Application of Compositional Data Analysis to Study the Relationship between Bacterial Diversity in Human Faeces and Sex, Age, and Weight
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Clinical Trial
2.2. Stool Sample Processing
2.3. Analysis of Bacterial Diversity
2.4. CoDa Analysis
3. Results
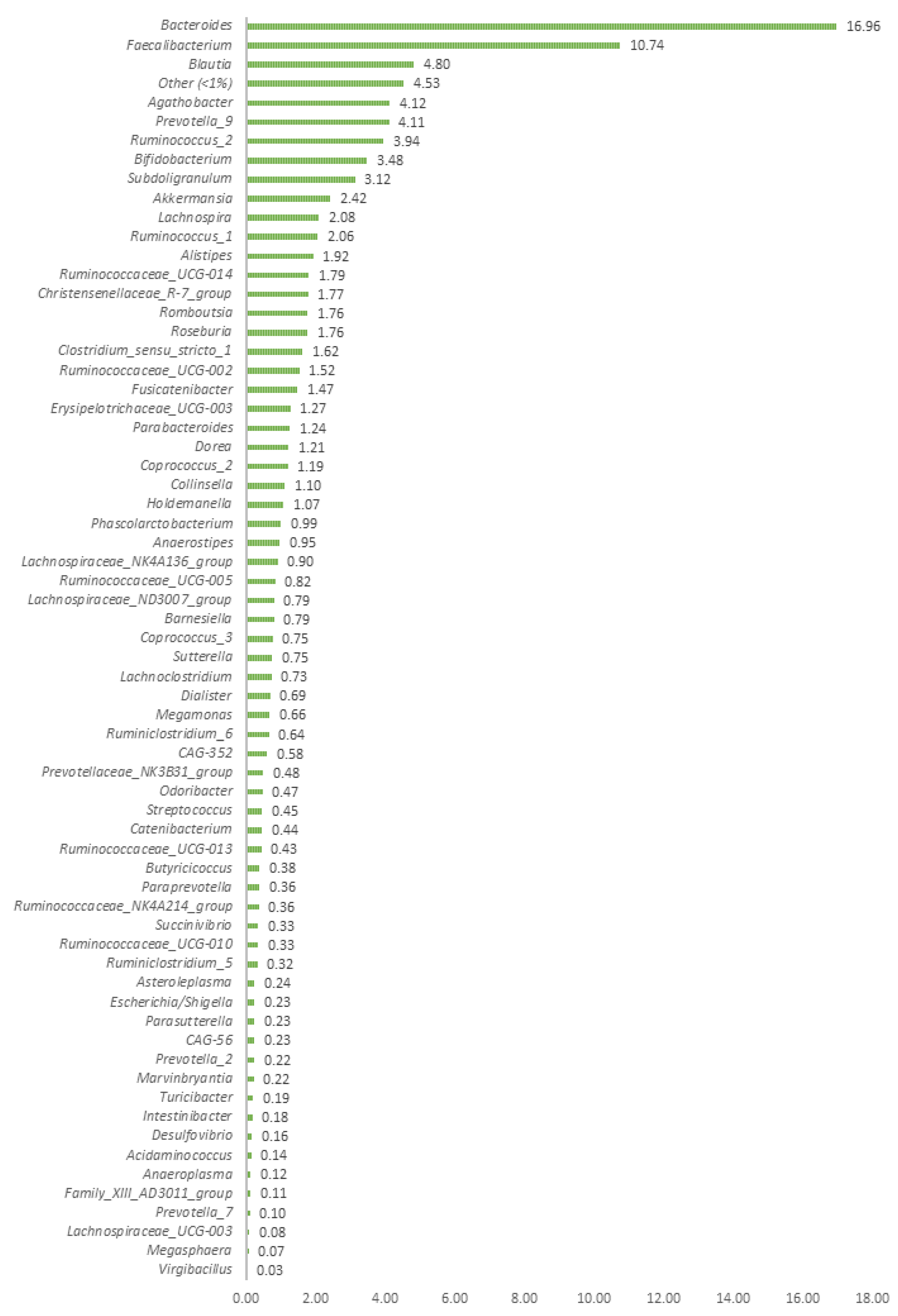
3.1. Study of Bacterial Diversity
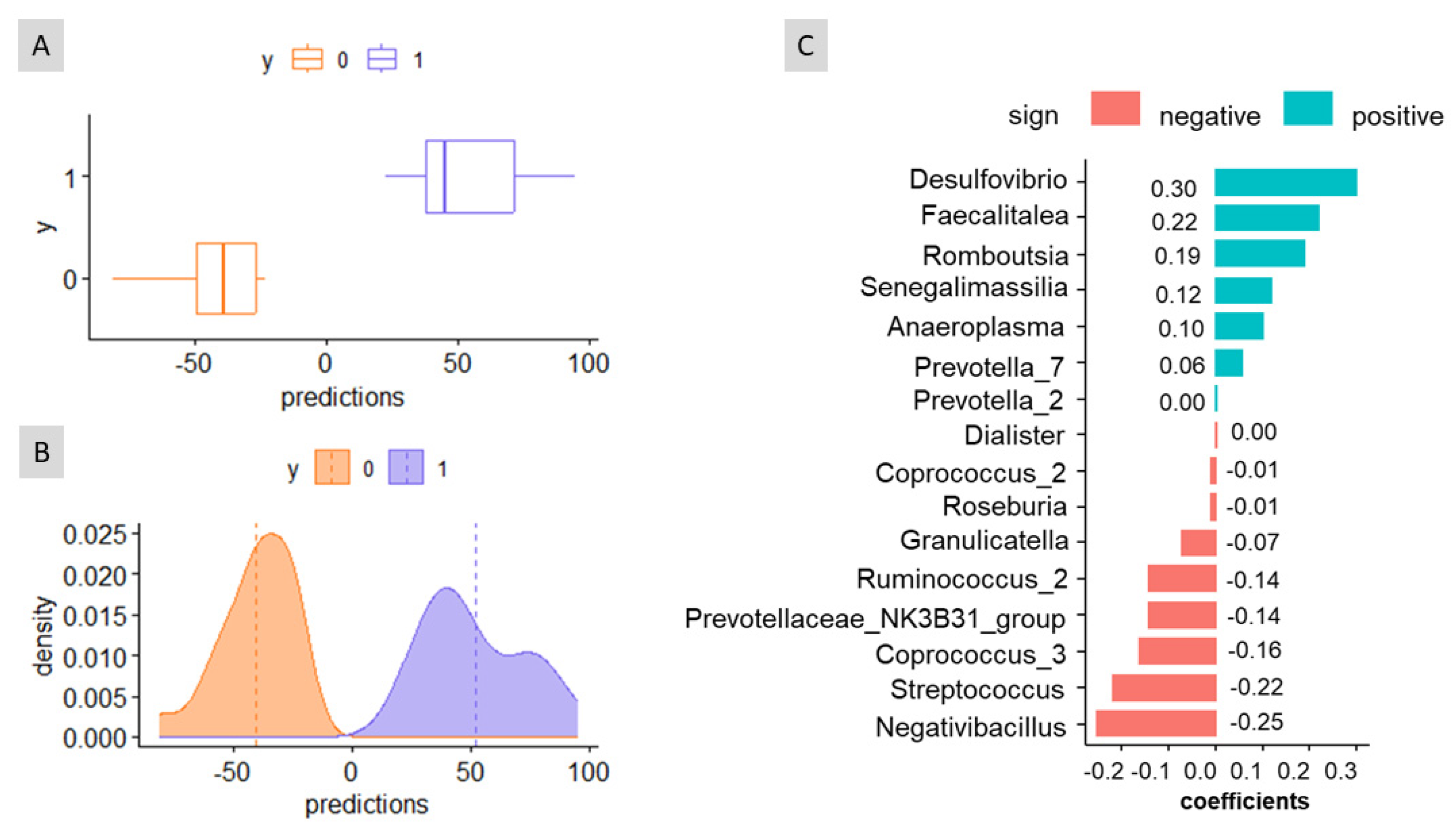
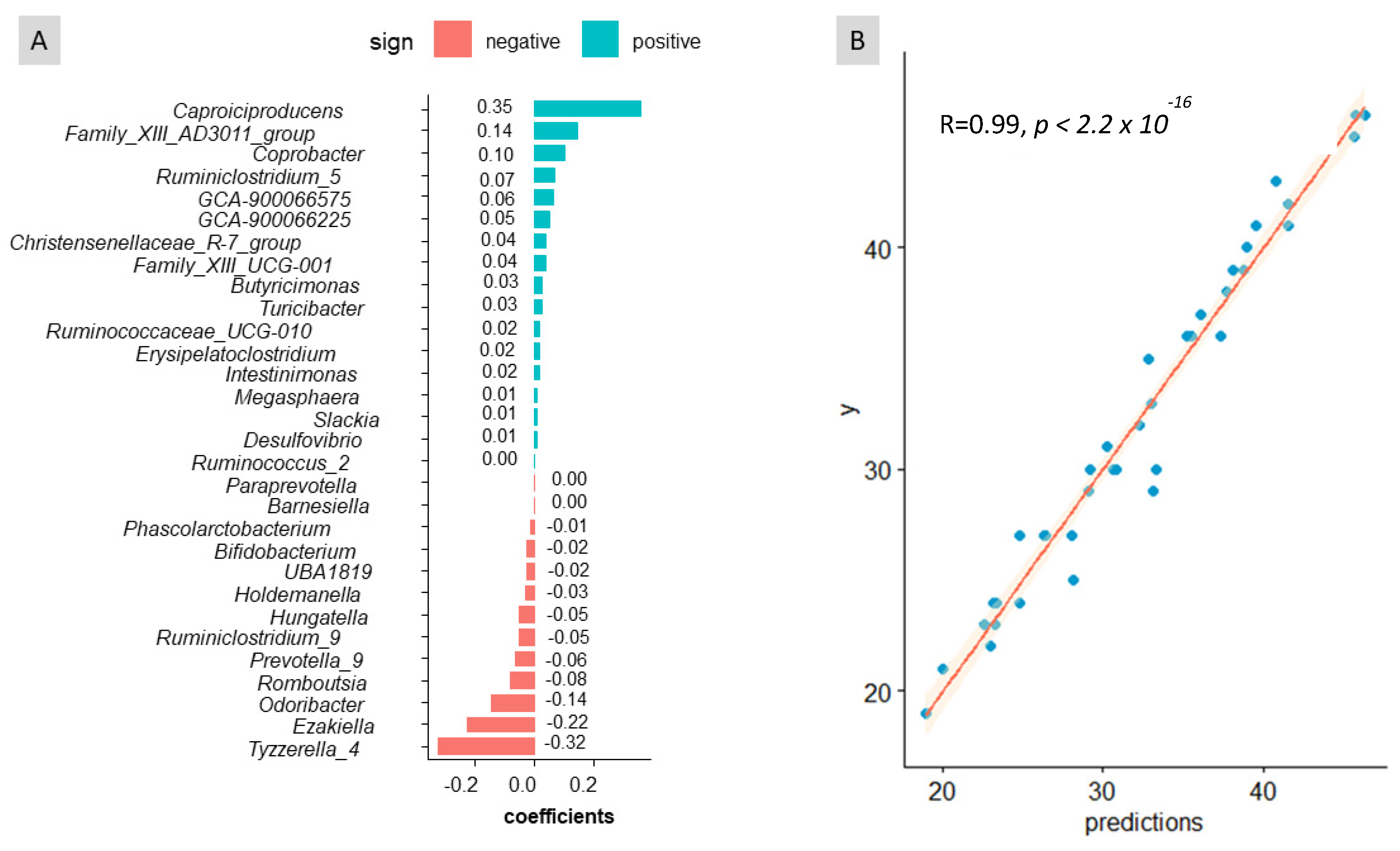
3.2. Association of Sex, Age, and Weight with Bacteria in Faeces
3.3. Bacterial Taxa Associated with Predictions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Pawlowsky-Glahn, V.; Egozcue, J.J. Compositional analysis of microbiome data. In Proceedings of the Conference: B-Debates on the Human Microbiome, Barcelona, Spain, 30 June–1 July 2016. [Google Scholar]
- Erb, I.; Notredame, C. How should we measure proportionality on relative gene expression data? Theory Biosci. 2016, 135, 21–36. [Google Scholar] [CrossRef]
- Gloor, G.B.; Wu, J.R.; Pawlowsky-Glahn, V.; Egozcue, J.J. It’s all relative: Analyzing microbiome data as compositions. Ann. Epidemiol. 2016, 26, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome datasets are compositional: And this is not optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef]
- Quinn, T.P.; Erb, I.; Richardson, M.F.; Crowley, T.M. Understanding sequencing data as compositions: An outlook and review. Bioinformatics 2018, 34, 2870–2878. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Pinto, J.; Egozcue, J.J.; Pawlowsky-Glahn, V.; Paredes, R.; Noguera-Julian, M.; Calle, M.L. Balances: A new perspective for microbiome analysis. MSystems 2018, 3, 53. [Google Scholar] [CrossRef]
- Calle, M.L. Statistical analysis of metagenomics data. Genom. Inform. 2019, 17, e6. [Google Scholar] [CrossRef]
