, Q.; Villanueva, N.; Nian, W.; Liu, X.; Huan, T. Metabolomics-Based Discovery of Molecular Signatures for Triple Negative Breast Cancer in Asian Female Population. Sci. Rep. 2020, 10, 370. [Google Scholar] [CrossRef]
- Whiteaker, J.R.; Halusa, G.N.; Hoofnagle, A.N.; Sharma, V.; MacLean, B.; Yan, P.; Wrobel, J.A.; Kennedy, J.; Mani, D.R.; Zimmerman, L.J.; et al. CPTAC Assay Portal: A repository of targeted proteomic assays. Nat. Methods 2014, 11, 703–704. [Google Scholar] [CrossRef]
- Gaither, C.; Popp, R.; Borchers, S.P.; Skarphedinsson, K.; Eiriksson, F.F.; Thorsteinsdóttir, M.; Mohammed, Y.; Borchers, C.H. Performance Assessment of a 125 Human Plasma Peptide Mixture Stored at Room Temperature for Multiple Reaction Monitoring-Mass Spectrometry. J. Proteome Res. 2021, 20, 4292–4302. [Google Scholar] [CrossRef]
- Starodubtseva, N.; Chagovets, V.; Borisova, A.; Salimova, D.; Aleksandrova, N.; Chingin, K.; Chen, H.; Frankevich, V. Identification of potential endometriosis biomarkers in peritoneal fluid and blood plasma via shotgun lipidomics. Clin. Mass Spectrom. 2019, 13, 21–26. [Google Scholar] [CrossRef]
- Chambers, M.C.; MacLean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Koelmel, J.P.; Kroeger, N.M.; Ulmer, C.Z.; Bowden, J.A.; Patterson, R.E.; Cochran, J.A.; Beecher, C.W.W.; Garrett, T.J.; Yost, R.A. LipidMatch: An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinform. 2017, 18, 331. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, 527–532. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Guyon, I.; Elisseeff, A. An Introduction of Variable and Feature Selection An Introduction to Variable and Feature Selection 1 Introduction. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar] [CrossRef]
- Vuskovic, M.; Xu, H.; Bovin, N.; Pass, H.; Huflejt, M. Processing and analysis of Printed Glycan Array data for early detection, diagnosis and prognosis of cancers. Int. J. Bioinform. Res. Appl. 2011, 7, 402–426. [Google Scholar] [CrossRef]
- Lomova, N.; Dolgushina, N.; Tokareva, A.; Chagovets, V.; Starodubtseva, N. Past COVID-19: The Impact on IVF Outcomes Based on Follicular Fluid Lipid Profile. Int. J. Mol. Sci. 2023, 24, 10. [Google Scholar] [CrossRef]
- Tokareva, A.O.; Chagovets, V.V.; Kononikhin, A.S.; Starodubtseva, N.L.; Nikolaev, E.N.; Frankevich, V.E. Comparison of the effectiveness of variable selection method for creating a diagnostic panel of biomarkers for mass spectrometric lipidome analysis. J. Mass Spectrom. 2021, 56, e4702. [Google Scholar] [CrossRef]
