Type II Transmembrane Serine Proteases as Modulators in Adipose Tissue Phenotype and Function
Abstract
1. Introduction
2. TTSPs: General Background
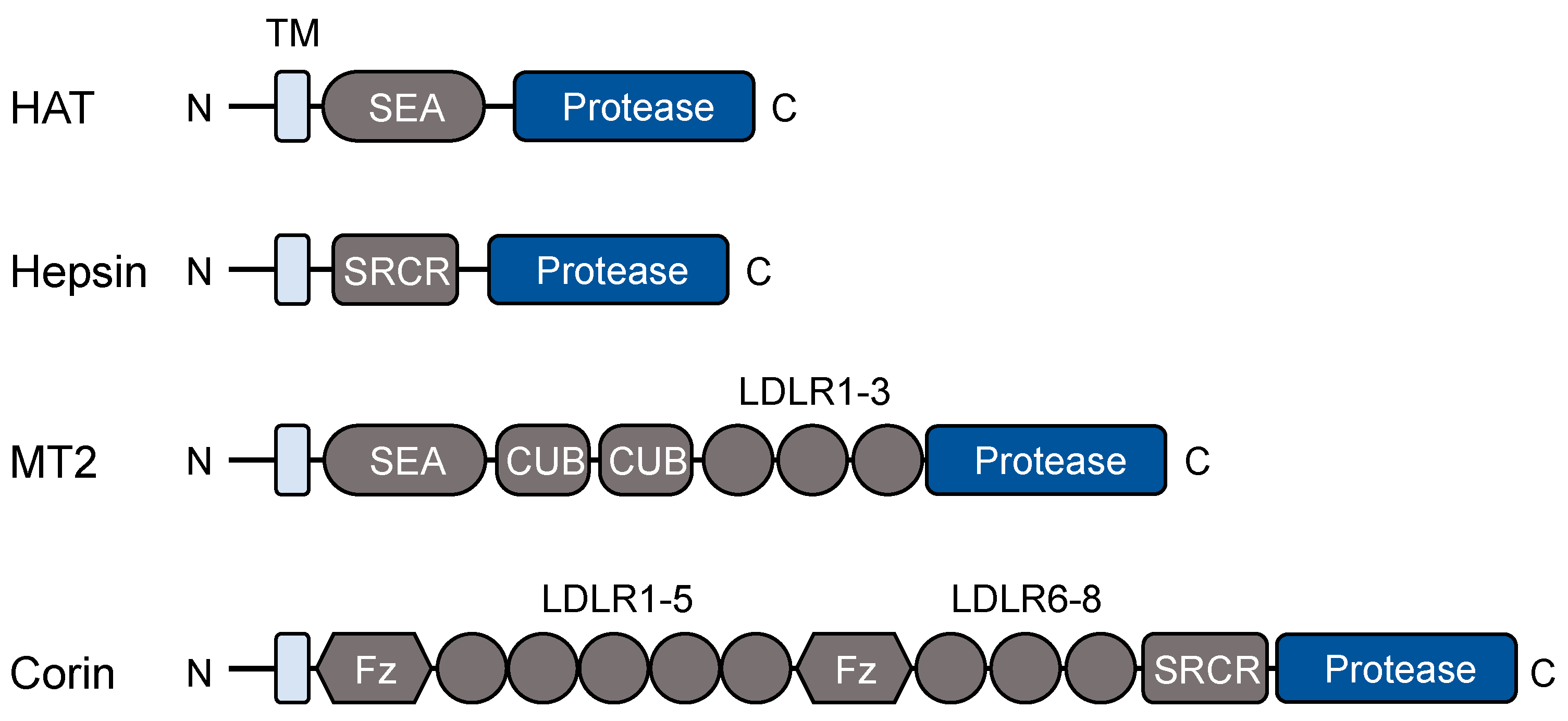
2.1. Protein Domains and Post-Translational Modifications
2.2. Physiological Functions
3. TTSP Expression in Adipose Tissues
4. Hepsin in Adipose Tissue Differentiation
4.1. Hepsin Protein and Function
4.2. Role of Hepsin in Adipose Tissue Browning
4.3. Regulation of Hepsin Expression in Adipose Tissue
5. Matriptase-2 in Iron Metabolism and Adiposity
5.1. Matriptase-2 in Iron Metabolism
5.2. Matriptase-2 in Lipolysis and Obesity
6. Corin in Adipose Tissue Phenotype and Thermogenesis
6.1. Corin in Pro-ANP Processing
6.2. Role of ANP in Lipid Metabolism in Adipose Tissue
6.3. Role of ANP in Adipose Tissue Browning and Thermogenesis
6.4. Role of ANP in Adipose Tissue Inflammation
6.5. Impaired Adipose Tissue Browning and Thermogenesis in Corin KO Mice
7. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sakers, A.; De Siqueira, M.K.; Seale, P.; Villanueva, C.J. Adipose-tissue plasticity in health and disease. Cell 2022, 185, 419–446. [Google Scholar] [CrossRef]
- Scheja, L.; Heeren, J. The endocrine function of adipose tissues in health and cardiometabolic disease. Nat. Rev. Endocrinol. 2019, 15, 507–524. [Google Scholar] [CrossRef] [PubMed]
- Perona, J.J.; Craik, C.S. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995, 4, 337–360. [Google Scholar] [CrossRef] [PubMed]
- Antalis, T.M.; Buzza, M.S.; Hodge, K.M.; Hooper, J.D.; Netzel-Arnett, S. The cutting edge: Membrane-anchored serine protease activities in the pericellular microenvironment. Biochem. J. 2010, 428, 325–346. [Google Scholar] [CrossRef] [PubMed]
- Szabo, R.; Bugge, T.H. Membrane-anchored serine proteases in vertebrate cell and developmental biology. Annu. Rev. Cell. Dev. Biol. 2011, 27, 213–235. [Google Scholar] [CrossRef] [PubMed]
- Antalis, T.M.; Conway, G.D.; Peroutka, R.J.; Buzza, M.S. Membrane-anchored proteases in endothelial cell biology. Curr. Opin. Hematol. 2016, 23, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.E.; List, K. Cell surface-anchored serine proteases in cancer progression and metastasis. Cancer Metastasis Rev. 2019, 38, 357–387. [Google Scholar] [CrossRef]
- Overall, C.M.; Blobel, C.P. In search of partners: Linking extracellular proteases to substrates. Nat. Rev. Mol. Cell. Biol. 2007, 8, 245–257. [Google Scholar] [CrossRef]
- Afar, D.E.; Vivanco, I.; Hubert, R.S.; Kuo, J.; Chen, E.; Saffran, D.C.; Raitano, A.B.; Jakobovits, A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001, 61, 1686–1692. [Google Scholar]
- Zhang, C.; Zhang, Y.; Zhang, S.; Wang, Z.; Sun, S.; Liu, M.; Chen, Y.; Dong, N.; Wu, Q. Intracellular autoactivation of TMPRSS11A, an airway epithelial transmembrane serine protease. J. Biol. Chem. 2020, 295, 12686–12696. [Google Scholar] [CrossRef]
- Murray, A.S.; Varela, F.A.; Hyland, T.E.; Schoenbeck, A.J.; White, J.M.; Tanabe, L.M.; Todi, S.V.; List, K. Phosphorylation of the type II transmembrane serine protease, TMPRSS13, in hepatocyte growth factor activator inhibitor-1 and -2-mediated cell-surface localization. J. Biol. Chem. 2017, 292, 14867–14884. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, C.; Sun, S.; Chen, Y.; Hu, Y.; Wang, H.; Liu, M.; Dong, N.; Wu, Q. Autoactivation and calpain-1-mediated shedding of hepsin in human hepatoma cells. Biochem. J. 2019, 476, 2355–2369. [Google Scholar] [CrossRef] [PubMed]
- Stirnberg, M.; Maurer, E.; Horstmeyer, A.; Kolp, S.; Frank, S.; Bald, T.; Arenz, K.; Janzer, A.; Prager, K.; Wunderlich, P.; et al. Proteolytic processing of the serine protease matriptase-2: Identification of the cleavage sites required for its autocatalytic release from the cell surface. Biochem. J. 2010, 430, 87–95. [Google Scholar] [CrossRef]
- Lu, D.; Yuan, X.; Zheng, X.; Sadler, J.E. Bovine proenteropeptidase is activated by trypsin, and the specificity of enteropeptidase depends on the heavy chain. J. Biol. Chem. 1997, 272, 31293–31300. [Google Scholar] [CrossRef]
- Chen, S.; Cao, P.; Dong, N.; Peng, J.; Zhang, C.; Wang, H.; Zhou, T.; Yang, J.; Zhang, Y.; Martelli, E.E.; et al. PCSK6-mediated corin activation is essential for normal blood pressure. Nat. Med. 2015, 21, 1048–1053. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.K.; Teng, I.J.; Lo, T.J.; Moore, S.; Yeo, Y.H.; Teng, Y.C.; Kaul, M.; Chen, C.C.; Zou, A.H.; Chou, F.P.; et al. Matriptase autoactivation is tightly regulated by the cellular chemical environments. PLoS ONE 2014, 9, e93899. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Thompson, H.A.; Barndt, R.B.; Chiu, Y.L.; Lee, M.J.; Lee, S.C.; Wang, J.K.; Tang, H.J.; Lin, C.Y.; Johnson, M.D. Mild acidity likely accelerates the physiological matriptase autoactivation process: A comparative study between spontaneous and acid-induced matriptase zymogen activation. Hum. Cell. 2020, 33, 1068–1080. [Google Scholar] [CrossRef]