- Calle, M.L.; Susin, A. coda4microbiome: Compositional data analysis for microbiome studies. bioRxiv 2022. bioRxiv:2022.06.09.495511. [Google Scholar] [CrossRef]
- Cappellato, M.; Baruzzo, G.; Di Camillo, B. Investigating differential abundance methods in microbiome data: A benchmark study. PLoS Comput. Biol. 2022, 18, e1010467. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Macklaim, J.M.; Linn, T.G.; Reid, G.; Gloor, G.B. ANOVA-Like Differential Expression (ALDEx) Analysis for Mixed Population RNA-Seq. PLoS ONE 2013, 8, e67019. [Google Scholar]
- Lin, H.; Eggesbø, M.; Peddada, S.D. Linear and nonlinear correlation estimators unveil undescribed taxa interactions in microbiome data. Nat. Commun. 2022, 13, 4946. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Peddada, S.D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef] [PubMed]
- Nearing, J.T.; Douglas, G.M.; Hayes, M.G.; MacDonald, J.; Desai, D.K.; Allward, N.; Jones, C.M.; Wright, R.J.; Dhanani, A.S.; Comeau, A.M. Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 2022, 13, 342. [Google Scholar] [CrossRef]
- López-García, E.; Benítez-Cabello, A.; Arenas-de Larriva, A.P.; Gutierrez-Mariscal, F.M.; Pérez-Martínez, P.; Yubero-Serrano, E.M.; Garrido-Fernández, A.; Arroyo-López, F.N. Oral intake of Lactiplantibacillus pentosus LPG1 Produces a Beneficial Regulation of Gut Microbiota in Healthy Persons: A Randomised, Placebo-Controlled, Single-Blind Trial. Nutrients 2023, 15, 1931. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Martín-Fernández, J.; Hron, K.; Templ, M.; Filzmoser, P.; Palarea-Albaladejo, J. Bayesian-multiplicative treatment of count zeros in compositional data sets. Stat. Model. 2015, 15, 134–158. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, R v 4.3.0; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 14 April 2023).
- RStudio Team. RStudio: Integrated Development for R, v0.98.1074; RStudio, Inc.: Boston, MA, USA, 2015; Available online: https://www.rstudio.com/products/rstudio (accessed on 14 April 2023).