- Lyman, G.H.; Temin, S.; Edge, S.B.; Newman, L.A.; Turner, R.R.; Weaver, D.L.; Benson, A.B.; Bosserman, L.D.; Burstein, H.J.; Cody, H.; et al. Sentinel lymph node biopsy for patients with early-stage breast cancer: American Society of Clinical Oncology clinical practice guideline update. J. Clin. Oncol. 2014, 32, 1365–1383. [Google Scholar] [CrossRef]
- Gentilini, O.; Veronesi, U. Staging the axilla in early breast cancer: Will imaging replace surgery? JAMA Oncol. 2015, 1, 1031–1032. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gao, Y.; Gong, H.; Feng, W.; Ma, Q.; Li, J.; Lu, X.; Wang, X.; Lei, J. Different Imaging Modalities for the Diagnosis of Axillary Lymph Node Metastases in Breast Cancer: A Systematic Review and Network Meta-Analysis of Diagnostic Test Accuracy. J. Magn. Reson. Imaging 2022, 57, 1392–1403. [Google Scholar] [CrossRef]
- Sukhikh, G.T.; Sencha, A.N. Multiparametric Ultrasound Diagnosis of Breast Diseases; Springer: Cham, Switzerland, 2018; ISBN 9783319750347. [Google Scholar]
- Wang, R.; Zhang, T.; MA, Z.; Wang, Y.; Cheng, Z.; Xu, H.; Li, W.; Wang, X. The interaction of coagulation factor XII and monocyte/macrophages mediating peritoneal metastasis of epithelial ovarian cancer. Gynecol. Oncol. 2010, 117, 460–466. [Google Scholar] [CrossRef]
- Renné, T.; Stavrou, E.X. Roles of factor XII in innate immunity. Front. Immunol. 2019, 10, 2011. [Google Scholar] [CrossRef] [PubMed]
- Todd, J.R.; Ryall, K.A.; Vyse, S.; Wong, J.P.; Natrajan, R.C.; Yuan, Y.; Tan, A.C.; Huang, P.H. Systematic analysis of tumour cell-extracellular matrix adhesion identifies independent prognostic factors in breast cancer. Oncotarget 2016, 7, 62939–62953. [Google Scholar] [CrossRef] [PubMed]
- Florea, G.; Tudorache, I.F.; Fuior, E.V.; Ionita, R.; Dumitrescu, M.; Fenyo, I.M.; Bivol, V.G.; Gafencu, A.V. Apolipoprotein A-II, a Player in Multiple Processes and Diseases. Biomedicines 2022, 10, 1578. [Google Scholar] [CrossRef]
- Furlaneto, C.J.; Ribeiro, F.P.; Hatanaka, E.; Souza, G.M.; Cassatella, M.A.; Campa, A. Apolipoproteins A-I and A-II downregulate neutrophil functions. Lipids 2002, 37, 925–928. [Google Scholar] [CrossRef]
- Wolska, A.; Dunbarb, R.L.; Freeman, L.A.; Ueda, M.; Amar, M.J.; Sviridov, D.O.; Remaleya, A.T. Apolipoprotein C-II: New findings related to genetics, biochemistry, and role in triglyceride metabolism. Atherosclerosis 2017, 267, 49–60. [Google Scholar] [CrossRef]