- Friis, S.; Uzzun Sales, K.; Godiksen, S.; Peters, D.E.; Lin, C.Y.; Vogel, L.K.; Bugge, T.H. A matriptase-prostasin reciprocal zymogen activation complex with unique features: Prostasin as a non-enzymatic co-factor for matriptase activation. J. Biol. Chem. 2013, 288, 19028–19039. [Google Scholar] [CrossRef]
- Szabo, R.; Lantsman, T.; Peters, D.E.; Bugge, T.H. Delineation of proteolytic and non-proteolytic functions of the membrane-anchored serine protease prostasin. Development 2016, 143, 2818–2828. [Google Scholar] [CrossRef]
- Buzza, M.S.; Martin, E.W.; Driesbaugh, K.H.; Désilets, A.; Leduc, R.; Antalis, T.M. Prostasin is required for matriptase activation in intestinal epithelial cells to regulate closure of the paracellular pathway. J. Biol. Chem. 2013, 288, 10328–10337. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chen, C.J.; Lai, C.H.; Wu, B.Y.; Lee, S.P.; Johnson, M.D.; Lin, C.Y.; Wang, J.K. Increased matriptase zymogen activation by UV irradiation protects keratinocyte from cell death. J. Dermatol. Sci. 2016, 83, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.J.; Desilets, A.; Lin, H.; Charlton, S.; Del Carmen Arques, M.; Falconer, A.; Bullock, C.; Hsu, Y.C.; Birchall, K.; Hawkins, A.; et al. The serine proteinase hepsin is an activator of pro-matrix metalloproteinases: Molecular mechanisms and implications for extracellular matrix turnover. Sci. Rep. 2017, 7, 16693. [Google Scholar] [CrossRef]
- Gaymon, D.O.; Barndt, R.; Stires, H.; Riggins, R.B.; Johnson, M.D. ROS is a master regulator of in vitro matriptase activation. PLoS ONE 2023, 18, e0267492. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef] [PubMed]
- Esmail, S.; Manolson, M.F. Advances in understanding N-glycosylation structure, function, and regulation in health and disease. Eur. J. Cell. Biol. 2021, 100, 151186. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, S.; Wang, J.; Chen, S.; Sun, X.L.; Wu, Q. N-glycosylation in the protease domain of trypsin-like serine proteases mediates calnexin-assisted protein folding. eLife 2018, 7, e35672. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, G.; Gehring, K. Calnexin cycle—Structural features of the ER chaperone system. FEBS J. 2020, 287, 4322–4340. [Google Scholar] [CrossRef] [PubMed]
- Kuribara, T.; Usui, R.; Totani, K. Glycan structure-based perspectives on the entry and release of glycoproteins in the calnexin/calreticulin cycle. Carbohydr. Res. 2021, 502, 108273. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, Y.; Sun, S.; Zhang, Y.; Wang, L.; Luo, Z.; Liu, M.; Dong, L.; Dong, N.; Wu, Q. A conserved LDL-receptor motif regulates corin and CD320 membrane targeting in polarized renal epithelial cells. eLife 2020, 9, e56059. [Google Scholar] [CrossRef]
- Gladysheva, I.P.; Robinson, B.R.; Houng, A.K.; Kováts, T.; King, S.M. Corin is co-expressed with pro-ANP and localized on the cardiomyocyte surface in both zymogen and catalytically active forms. J. Mol. Cell. Cardiol. 2008, 44, 131–142. [Google Scholar] [CrossRef]
- Lichtenthaler, S.F.; Lemberg, M.K.; Fluhrer, R. Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments. EMBO J. 2018, 37, e99456. [Google Scholar] [CrossRef]
- Rosenbaum, D.; Saftig, P. New insights into the function and pathophysiology of the ectodomain sheddase A Disintegrin And Metalloproteinase 10 (ADAM10). FEBS J. 2023. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.C.; Jia, B.; Barndt, R.; Gu, Y.; Chen, C.Y.; Tseng, I.C.; Su, S.F.; Wang, J.K.; Johnson, M.D.; Lin, C.Y. Matriptase shedding is closely coupled with matriptase zymogen activation and requires de novo proteolytic cleavage likely involving its own activity. PLoS ONE 2017, 12, e0183507. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Cho, Y.; Kim, K.Y.; Yoon, M.J.; Lee, H.S.; Jeon, S.D.; Cho, Y.; Kim, C.; Kim, M.G. AJUNN-terminal kinase inhibitor induces ectodomain shedding of the cancer-associated membrane protease Prss14/epithin via protein kinase CβII. J. Biol. Chem. 2020, 295, 7168–7177. [Google Scholar] [CrossRef]
- Najy, A.J.; Dyson, G.; Jena, B.P.; Lin, C.Y.; Kim, H.R. Matriptase activation and shedding through PDGF-D-mediated extracellular acidosis. Am. J. Physiol. Cell. Physiol. 2016, 310, C293–C304. [Google Scholar] [CrossRef]
- Friis, S.; Sales, K.U.; Schafer, J.M.; Vogel, L.K.; Kataoka, H.; Bugge, T.H. The protease inhibitor HAI-2, but not HAI-1, regulates matriptase activation and shedding through prostasin. J. Biol. Chem. 2014, 289, 22319–22332. [Google Scholar] [CrossRef] [PubMed]
- Larsen, B.R.; Steffensen, S.D.; Nielsen, N.V.; Friis, S.; Godiksen, S.; Bornholdt, J.; Soendergaard, C.; Nonboe, A.W.; Anderson, M.N.; Poulsen, S.S.; et al. Hepatocyte growth factor activator inhibitor-2 prevents shedding of matriptase. Exp. Cell. Res. 2013, 319, 918–929. [Google Scholar] [CrossRef]