- Kotite, L.; Zhang, L.H.; Yu, Z.; Burlingame, A.L.; Havel, R.J. Human apoC-IV: Isolation, characterization, and immunochemical quantification in plasma and plasma lipoproteins. J. Lipid Res. 2003, 44, 1387–1394. [Google Scholar] [CrossRef]
- Harima, Y.; Ariga, T.; Kaneyasu, Y.; Ikushima, H.; Tokumaru, S.; Shimamoto, S.; Takahashi, T.; Ii, N.; Tsujino, K.; Saito, A.I.; et al. Clinical value of serum biomarkers, squamous cell carcinoma antigen and apolipoprotein C-II in follow-up of patients with locally advanced cervical squamous cell carcinoma treated with radiation: A multicenter prospective cohort study. PLoS ONE 2021, 16, e0259235. [Google Scholar] [CrossRef]
- Xue, A.; Chang, J.W.; Chung, L.; Samra, J.; Hugh, T.; Gill, A.; Butturini, G.; Baxter, R.C.; Smith, R.C. Serum apolipoprotein C-II is prognostic for survival after pancreatic resection for adenocarcinoma. Br. J. Cancer 2012, 107, 1883–1891. [Google Scholar] [CrossRef] [PubMed]
- Güven, H.E.; Doğan, L.; Gülçelik, M.A.; Ersöz Gülçelik, N. Adiponectin: A Predictor for Breast Cancer Survival? Eur. J. Breast Health 2019, 15, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Pham, D.V.; Park, P.H. Adiponectin triggers breast cancer cell death via fatty acid metabolic reprogramming. J. Exp. Clin. Cancer Res. 2022, 41, 9. [Google Scholar] [CrossRef]
- Lira, L.G.; Justa, R.M.D.E.; Carioca, A.A.F.; Verde, S.M.M.L.; Sampaio, G.R.; da Torres, E.A.F.S.; Damasceno, N.R.T. Plasma and erythrocyte ω-3 and ω-6 fatty acids are associated with multiple inflammatory and oxidative stress biomarkers in breast cancer. Nutrition 2019, 58, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, Y.; Funahashi, T.; Kihara, S.; Taguchi, T.; Tamaki, Y.; Matsuzawa, Y.; Noguchi, S. Association of Serum Adiponectin Levels with Breast Cancer Risk. Clin. Cancer Res. 2003, 9, 5699–5704. [Google Scholar]
- Jemmerson, R.; Staskus, K.; Higgins, L.; Conklin, K.; Kelekar, A. Intracellular leucine-rich alpha-2-glycoprotein-1 competes with Apaf-1 for binding cytochrome c in protecting MCF-7 breast cancer cells from apoptosis. Apoptosis 2021, 26, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, K.; Narumi, R.; Nagayama, S.; Masuda, K.; Esaki, T.; Obama, K.; Tomonaga, T.; Sakai, Y.; Shimizu, Y.; Adachi, J. A large-scale targeted proteomics of plasma extracellular vesicles shows utility for prognosis prediction subtyping in colorectal cancer. Cancer Med. 2022, 12, 7616–7626. [Google Scholar] [CrossRef]