- Zhao, N.; Nizzi, C.P.; Anderson, S.A.; Wang, J.; Ueno, A.; Tsukamoto, H.; Eisenstein, R.S.; Enns, C.A.; Zhang, A.S. Low intracellular iron increases the stability of matriptase-2. J. Biol. Chem. 2015, 290, 4432–4446. [Google Scholar] [CrossRef]
- Martin, C.E.; Murray, A.S.; Mackinder, J.R.; Sala-Hamrick, K.E.; Flynn, M.G.; Lundgren, J.G.; Varela, F.A.; List, K. TMPRSS13 zymogen activation, surface localization, and shedding is regulated by proteolytic cleavage within the non-catalytic stem region. Biol. Chem. 2022, 403, 969–982. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, Q. Corin in natriuretic peptide processing and hypertension. Curr. Hypertens. Rep. 2014, 16, 415. [Google Scholar] [CrossRef]
- Béliveau, F.; Brulé, C.; Désilets, A.; Zimmerman, B.; Laporte, S.A.; Lavoie, C.L.; Leduc, R. Essential role of endocytosis of the type II transmembrane serine protease TMPRSS6 in regulating its functionality. J. Biol. Chem. 2011, 286, 29035–29043. [Google Scholar] [CrossRef]
- Zheng, X.L.; Kitamoto, Y.; Sadler, J.E. Enteropeptidase, a type II transmembrane serine protease. Front. Biosci. (Elite Ed.) 2009, 1, 242–249. [Google Scholar]
- List, K.; Kosa, P.; Szabo, R.; Bey, A.L.; Wang, C.B.; Molinolo, A.; Bugge, T.H. Epithelial integrity is maintained by a matriptase-dependent proteolytic pathway. Am. J. Pathol. 2009, 175, 1453–1463. [Google Scholar] [CrossRef] [PubMed]
- Buzza, M.S.; Netzel-Arnett, S.; Shea-Donohue, T.; Zhao, A.; Lin, C.Y.; List, K.; Szabo, R.; Fasano, A.; Bugge, T.H.; Antalis, T.M. Membrane-anchored serine protease matriptase regulates epithelial barrier formation and permeability in the intestine. Proc. Natl. Acad. Sci. USA 2010, 107, 4200–4205. [Google Scholar] [CrossRef] [PubMed]
- Szabo, R.; Bugge, T.H. Membrane-anchored serine proteases as regulators of epithelial function. Biochem. Soc. Trans. 2020, 48, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.J.; Feng, X.; Lu, M.; Morimura, S.; Udey, M.C. Matriptase-mediated cleavage of EpCAM destabilizes claudins and dysregulates intestinal epithelial homeostasis. J. Clin. Investig. 2017, 127, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Szabo, R.; Callies, L.K.; Bugge, T.H. Matriptase drives early-onset intestinal failure in a mouse model of congenital tufting enteropathy. Development 2019, 146, dev183392. [Google Scholar] [CrossRef]
- Fasquelle, L.; Scott, H.S.; Lenoir, M.; Wang, J.; Rebillard, G.; Gaboyard, S.; Venteo, S.; Francois, F.; Mausset-Bonnofont, A.L.; Antonarakis, S.E.; et al. Tmprss3, a transmembrane serine protease deficient in human DFNB8/10 deafness, is critical for cochlear hair cell survival at the onset of hearing. J. Biol. Chem. 2011, 286, 17383–17397. [Google Scholar] [CrossRef]
- Molina, L.; Fasquelle, L.; Nouvian, R.; Salvetat, N.; Scott, H.S.; Guipponi, M.; Molina, F.; Puel, J.L.; Delprat, B. Tmprss3 loss of function impairs cochlear inner hair cell Kcnma1 channel membrane expression. Hum. Mol. Genet. 2013, 22, 1289–1299. [Google Scholar] [CrossRef]
- Battelino, S.; Klancar, G.; Kovac, J.; Battelino, T.; Trebusak Podkrajsek, K. TMPRSS3 mutations in autosomal recessive nonsyndromic hearing loss. Eur. Arch. Otorhinolaryngol. 2016, 273, 1151–1154. [Google Scholar] [CrossRef]
- Menou, A.; Duitman, J.; Flajolet, P.; Sallenave, J.M.; Mailleux, A.A.; Crestani, B. Human airway trypsin-like protease, a serine protease involved in respiratory diseases. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 312, L657–L668. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, R.; Takahashi, A.; Nakaya, Y.; Maezawa, H.; Miki, M.; Nakamura, Y.; Ohgushi, F.; Yasuoka, S. Human airway trypsin-like protease stimulates human bronchial fibroblast proliferation in a protease-activated receptor-2-dependent pathway. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006, 290, L385–L395. [Google Scholar] [CrossRef] [PubMed]
- Miki, M.; Nakamura, Y.; Takahashi, A.; Nakaya, Y.; Eguchi, H.; Masegi, T.; Yoneda, K.; Yasuoka, S.; Sone, S. Effect of human airway trypsin-like protease on intracellular free Ca2+ concentration in human bronchial epithelial cells. J. Med. Investig. 2003, 50, 95–107. [Google Scholar]
- Beaufort, N.; Leduc, D.; Eguchi, H.; Mengele, K.; Hellmann, D.; Masegi, T.; Kamimura, T.; Yasuoka, S.; Fend, F.; Chignard, M.; et al. The human airway trypsin-like protease modulates the urokinase receptor (uPAR, CD87) structure and functions. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 292, L1263–L1272. [Google Scholar] [CrossRef]
- Fernandez, C.; Burgos, A.; Morales, D.; Rosales-Rojas, R.; Canelo, J.; Vergara-Jaque, A.; Vieira, G.V.; Alves da Silva, R.A.; Sales, K.U.; Conboy, M.J.; et al. TMPRSS11a is a novel age-altered, tissue specific regulator of migration and wound healing. FASEB J. 2021, 35, e21597. [Google Scholar] [CrossRef]