- Zhou, Q.; Andersson, R.; Hu, D.; Bauden, M.; Sasor, A.; Bygott, T.; PawŁowski, K.; Pla, I.; Marko-Varga, G.; Ansari, D. Alpha-1-acid glycoprotein 1 is upregulated in pancreatic ductal adenocarcinoma and confers a poor prognosis. Transl. Res. 2019, 212, 67–79. [Google Scholar] [CrossRef]
- Otsuru, T.; Kobayashi, S.; Wada, H.; Takahashi, T.; Gotoh, K.; Iwagami, Y.; Yamada, D.; Noda, T.; Asaoka, T.; Serada, S.; et al. Epithelial-mesenchymal transition via transforming growth factor beta in pancreatic cancer is potentiated by the inflammatory glycoprotein leucine-rich alpha-2 glycoprotein. Cancer Sci. 2019, 110, 985–996. [Google Scholar] [CrossRef]
- Kwan, Y.P.; Teo, M.H.Y.; Lim, J.C.W.; Tan, M.S.; Rosellinny, G.; Wahli, W.; Wang, X. Lrg1 promotes metastatic dissemination of melanoma through regulating egfr/stat3 signalling. Cancers 2021, 13, 3279. [Google Scholar] [CrossRef]
- Zhong, B.; Cheng, B.; Huang, X.; Xiao, Q.; Niu, Z.; Chen, Y.; Yu, Q.; Wang, W.; Wu, X.J. Colorectal cancer-associated fibroblasts promote metastasis by up-regulating LRG1 through stromal IL-6/STAT3 signaling. Cell Death Dis. 2022, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Verathamjamras, C.; Chantaraamporn, J.; Sornprachum, T.; Mutapat, P.; Chokchaichamnankit, D.; Mingkwan, K.; Luevisadpibul, V.; Srisomsap, C.; Chutipongtanate, S.; Svasti, J.; et al. Label-free quantitative proteomics reveals aberrant expression levels of LRG, C9, FN, A1AT and AGP1 in the plasma of patients with colorectal cancer. Clin. Proteom. 2023, 20, 15. [Google Scholar] [CrossRef] [PubMed]
- Icer, M.A.; Yıldıran, H. Effects of fetuin-A with diverse functions and multiple mechanisms on human health. Clin. Biochem. 2021, 88, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.H.; Akhtar, J.; Arora, J.; Saran, R.K.; Mishra, N.; Polisetty, R.V.; Sirdeshmukh, R.; Gautam, P. Quantitative proteomic analysis of GnRH agonist treated GBM cell line LN229 revealed regulatory proteins inhibiting cancer cell proliferation. BMC Cancer 2022, 22, 133. [Google Scholar] [CrossRef] [PubMed]
- Misra, U.K.; Pizzo, S.V. Activated α2-macroglobulin binding to human prostate cancer cells triggers insulin-like responses. J. Biol. Chem. 2015, 290, 9571–9587. [Google Scholar] [CrossRef]
- Roeise, O.; Sivertsen, S.; Ruud, T.E.; Bouma, B.N.; Stadaas, J.O.; Aasen, A.O. Studies on components of the contact phase system in patients with advanced gastrointestinal cancer. Cancer 1990, 65, 1355–1359. [Google Scholar] [CrossRef]
- Chantaraamporn, J.; Champattanachai, V.; Khongmanee, A.; Verathamjamras, C.; Prasongsook, N.; Mingkwan, K.; Luevisadpibul, V.; Chutipongtanate, S.; Svasti, J. Glycoproteomic analysis reveals aberrant expression of complement C9 and fibronectin in the plasma of patients with colorectal cancer. Proteomes 2020, 8, 26. [Google Scholar] [CrossRef]
- Tsumita, T.; Maishi, N.; Annan, D.A.M.; Towfik, M.A.; Matsuda, A.; Onodera, Y.; Nam, J.M.; Hida, Y.; Hida, K. The oxidized-LDL/LOX-1 axis in tumor endothelial cells enhances metastasis by recruiting neutrophils and cancer cells. Int. J. Cancer 2022, 151, 944–956. [Google Scholar] [CrossRef]
- He, X.; Deng, T.; Li, J.; Guo, R.; Wang, Y.; Li, T.; Zang, S.; Li, J.; Zhang, L.; Li, M.; et al. A core-satellite micellar system against primary tumors and their lymphatic metastasis through modulation of fatty acid metabolism blockade and tumor-associated macrophages. Nanoscale 2023, 15, 8320–8336. [Google Scholar] [CrossRef]
- Cao, W.; Ramakrishnan, R.; Tuyrin, V.A.; Veglia, F.; Condamine, T.; Amoscato, A.; Mohammadyani, D.; Johnson, J.J.; Zhang, L.M.; Klein-Seetharaman, J.; et al. Oxidized Lipids Block Antigen Cross-Presentation by Dendritic Cells in Cancer. J. Immunol. 2014, 192, 4935. [Google Scholar] [CrossRef]
- Tallima, H.; Azzazy, H.M.E.; El Ridi, R. Cell surface sphingomyelin: Key role in cancer initiation, progression, and immune evasion. Lipids Health Dis. 2021, 20, 150. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.M.; Mahon, K.L.; Weir, J.M.; Mundra, P.A.; Spielman, C.; Briscoe, K.; Gurney, H.; Mallesara, G.; Marx, G.; Stockler, M.R.; et al. A distinct plasma lipid signature associated with poor prognosis in castration-resistant prostate cancer. Int. J. Cancer 2017, 141, 2112–2120. [Google Scholar] [CrossRef] [PubMed]
- Feingold, K.R. Introduction to Lipids and Lipoproteins. In Endotext; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Hofland, J., et al., Eds.; MDText. com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Brown, A.J. Cholesterol, statins and cancer. Clin. Exp. Pharmacol. Physiol. 2007, 34, 135–141. [Google Scholar] [CrossRef]