- Zhang, Z.; Hu, Y.; Yan, R.; Dong, L.; Jiang, Y.; Zhou, Z.; Liu, M.; Zhou, T.; Dong, N.; Wu, Q. The Transmembrane Serine Protease HAT-like 4 Is Important for Epidermal Barrier Function to Prevent Body Fluid Loss. Sci. Rep. 2017, 7, 45262. [Google Scholar] [CrossRef]
- Callies, L.K.; Tadeo, D.; Simper, J.; Bugge, T.H.; Szabo, R. Iterative, multiplexed CRISPR-mediated gene editing for functional analysis of complex protease gene clusters. J. Biol. Chem. 2019, 294, 15987–15996. [Google Scholar] [CrossRef]
- Madsen, D.H.; Szabo, R.; Molinolo, A.A.; Bugge, T.H. TMPRSS13 deficiency impairs stratum corneum formation and epidermal barrier acquisition. Biochem. J. 2014, 461, 487–495. [Google Scholar] [CrossRef]
- Li, S.; Wang, L.; Sun, S.; Wu, Q. Hepsin: A multifunctional transmembrane serine protease in pathobiology. FEBS J. 2021, 288, 5252–5264. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.C.; Huang, H.P.; Yu, I.S.; Su, K.Y.; Lin, S.R.; Lin, W.C.; Wu, H.L.; Shi, G.Y.; Tao, M.H.; Kao, C.H.; et al. Serine protease hepsin regulates hepatocyte size and hemodynamic retention of tumor cells by hepatocyte growth factor signaling in mice. Hepatology 2012, 56, 1913–1923. [Google Scholar] [CrossRef]
- Stirnberg, M.; Gütschow, M. Matriptase-2, a regulatory protease of iron homeostasis: Possible substrates, cleavage sites and inhibitors. Curr. Pharm. Des. 2013, 19, 1052–1061. [Google Scholar] [PubMed]
- Wahedi, M.; Wortham, A.M.; Kleven, M.D.; Zhao, N.; Jue, S.; Enns, C.A.; Zhang, A.S. Matriptase-2 suppresses hepcidin expression by cleaving multiple components of the hepcidin induction pathway. J. Biol. Chem. 2017, 292, 18354–18371. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, R.D.; Houng, A.K.; Gladysheva, I.P.; Fan, T.M.; Tripathi, R.; Reed, G.L.; Wang, D. Corin Overexpression Reduces Myocardial Infarct Size and Modulates Cardiomyocyte Apoptotic Cell Death. Int. J. Mol. Sci. 2020, 21, 3456. [Google Scholar] [CrossRef]
- Kim, T.S.; Heinlein, C.; Hackman, R.C.; Nelson, P.S. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol. Cell. Biol. 2006, 26, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Sales, K.U.; Hobson, J.P.; Wagenaar-Miller, R.; Szabo, R.; Rasmussen, A.L.; Bey, A.; Shah, M.F.; Molinolo, A.A.; Bugge, T.H. Expression and genetic loss of function analysis of the HAT/DESC cluster proteases TMPRSS11A and HAT. PLoS ONE 2011, 6, e23261. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Ko, C.J.; Lo, T.Y.; Wu, S.R.; Lan, S.W.; Huang, C.A.; Lin, Y.C.; Lin, H.H.; Tu, H.F.; Lee, C.F.; et al. Matriptase-2/NR4A3 axis switches TGF-β action toward suppression of prostate cancer cell invasion, tumor growth, and metastasis. Oncogene 2022, 41, 2833–2845. [Google Scholar] [CrossRef]
- Murray, A.S.; Hyland, T.E.; Sala-Hamrick, K.E.; Mackinder, J.R.; Martin, C.E.; Tanabe, L.M.; Varela, F.A.; List, K. The cell-surface anchored serine protease TMPRSS13 promotes breast cancer progression and resistance to chemotherapy. Oncogene 2020, 39, 6421–6436. [Google Scholar] [CrossRef]
- Mukai, S.; Yamasaki, K.; Fujii, M.; Nagai, T.; Terada, N.; Kataoka, H.; Kamoto, T. Dysregulation of Type II Transmembrane Serine Proteases and Ligand-Dependent Activation of MET in Urological Cancers. Int. J. Mol. Sci. 2020, 21, 2663. [Google Scholar] [CrossRef]
- Fuentes-Prior, P. Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection. J. Biol. Chem. 2021, 296, 100135. [Google Scholar] [CrossRef]
- Harbig, A.; Mernberger, M.; Bittel, L.; Pleschka, S.; Schughart, K.; Steinmetzer, T.; Stiewe, T.; Nist, A.; Bottcher-Friebershauser, E. Transcriptome profiling and protease inhibition experiments identify proteases that activate H3N2 influenza A and influenza B viruses in murine airways. J. Biol. Chem. 2020, 295, 11388–11407. [Google Scholar] [CrossRef]
- Murza, A.; Dion, S.P.; Boudreault, P.L.; Désilets, A.; Leduc, R.; Marsault, É. Inhibitors of type II transmembrane serine proteases in the treatment of diseases of the respiratory tract—A review of patent literature. Expert. Opin. Ther. Pat. 2020, 30, 807–824. [Google Scholar] [CrossRef]
- Forni, D.; Sironi, M.; Cagliani, R. Evolutionary history of type II transmembrane serine proteases involved in viral priming. Hum. Genet. 2022, 141, 1705–1722. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, H.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef] [PubMed]
- Shapira, T.; Monreal, I.A.; Dion, S.P.; Buchholz, D.W.; Imbiakha, B.; Olmstead, A.D.; Jager, M.; Desilets, A.; Gao, G.; Martins, M.; et al. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 2022, 605, 340–348. [Google Scholar] [CrossRef]
- Hsin, F.; Hsu, Y.C.; Tsai, Y.F.; Lin, S.W.; Liu, H.M. The transmembrane serine protease hepsin suppresses type I interferon induction by cleaving STING. Sci. Signal. 2021, 14, abb4752. [Google Scholar] [CrossRef]