- Menendez, J.A. Fine-tuning the lipogenic/lipolytic balance to optimize the metabolic requirements of cancer cell growth: Molecular mechanisms and therapeutic perspectives. BBA Mol. Cell Biol. Lipids 2010, 1801, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ren, S.; Piao, H.; Wang, F.; Yin, P.; Xu, C.; Lu, X.; Ye, G.; Shao, Y.; Yan, M.; et al. Integration of lipidomics and transcriptomics unravels aberrant lipid metabolism and defines cholesteryl oleate as potential biomarker of prostate cancer. Nat. Publ. Gr. 2016, 6, 20984. [Google Scholar] [CrossRef]
- Rousseau, A.; Larsen, A.K.; Van Dreden, P.; Sabbah, M.; Elalamy, I.; Gerotziafas, G.T. Differential contribution of tissue factor and Factor XII to thrombin generation triggered by breast and pancreatic cancer cells. Int. J. Oncol. 2017, 51, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Campello, E.; Henderson, M.W.; Noubouossie, D.F.; Simioni, P.; Key, N.S. Contact System Activation and Cancer: New Insights in the Pathophysiology of Cancer-Associated Thrombosis. Thromb. Haemost. 2018, 118, 251–265. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, M.; Zhang, C.; Zhou, S.; Ji, G. Molecular Functions of Ceruloplasmin in Metabolic Disease Pathology. Diabetes Metab. Syndr. Obes. 2022, 15, 695–711. [Google Scholar] [CrossRef]
- Kim, S.Y.; Choi, Y.; Kim, Y.S.; Ha, S.M.; Lee, S.H.; Han, W.; Kim, H.-K.; Cho, N.; Moon, W.K.; Chang, J.M. Use of imaging prediction model for omission of axillary surgery in early-stage breast cancer patients. Breast Cancer Res. Treat. 2023, 199, 489–499. [Google Scholar] [CrossRef]
- Bode, M.; Schrading, S.; Masoumi, A.; Morscheid, S.; Schacht, S.; Dirrichs, T.; Gaisa, N.; Stickeler, E.; Kuhl, C.K. Abbreviated MRI for Comprehensive Regional Lymph Node Staging during Pre-Operative Breast MRI. Cancers 2023, 15, 1859. [Google Scholar] [CrossRef]
- Gao, J.; Zhong, X.; Li, W.; Li, Q.; Shao, H.; Wang, Z.; Dai, Y.; Ma, H.; Shi, Y.; Zhang, H.; et al. Attention-based Deep Learning for the Preoperative Differentiation of Axillary Lymph Node Metastasis in Breast Cancer on DCE-MRI. J. Magn. Reson. Imaging 2022, 57, 1842–1853. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, X.; Lan, X.; Yu, T.; Li, L.; Tang, S.; Liu, S.; Jiang, F.; Wang, L.; Zhang, J. A radiomics model development via the associations with genomics features in predicting axillary lymph node metastasis of breast cancer: A study based on a public database and single-centre verification. Clin. Radiol. 