- Massier, L.; Jalkanen, J.; Elmastas, M.; Zhong, J.; Wang, T.; Nono Nankam, P.A.; Frendo-Cumbo, S.; Backdah, J.; Subramanian, N.; Sekine, T.; et al. An integrated single cell and spatial transcriptomic map of human white adipose tissue. Nat. Commun. 2023, 14, 1438. [Google Scholar] [CrossRef] [PubMed]
- Hildreth, A.D.; Ma, F.; Wong, Y.Y.; Sun, R.; Pellegrini, M.; O’Sullivan, T.E. Single-cell sequencing of human white adipose tissue identifies new cell states in health and obesity. Nat. Immunol. 2021, 22, 639–653. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef] [PubMed]
- Leytus, S.P.; Loeb, K.R.; Hagen, F.S.; Kurachi, K.; Davie, E.W. A novel trypsin-like serine protease (hepsin) with a putative transmembrane domain expressed by human liver and hepatoma cells. Biochemistry 1988, 27, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Herter, S.; Piper, D.E.; Aaron, W.; Gabriele, T.; Cutler, G.; Cao, P.; Bhatt, A.S.; Choe, Y.; Craik, C.S.; Walker, N.; et al. Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membrane-anchored serine protease implicated in prostate and ovarian cancers. Biochem. J. 2005, 390, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.C.; Yu, I.S.; Tsai, Y.F.; Wu, Y.M.; Chen, Y.T.; Sheu, J.C.; Lin, S.W. A Preconditioning Strategy to Augment Retention and Engraftment Rate of Donor Cells During Hepatocyte Transplantation. Transplantation 2021, 105, 785–795. [Google Scholar] [CrossRef]
- Li, S.; Peng, J.; Wang, H.; Zhang, W.; Brown, J.M.; Zhou, Y.; Wu, Q. Hepsin enhances liver metabolism and inhibits adipocyte browning in mice. Proc. Natl. Acad. Sci. USA 2020, 117, 12359–12367. [Google Scholar] [CrossRef] [PubMed]
- Aljakna, A.; Choi, S.; Savage, H.; Hageman Blair, R.; Gu, T.; Svenson, K.L.; Churchill, G.A.; Hibbs, M.; Korstanje, R. Pla2g12b and Hpn are genes identified by mouse ENU mutagenesis that affect HDL cholesterol. PLoS ONE 2012, 7, e43139. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, N.; van Rooij, F.J.; Prins, B.P.; Feitosa, M.F.; Karakas, M.; Eckfeldt, J.H.; Folsom, A.R.; Kopp, J.; Vaez, A.; Andrews, J.S.; et al. Discovery and fine mapping of serum protein loci through transethnic meta-analysis. Am. J. Hum. Genet. 2012, 91, 744–753. [Google Scholar] [CrossRef]
- Koch, J.P.; Aebersold, D.M.; Zimmer, Y.; Medová, M. MET targeting: Time for a rematch. Oncogene 2020, 39, 2845–2862. [Google Scholar] [CrossRef]
- Tervonen, T.A.; Belitškin, D.; Pant, S.M.; Englund, J.I.; Marques, E.; Ala-Hongisto, H.; Nevalaita, L.; Sihto, H.; Heikkila, P.; Leidenius, M.; et al. Deregulated hepsin protease activity confers oncogenicity by concomitantly augmenting HGF/MET signalling and disrupting epithelial cohesion. Oncogene 2016, 35, 1832–1846. [Google Scholar] [CrossRef] [PubMed]
- Brunati, M.; Perucca, S.; Han, L.; Cattaneo, A.; Consolato, F.; Andolfo, A.; Schaeffer, C.; Olinger, E.; Peng, J.; Santambrogio, S.; et al. The serine protease hepsin mediates urinary secretion and polymerisation of Zona Pellucida domain protein uromodulin. eLife 2015, 4, e08887. [Google Scholar] [CrossRef]
- Olinger, E.; Lake, J.; Sheehan, S.; Schiano, G.; Takata, T.; Tokonami, N.; Debaix, H.; Consolato, F.; Rampoldi, L.; Korstanje, R.; et al. Hepsin-mediated Processing of Uromodulin is Crucial for Salt-sensitivity and Thick Ascending Limb Homeostasis. Sci. Rep. 2019, 9, 12287. [Google Scholar] [CrossRef]
- Guipponi, M.; Tan, J.; Cannon, P.Z.; Donley, L.; Crewther, P.; Clarke, M.; Wu, Q.; Shepherd, R.K.; Scott, H.S. Mice deficient for the type II transmembrane serine protease, TMPRSS1/hepsin, exhibit profound hearing loss. Am. J. Pathol. 2007, 171, 608–616. [Google Scholar] [CrossRef]
- Naz, S. Molecular genetic landscape of hereditary hearing loss in Pakistan. Hum. Genet. 2022, 141, 633–648. [Google Scholar] [CrossRef] [PubMed]
- Belitškin, D.; Pant, S.M.; Munne, P.; Suleymanova, I.; Belitškina, K.; Hongisto, H.A.; Englund, J.; Raatikainen, T.; Klezovitch, O.; Vasioukhin, V.; et al. Hepsin regulates TGFβ signaling via fibronectin proteolysis. EMBO Rep. 2021, 22, e52532. [Google Scholar] [CrossRef] [PubMed]
- Puigserver, P.; Spiegelman, B.M. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): Transcriptional coactivator and metabolic regulator. Endocr. Rev. 2003, 24, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Tervonen, T.A.; Pant, S.M.; Belitškin, D.; Englund, J.I.; Närhi, K.; Haglund, C.; Kovanen, P.E.; Verschuren, E.W.; Klefström, J. Oncogenic Ras Disrupts Epithelial Integrity by Activating the Transmembrane Serine Protease Hepsin. Cancer Res. 2021, 81, 1513–1527. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, A.J.; Hooper, J.D.; Folgueras, A.R.; Velasco, G.; López-Otín, C. Matriptase-2 (TMPRSS6): A proteolytic regulator of iron homeostasis. Haematologica 2009, 94, 840–849. [Google Scholar] [CrossRef]
- Finberg, K.E.; Heeney, M.M.; Campagna, D.R.; Aydinok, Y.; Pearson, H.A.; Hartman, K.R.; Mayo, M.M.; Samuel, S.M.; Strouse, J.J.; Markianos, K.; et al. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat. Genet. 2008, 40, 569–571. [Google Scholar] [CrossRef]