2023, 78, e279–e287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, T.; Zhang, S.; Sun, J.; Zhang, F.; Li, X.; Ni, X. Prediction of Axillary Lymph Node Metastatic Load of Breast Cancer Based on Ultrasound Deep Learning Radiomics Nomogram. Technol. Cancer Res. Treat. 2023, 22, 15330338231166218. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cao, G.; Pang, H.; Li, J.; Yao, X. Development and validation of radiomics machine learning model based on contrast-enhanced computed tomography to predict axillary lymph node metastasis in breast cancer. Bosn. J. Basic Med. Sci. 2022, 23, 317–326. [Google Scholar] [CrossRef]
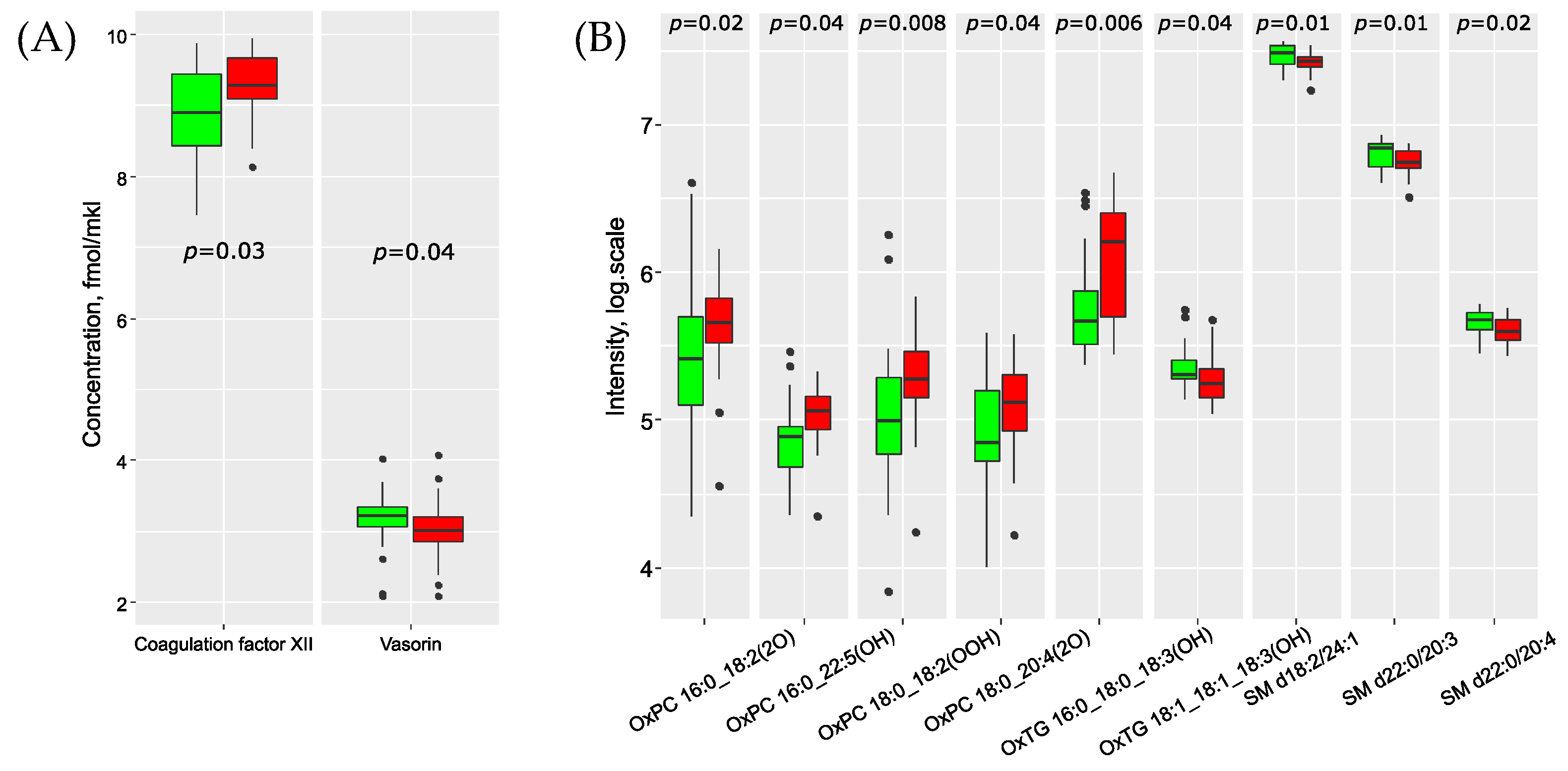
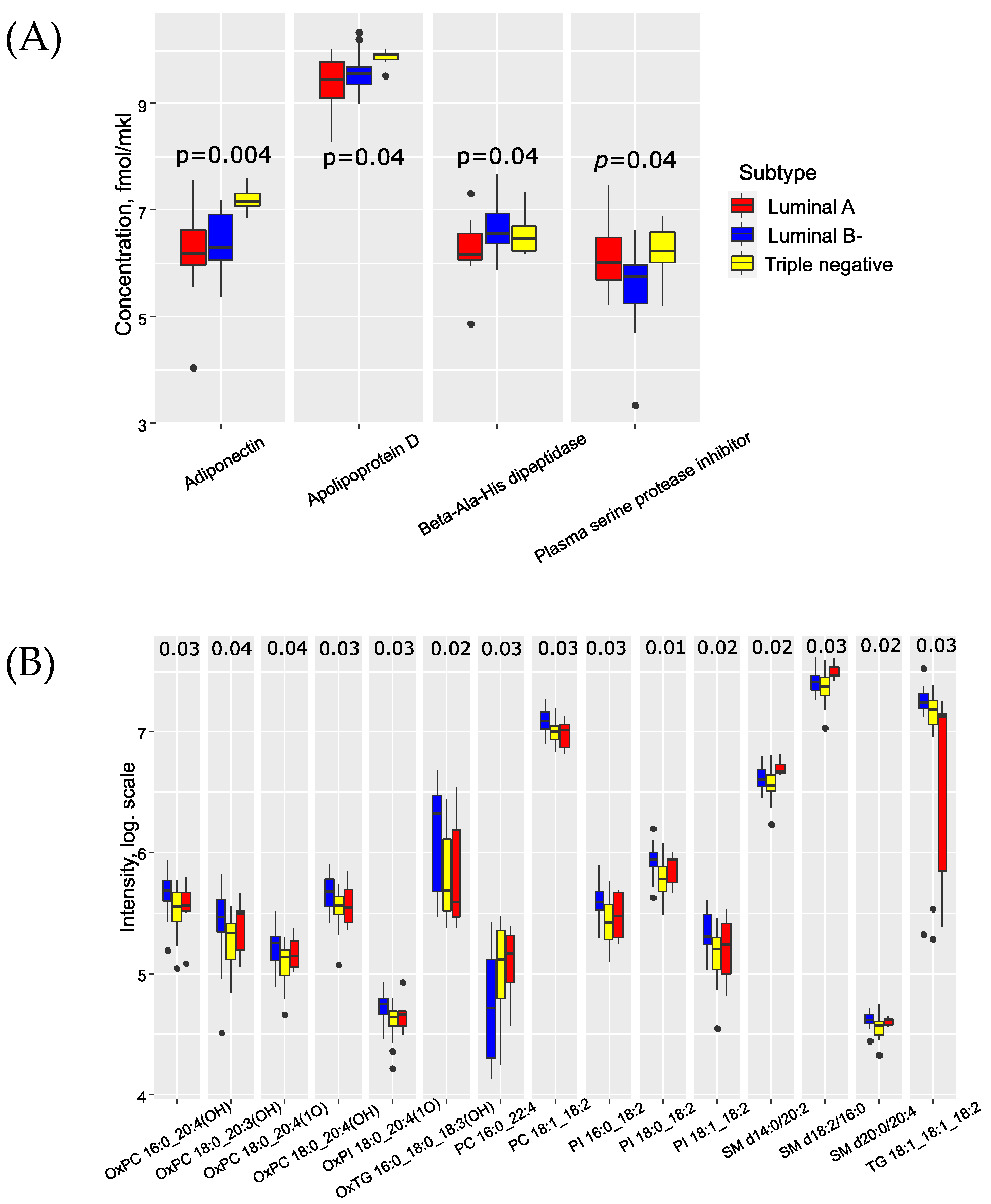
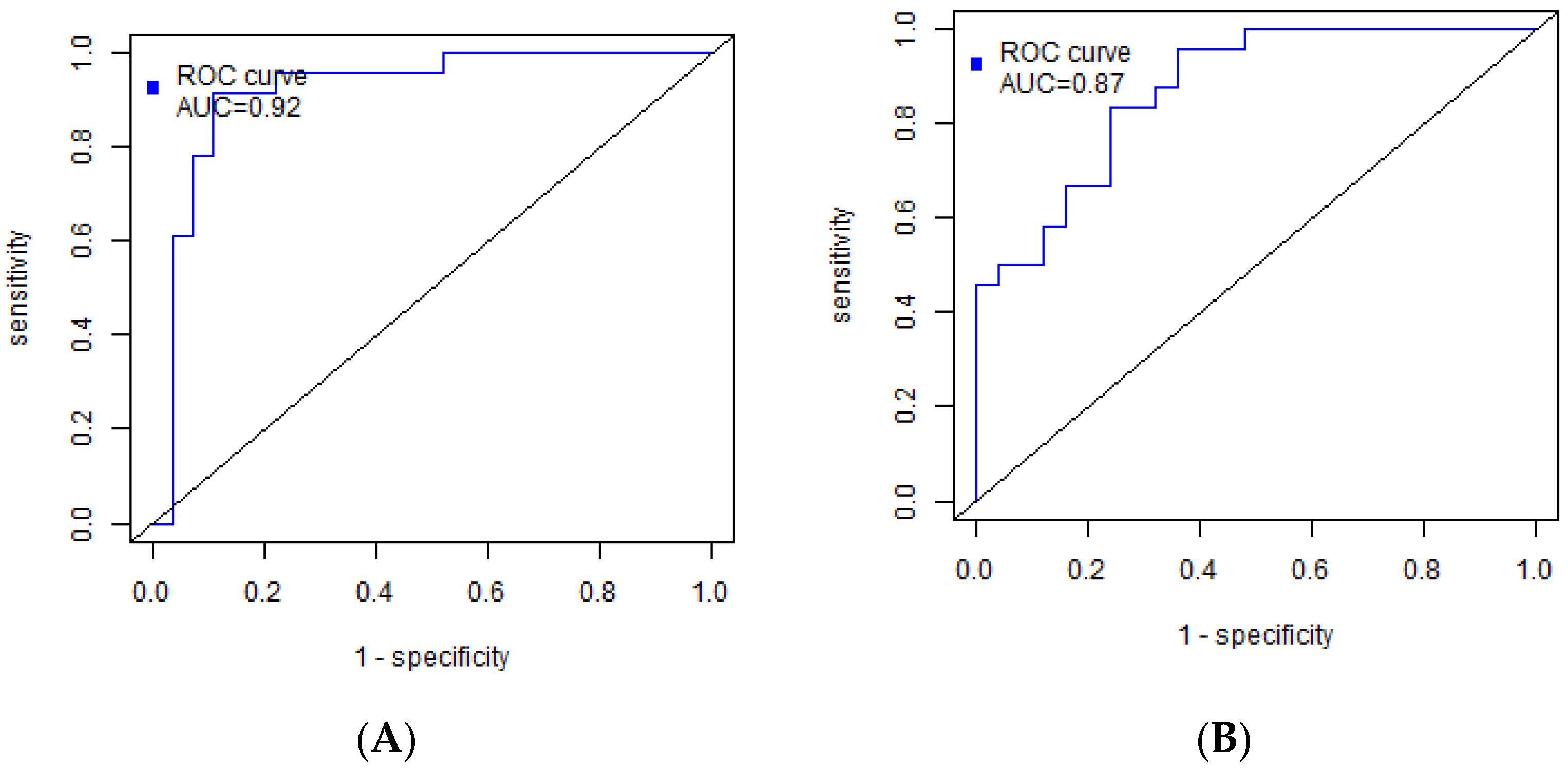
| Parameter | Metastases-Free Group (n = 25) | Group with Metastases (n = 25) | p-Value |
|---|---|---|---|
| Age (years) | 60 (52; 63) | 56 (44; 60) | 0.11 |
| Length of tumor (cm) | 2.1 (1.6; 2.4) | 2.4 (1.9; 3.0) | 0.13 |
| Biological subtype: | 0.47 | ||
| Luminal A | 8 (32.0%) | 10 (40.0%) | |
| Luminal B− | 11 (44.0%) | 12 (48.0%) | |
| Luminal B+ | 1 (4.0%) | 0 (0.0%) | |
| Her2+ | 0 (0.0%) | 1 (4.0%) | |
| TNBC | 5 (20.0%) | 2 (8.0%) | |
| Histological type: | 0.70 | ||
| 6 (24.0%) | 6 (24.0%) | |
| 5 (20.0%) | 4 (16.0%) | |
| 11 (44.0%) | 14 (56.0%) | |
| 3 (12.0%) | 1 (4.0%) | |
| Grade, G: | 0.20 | ||
| 3 (12.0%) | 1 (4.0%) | |
| 13 (52.0%) | 19 (76.0%) | |
| 9 (36.0%) | 5 (20.0%) | |
| Multifocality (>1 tumor): | 0.66 | ||
| 4 (16.0%) | 2 (8.0%) | |
| 21 (84.0%) | 23 (92.0%) | |
| Stage: | <0.001 | ||
| Ia: | 13 (52.0%) | 0 | |
| Ib: | 0 | 1 (4.0%) | |
| IIa | 11 (44.0%) | 3 (12.0%) | |
| IIb | 1 (4.0%) | 13 (52.0%) | |
| IIIa | 0 | 4 (16.0%) | |
| IIIb | 0 | 4 (16.0%) | |
| Total malignancy score (TMS) | 15 (13; 16) | 15 (14; 16) | 0.15 |
| Nottingham predictive index (NPI) | 3.4 (3.3; 4.4) | 4.7 (4.5; 5.4) | <0.001 |
| Number of metastases to regional lymph nodes | 0 | 2 (1; 5) | <0.001 |
| Estrogen receptor (ER) expression: | 8 (7; 8) | 8 (7; 8) | 0.60 |
| Progesterone receptor (PR) expression: | 7 (0; 7) | 4 (2; 8) | 1.00 |
| HER2 expression: | 1.00 | ||
| 2 (8.0%) | 1 (4.0%) | |
| 23 (92.0%) | 24 (96.0%) | |
| Level of Ki67, % | 28.0 (14.0; 45.0) | 22.0 (15.0; 38.0) | 0.96 |
| Variable | β | CI β | Z | p |
|---|---|---|---|---|
| Intercept | −32.32 | −75.78–−8.25 | −2.00 | 0.04 |
| Alpha-2-macroglobulin × Coagulation factor XII | 0.69 | 0.33–1.33 | 2.87 | 0.004 |
| Adiponectin × Leucine-rich alpha-2-glycoprotein | −1.71 | −3.81–−0.75 | −2.45 | 0.01 |