- Du, X.; She, E.; Gelbart, T.; Truksa, J.; Lee, P.; Xia, Y.; Khovananth, K.; Mudd, S.; Mann, N.; Moresco, E.M.Y.; et al. The serine protease TMPRSS6 is required to sense iron deficiency. Science 2008, 320, 1088–1092. [Google Scholar] [CrossRef]
- Babitt, J.L.; Huang, F.W.; Wrighting, D.M.; Xia, Y.; Sidis, Y.; Samad, T.A.; Campagna, J.A.; Chung, R.T.; Schneyer, A.L.; Woolf, C.J.; et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat. Genet. 2006, 38, 531–539. [Google Scholar] [CrossRef]
- Muckenthaler, M.U.; Rivella, S.; Hentze, M.W.; Galy, B. A Red Carpet for Iron Metabolism. Cell 2017, 168, 344–361. [Google Scholar] [CrossRef]
- Silvestri, L.; Pagani, A.; Nai, A.; De Domenico, I.; Kaplan, J.; Camaschella, C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008, 8, 502–511. [Google Scholar] [CrossRef]
- Enns, C.A.; Jue, S.; Zhang, A.S. The ectodomain of matriptase-2 plays an important nonproteolytic role in suppressing hepcidin expression in mice. Blood 2020, 136, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Friis, S.; Tadeo, D.; Le-Gall, S.M.; Jürgensen, H.J.; Sales, K.U.; Camerer, E.; Bugge, T.H. Matriptase zymogen supports epithelial development, homeostasis and regeneration. BMC Biol. 2017, 15, 46. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.; Sullivan, R.D.; Fan, T.M.; Houng, A.K.; Mehta, R.M.; Reed, G.L.; Gladysheva, I.P. Cardiac-Specific Overexpression of Catalytically Inactive Corin Reduces Edema, Contractile Dysfunction, and Death in Mice with Dilated Cardiomyopathy. Int. J. Mol. Sci. 2019, 21, 203. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, A.M.; Maurer, E.; Lülsdorff, V.; Wilms, A.; Furtmann, N.; Bajorath, J.; Gütschow, M.; Stirnberg, M. En Route to New Therapeutic Options for Iron Overload Diseases: Matriptase-2 as a Target for Kunitz-Type Inhibitors. Chembiochem 2016, 17, 595–604. [Google Scholar] [CrossRef]
- McClung, J.P.; Karl, J.P. Iron deficiency and obesity: The contribution of inflammation and diminished iron absorption. Nutr. Rev. 2009, 67, 100–104. [Google Scholar] [CrossRef]
- Del Giudice, E.M.; Santoro, N.; Amato, A.; Brienza, C.; Calabrò, P.; Wiegerinck, E.T.; Cirillo, G.; Tartaglino, N.; Grandone, A.; Swinkels, D.W.; et al. Hepcidin in obese children as a potential mediator of the association between obesity and iron deficiency. J. Clin. Endocrinol. Metab. 2009, 94, 5102–5107. [Google Scholar] [CrossRef]
- Stoffel, N.U.; El-Mallah, C.; Herter-Aeberli, I.; Bissani, N.; Wehbe, N.; Obeid, O.; Zimmermann, M.B. The effect of central obesity on inflammation, hepcidin, and iron metabolism in young women. Int. J. Obes. 2020, 44, 1291–1300. [Google Scholar] [CrossRef]
- Bekri, S.; Gual, P.; Anty, R.; Luciani, N.; Dahman, M.; Ramesh, B.; Iannelli, A.; Staccini-Myx, A.; Casanova, D.; Amor, I.B.; et al. Increased adipose tissue expression of hepcidin in severe obesity is independent from diabetes and NASH. Gastroenterology 2006, 131, 788–796. [Google Scholar] [CrossRef]
- Dongiovanni, P.; Ruscica, M.; Rametta, R.; Recalcati, S.; Steffani, L.; Gatti, S.; Girelli, D.; Cairo, G.; Magni, P.; Fargion, S.; et al. Dietary iron overload induces visceral adipose tissue insulin resistance. Am. J. Pathol. 2013, 182, 2254–2263. [Google Scholar] [CrossRef]
- Folgueras, A.R.; Freitas-Rodríguez, S.; Ramsay, A.J.; Garabaya, C.; Rodríguez, F.; Velasco, G.; López-Otín, C. Matriptase-2 deficiency protects from obesity by modulating iron homeostasis. Nat. Commun. 2018, 9, 1350. [Google Scholar] [CrossRef]
- Goetze, J.P.; Bruneau, B.G.; Ramos, H.R.; Ogawa, T.; de Bold, M.K.; de Bold, A.J. Cardiac natriuretic peptides. Nat. Rev. Cardiol. 2020, 17, 698–717. [Google Scholar] [CrossRef]
- Kuhn, M. Cardiac actions of atrial natriuretic peptide: New visions of an old friend. Circ. Res. 2015, 116, 1278–1280. [Google Scholar] [CrossRef] [PubMed]
- Pang, A.; Hu, Y.; Zhou, P.; Long, G.; Tian, X.; Men, L.; Shen, Y.; Liu, Y.; Cui, Y. Corin is down-regulated and exerts cardioprotective action via activating pro-atrial natriuretic peptide pathway in diabetic cardiomyopathy. Cardiovasc. Diabetol. 2015, 14, 134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, S.; Lou, J.; Li, H.; Liu, M.; Dong, N.; Wu, Q. Atrial natriuretic peptide promotes uterine decidualization and a TRAIL-dependent mechanism in spiral artery remodeling. J. Clin. Investig. 2021, 131, e151053. [Google Scholar] [CrossRef] [PubMed]
- Della Corte, V.; Pacinella, G.; Todaro, F.; Pecoraro, R.; Tuttolomondo, A. The Natriuretic Peptide System: A Single Entity, Pleiotropic Effects. Int. J. Mol. Sci. 2023, 24, 9642. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Zhang, S.; Du, C.; Wang, K.; Gu, X.; Sun, S.; Zhang, X.; Niu, Y.; Wang, C.; Liu, M.; et al. Renal Corin Is Essential for Normal Blood Pressure and Sodium Homeostasis. Int. J. Mol. Sci. 2022, 23, 11251. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhou, T.; Niu, Y.; Feng, W.; Gu, X.; Xu, W.; Zhang, S.; Wang, Z.; Zhang, Y.; Wang, C.; et al. The protease corin regulates electrolyte homeostasis in eccrine sweat glands. PLoS Biol. 2021, 19, e3001090. [Google Scholar] [CrossRef]
- Rame, J.E.; Drazner, M.H.; Post, W.; Peshock, R.; Lima, J.; Cooper, R.S.; Dries, D.L. Corin I555(P568) allele is associated with enhanced cardiac hypertrophic response to increased systemic afterload. Hypertension 2007, 49, 857–864. [Google Scholar] [CrossRef]
- Rame, J.E.; Tam, S.W.; McNamara, D.; Worcel, M.; Sabolinski, M.L.; Wu, A.H.; Dries, D.L. Dysfunctional corin i555(p568) allele is associated with impaired brain natriuretic peptide processing and adverse outcomes in blacks with systolic heart failure: Results from the Genetic Risk Assessment in Heart Failure substudy. Circ. Heart Fail. 2009, 2, 541–548. [Google Scholar] [CrossRef]
- Dong, N.; Zhou, T.; Zhang, Y.; Liu, M.; Li, H.; Huang, X.; Liu, Z.; Wu, Y.; Fukuda, K.; Qin, J.; et al. Corin mutations K317E and S472G from preeclamptic patients alter zymogen activation and cell surface targeting. J. Biol. Chem. 2014, 289, 17909–17916. [Google Scholar] [CrossRef]
- Zhao, Y.; Yuan, X.; Zhong, Y.; Zhang, Y.; Zhang, S.; Li, S.; Zheng, W.; Zheng, W.; Liu, J.; Xia, Y.; et al. Single-Nucleotide Polymorphisms in the 3′ Untranslated Region of CORIN Associated With Cardiovascular Diseases in a Chinese Han Population: A Case-Control Study. Front. Cardiovasc. Med. 2021, 8, 625072. [Google Scholar] [CrossRef]
- Collins, S. A heart-adipose tissue connection in the regulation of energy metabolism. Nat. Rev. Endocrinol. 2014, 10, 157–163. [Google Scholar] [CrossRef]
- Sengenes, C.; Bouloumie, A.; Hauner, H.; Berlan, M.; Busse, R.; Lafontan, M.; Galitzky, J. Involvement of a cGMP-dependent pathway in the natriuretic peptide-mediated hormone-sensitive lipase phosphorylation in human adipocytes. J. Biol. Chem. 2003, 278, 48617–48626. [Google Scholar] [CrossRef] [PubMed]
- Birkenfeld, A.L.; Budziarek, P.; Boschmann, M.; Moro, C.; Adams, F.; Franke, G.; Berlan, M.; Marques, M.A.; Sweep, F.C.G.J.; Luft, F.D.; et al. Atrial natriuretic peptide induces postprandial lipid oxidation in humans. Diabetes 2008, 57, 3199–3204. [Google Scholar] [CrossRef]
- Wu, W.; Shi, F.; Liu, D.; Ceddia, R.P.; Gaffin, R.; Wei, W.; Fang, H.; Lewandoski, E.D.; Collins, S. Enhancing natriuretic peptide signaling in adipose tissue, but not in muscle, protects against diet-induced obesity and insulin resistance. Sci. Signal. 2017, 10, eaam6870. [Google Scholar] [CrossRef] [PubMed]
- Cabiati, M.; Raucci, S.; Liistro, T.; Belcastro, E.; Prescimone, T.; Caselli, C.; Matteucci, M.; Iozzo, P.; Mattii, L.; Giannessi, D.; et al. Impact of obesity on the expression profile of natriuretic peptide system in a rat experimental model. PLoS ONE 2013, 8, e72959. [Google Scholar] [CrossRef] [PubMed]
- Bartels, E.D.; Nielsen, J.M.; Bisgaard, L.S.; Goetze, J.P.; Nielsen, L.B. Decreased expression of natriuretic peptides associated with lipid accumulation in cardiac ventricle of obese mice. Endocrinology 2010, 151, 5218–5225. [Google Scholar] [CrossRef]
- Galitzky, J.; Sengenès, C.; Thalamas, C.; Marques, M.A.; Senard, J.M.; Lafontan, M.; Berlan, M. The lipid-mobilizing effect of atrial natriuretic peptide is unrelated to sympathetic nervous system activation or obesity in young men. J. Lipid Res. 2001, 42, 536–544. [Google Scholar] [CrossRef]
- Engeli, S.; Birkenfeld, A.L.; Badin, P.M.; Bourlier, V.; Louche, K.; Viguerie, N.; Thalamas, C.; Montastier, E.; Larrouy, D.; Harant, I.; et al. Natriuretic peptides enhance the oxidative capacity of human skeletal muscle. J. Clin. Investig. 2012, 122, 4675–4679. [Google Scholar] [CrossRef]
- Bordicchia, M.; Liu, D.; Amri, E.Z.; Ailhaud, G.; Dessì-Fulgheri, P.; Zhang, C.; Takahashi, N.; Sarzani, R.; Collins, S. Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J. Clin. Investig. 2012, 122, 1022–1036. [Google Scholar] [CrossRef]
- Coué, M.; Moro, C. Natriuretic peptide control of energy balance and glucose homeostasis. Biochimie 2016, 124, 84–91. [Google Scholar] [CrossRef]
- Kimura, H.; Nagoshi, T.; Oi, Y.; Yoshii, A.; Tanaka, Y.; Takahashi, H.; Kashiwagi, Y.; Tanaka, T.; Yoshimura, M. Treatment with atrial natriuretic peptide induces adipose tissue browning and exerts thermogenic actions in vivo. Sci. Rep. 2021, 11, 17466. [Google Scholar] [CrossRef]
- Liu, D.; Ceddia, R.P.; Collins, S. Cardiac natriuretic peptides promote adipose ‘browning’ through mTOR complex-1. Mol. Metab. 2018, 9, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Carper, D.; Coué, M.; Nascimento, E.B.M.; Barquissau, V.; Lagarde, D.; Pestourie, C.; Laurens, C.; Petit, J.V.; Soty, M.; Monbrun, L.; et al. Atrial Natriuretic Peptide Orchestrates a Coordinated Physiological Response to Fuel Non-shivering Thermogenesis. Cell. Rep. 2020, 32, 108075. [Google Scholar] [CrossRef]