| Alpha-2-HS-glycoprotein × Ig mu chain C region | 0.34 | 0.14–0.81 | 2.16 | 0.03 |
| Apolipoprotein C-IV × Carbonic anhydrase 1 | −0.39 | −0.86–−0.15 | −2.33 | 0.02 |
| Apolipoprotein A-II × Apolipoprotein C-II | −0.09 | −0.20–−0.03 | −2.41 | 0.02 |
| Adiponectin × Alpha-1-acid glycoprotein 1 | 0.79 | 0.24–1.86 | 2.09 | 0.04 |
| Variable | β | CI β | Z | p |
|---|---|---|---|---|
| Intercept | 9.52 | 3.47–19.42 | 2.43 | 0.02 |
| OxTG 16:0_18:0_18:3(OH) × OxTG 18:1_18:1_18:1(Ke,OH) | 1.38 × 10−12 | 4.65 × 10−13–3.09 × 10−12 | 2.05 | 0.04 |
| SM d18:2/24:1 × TG 16:0_16:1_18:1 | −2.06 × 10−14 | −4.17 × 10−14–−7.73 × 10−15 | −2.46 | 0.01 |
| PC 18:0_22:6 × TG 18:1_18:1_18:2 | −1.12 × 10−14 | −2.26 × 10−14–−3.34 × 10−15 | −2.41 | 0.02 |
| OxTG 18:1_18:1_18:2(OOH) × PC 16:1_20:4 | 3.41 × 10−14 | 1.03 × 10−14–6.68 × 10−14 | 2.45 | 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Starodubtseva, N.L.; Tokareva, A.O.; Rodionov, V.V.; Brzhozovskiy, A.G.; Bugrova, A.E.; Chagovets, V.V.; Kometova, V.V.; Kukaev, E.N.; Soares, N.C.; Kovalev, G.I.; et al. Integrating Proteomics and Lipidomics for Evaluating the Risk of Breast Cancer Progression: A Pilot Study. Biomedicines 2023, 11, 1786. https://doi.org/10.3390/biomedicines11071786
Starodubtseva NL, Tokareva AO, Rodionov VV, Brzhozovskiy AG, Bugrova AE, Chagovets VV, Kometova VV, Kukaev EN, Soares NC, Kovalev GI, et al. Integrating Proteomics and Lipidomics for Evaluating the Risk of Breast Cancer Progression: A Pilot Study. Biomedicines. 2023; 11(7):1786. https://doi.org/10.3390/biomedicines11071786
Chicago/Turabian StyleStarodubtseva, Natalia L., Alisa O. Tokareva, Valeriy V. Rodionov, Alexander G. Brzhozovskiy, Anna E. Bugrova, Vitaliy V. Chagovets, Vlada V. Kometova, Evgenii N. Kukaev, Nelson C. Soares, Grigoriy I. Kovalev, and et al. 2023. "Integrating Proteomics and Lipidomics for Evaluating the Risk of Breast Cancer Progression: A Pilot Study" Biomedicines 11, no. 7: 1786. https://doi.org/10.3390/biomedicines11071786
APA StyleStarodubtseva, N. L., Tokareva, A. O., Rodionov, V. V., Brzhozovskiy, A. G., Bugrova, A. E., Chagovets, V. V., Kometova, V. V., Kukaev, E. N., Soares, N. C., Kovalev, G. I., Kononikhin, A. S., Frankevich, V. E., Nikolaev, E. N., & Sukhikh, G. T. (2023). Integrating Proteomics and Lipidomics for Evaluating the Risk of Breast Cancer Progression: A Pilot Study. Biomedicines, 11(7), 1786. https://doi.org/10.3390/biomedicines11071786