- Inoue, K.; Sakamoto, T.; Yuge, S.; Iwatani, H.; Yamagami, S.; Tsutsumi, M.; Hori, H.; Cerra, M.C.; Tota, B.; Suzuki, N.; et al. Structural and functional evolution of three cardiac natriuretic peptides. Mol. Biol. Evol. 2005, 22, 2428–2434. [Google Scholar] [CrossRef]
- Loretz, C.A.; Pollina, C. Natriuretic peptides in fish physiology. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2000, 125, 169–187. [Google Scholar] [CrossRef]
- Chait, A.; den Hartigh, L.J. Adipose Tissue Distribution, Inflammation and Its Metabolic Consequences, Including Diabetes and Cardiovascular Disease. Front. Cardiovasc. Med. 2020, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Zatterale, F.; Longo, M.; Naderi, J.; Raciti, G.A.; Desiderio, A.; Miele, C.; Beguinot, F. Chronic Adipose Tissue Inflammation Linking Obesity to Insulin Resistance and Type 2 Diabetes. Front. Physiol. 2019, 10, 1607. [Google Scholar] [CrossRef] [PubMed]
- Staedtke, V.; Bai, R.Y.; Kim, K.; Darvas, M.; Davila, M.L.; Riggins, G.J.; Rothman, P.B.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B.; et al. Disruption of a self-amplifying catecholamine loop reduces cytokine release syndrome. Nature 2018, 564, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Mezzasoma, L.; Antognelli, C.; Talesa, V.N. Atrial natriuretic peptide down-regulates LPS/ATP-mediated IL-1β release by inhibiting NF-kB, NLRP3 inflammasome and caspase-1 activation in THP-1 cells. Immunol. Res. 2016, 64, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Moro, C.; Klimcakova, E.; Lolmède, K.; Berlan, M.; Lafontan, M.; Stich, V.; Bouloumié, A.; Galitzky, J.; Arner, P.; Langin, D. Atrial natriuretic peptide inhibits the production of adipokines and cytokines linked to inflammation and insulin resistance in human subcutaneous adipose tissue. Diabetologia 2007, 50, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Xue, F.; Sui, W.; Meng, L.; Xie, L.; Zhang, C.; Yang, J.; Zhang, Y. Deletion of natriuretic peptide receptor C alleviates adipose tissue inflammation in hypercholesterolemic Apolipoprotein E knockout mice. J. Cell. Mol. Med. 2021, 25, 9837–9850. [Google Scholar] [CrossRef] [PubMed]
- Sarzani, R.; Strazzullo, P.; Salvi, F.; Iacone, R.; Pietrucci, F.; Siani, A.; Barba, G.; Gerardi, M.C.; Dessi-Fulgheri, P.; Rappelli, A. Natriuretic peptide clearance receptor alleles and susceptibility to abdominal adiposity. Obes. Res. 2004, 12, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, W.; Zhou, T.; Liu, M.; Wu, Q.; Dong, N. Corin Deficiency Alters Adipose Tissue Phenotype and Impairs Thermogenesis in Mice. Biology 2022, 11, 1101. [Google Scholar] [CrossRef]
- Garruti, G.; Giusti, V.; Nussberger, J.; Darimont, C.; Verdumo, C.; Amstutz, C.; Puglisi, F.; Giorgino, F.; Giorgio, R.; Cotecchia, S. Expression and secretion of the atrial natriuretic peptide in human adipose tissue and preadipocytes. Obesity 2007, 15, 2181–2189. [Google Scholar] [CrossRef]
- Kamberov, Y.G.; Karlsson, E.K.; Kamberova, G.L.; Lieberman, D.E.; Sabeti, P.C.; Morgan, B.A.; Tabin, C.J. A genetic basis of variation in eccrine sweat gland and hair follicle density. Proc. Natl. Acad. Sci. USA 2015, 112, 9932–9937. [Google Scholar] [CrossRef]
- Best, A.; Kamilar, J.M. The evolution of eccrine sweat glands in human and nonhuman primates. J. Hum. Evol. 2018, 117, 33–43. [Google Scholar] [CrossRef]
- Bramble, D.M.; Lieberman, D.E. Endurance running and the evolution of Homo. Nature 2004, 432, 345–352. [Google Scholar] [CrossRef]
- Hooper, J.D.; Clements, J.A.; Quigley, J.P.; Antalis, T.M. Type II transmembrane serine proteases. Insights into an emerging class of cell surface proteolytic enzymes. J. Biol. Chem. 2001, 276, 857–860. [Google Scholar] [CrossRef]
- Wu, Q. Type II transmembrane serine proteases. Curr. Top. Dev. Biol. 2003, 54, 167–206. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Li, S.; Zhang, X.; Dong, N. Type II Transmembrane Serine Proteases as Modulators in Adipose Tissue Phenotype and Function. Biomedicines 2023, 11, 1794. https://doi.org/10.3390/biomedicines11071794
Wu Q, Li S, Zhang X, Dong N. Type II Transmembrane Serine Proteases as Modulators in Adipose Tissue Phenotype and Function. Biomedicines. 2023; 11(7):1794. https://doi.org/10.3390/biomedicines11071794
Chicago/Turabian StyleWu, Qingyu, Shuo Li, Xianrui Zhang, and Ningzheng Dong. 2023. "Type II Transmembrane Serine Proteases as Modulators in Adipose Tissue Phenotype and Function" Biomedicines 11, no. 7: 1794. https://doi.org/10.3390/biomedicines11071794
APA StyleWu, Q., Li, S., Zhang, X., & Dong, N. (2023). Type II Transmembrane Serine Proteases as Modulators in Adipose Tissue Phenotype and Function. Biomedicines, 11(7), 1794. https://doi.org/10.3390/biomedicines11071794