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Brooks, C.N.; Wight, M.E.; Azeez, O.E.; Bleich, R.M.; Zwetsloot, K.A. Growing old together: What we know about the influence of diet and exercise on the aging host’s gut microbiome. Front. Sports Act. Living 2023, 5, 1168731. [Google Scholar] [CrossRef]
- Fuhrman, B.J.; Feigelson, H.S.; Flores, R.; Gail, M.H.; Xu, X.; Ravel, J.; Goedert, J.J. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J. Clin. Endocrinol. Metab. 2014, 99, 4632–4640. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Shi, J.; Fuhrman, B.; Xu, X.; Veenstra, T.D.; Gail, M.H.; Gajer, P.; Ravel, J.; Goedert, J.J. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: A cross-sectional study. J. Transl. Med. 2012, 10, 253. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.; Kim, N. Roles of sex hormones and gender in the gut microbiota. J. Neurogastroenterol. Motil. 2021, 27, 314. [Google Scholar] [CrossRef] [PubMed]
- Carmody, R.N.; Bisanz, J.E. Roles of the gut microbiome in weight management. Nat. Rev. Microbiol. 2023, 21, 535–550. [Google Scholar] [CrossRef]
- de la Cuesta-Zuluaga, J.; Kelley, S.T.; Chen, Y.; Escobar, J.S.; Mueller, N.T.; Ley, R.E.; McDonald, D.; Huang, S.; Swafford, A.D.; Knight, R. Age-and sex-dependent patterns of gut microbial diversity in human adults. Msystems 2019, 4, 261. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Hugon, P.; Dufour, J.; Colson, P.; Fournier, P.; Sallah, K.; Raoult, D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect. Dis. 2015, 15, 1211–1219. [Google Scholar] [CrossRef]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef]
- Zhou, Y.; Mihindukulasuriya, K.A.; Gao, H.; La Rosa, P.S.; Wylie, K.M.; Martin, J.C.; Kota, K.; Shannon, W.D.; Mitreva, M.; Sodergren, E. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014, 15, 1–18. [Google Scholar] [CrossRef]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia—A new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1875796. [Google Scholar] [CrossRef]
- Zafar, H.; Saier, M.H., Jr. Gut Bacteroides species in health and disease. Gut Microbes 2021, 13, 1848158. [Google Scholar] [CrossRef]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Markle, J.G.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; Von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Park, Y.; Sim, M.; Kim, S.; Joung, H.; Shin, D. Serum level of sex steroid hormone is associated with diversity and profiles of human gut microbiome. Res. Microbiol. 2019, 170, 192–201. [Google Scholar] [CrossRef]
- Maslennikov, R.; Efremova, I.; Ivashkin, V.; Zharkova, M.; Poluektova, E.; Shirokova, E.; Ivashkin, K. Effect of probiotics on hemodynamic changes and complications associated with cirrhosis: A pilot randomized controlled trial. World J. Hepatol. 2022, 14, 1667–1677. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jing, Q.; Chen, F.; Zheng, H.; Chen, L.; Yang, Z. Desulfovibrio is not always associated with adverse health effects in the Guangdong Gut Microbiome Project. PeerJ 2021, 9, e12033. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fu, R.; Li, R.; Zeng, J.; Liu, T.; Li, X.; Jiang, W. Causality of gut microbiome and hypertension: A bidirectional mendelian randomization study. Front. Cardiovasc. Med. 2023, 10, 1167346. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Havulinna, A.S.; Liu, Y.; Jousilahti, P.; Ritchie, S.C.; Tokolyi, A.; Sanders, J.G.; Valsta, L.; Brożyńska, M.; Zhu, Q. Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat. Genet. 2022, 54, 134–142. [Google Scholar] [CrossRef]
- Kelly, T.N.; Bazzano, L.A.; Ajami, N.J.; He, H.; Zhao, J.; Petrosino, J.F.; Correa, A.; He, J. Gut microbiome associates with lifetime cardiovascular disease risk profile among bogalusa heart study participants. Circ. Res. 2016, 119, 956–964. [Google Scholar] [CrossRef]

| Variable | Type | Mean | Confidence −95% | Confidence +95% | Standard Deviation | Median | Minimum | Maximum |
|---|---|---|---|---|---|---|---|---|
| Sex | Qualitative * | - | - | - | - | - | - | - |
| Age | Quantitative | 32.51 | 30.03 | 34.99 | 7.64 | 31.00 | 19.00 | 46.00 |
| Weight | Quantitative | 69.48 | 65.35 | 73.62 | 12.74 | 66.40 | 48.60 | 99.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-García, E.; Benítez-Cabello, A.; Arenas-de Larriva, A.P.; Gutierrez-Mariscal, F.M.; Pérez-Martínez, P.; Yubero-Serrano, E.M.; Arroyo-López, F.N.; Garrido-Fernández, A. Application of Compositional Data Analysis to Study the Relationship between Bacterial Diversity in Human Faeces and Sex, Age, and Weight. Biomedicines 2023, 11, 2134. https://doi.org/10.3390/biomedicines11082134
López-García E, Benítez-Cabello A, Arenas-de Larriva AP, Gutierrez-Mariscal FM, Pérez-Martínez P, Yubero-Serrano EM, Arroyo-López FN, Garrido-Fernández A. Application of Compositional Data Analysis to Study the Relationship between Bacterial Diversity in Human Faeces and Sex, Age, and Weight. Biomedicines. 2023; 11(8):2134. https://doi.org/10.3390/biomedicines11082134
Chicago/Turabian StyleLópez-García, Elio, Antonio Benítez-Cabello, Antonio Pablo Arenas-de Larriva, Francisco Miguel Gutierrez-Mariscal, Pablo Pérez-Martínez, Elena María Yubero-Serrano, Francisco Noé Arroyo-López, and Antonio Garrido-Fernández. 2023. "Application of Compositional Data Analysis to Study the Relationship between Bacterial Diversity in Human Faeces and Sex, Age, and Weight" Biomedicines 11, no. 8: 2134. https://doi.org/10.3390/biomedicines11082134
APA StyleLópez-García, E., Benítez-Cabello, A., Arenas-de Larriva, A. P., Gutierrez-Mariscal, F. M., Pérez-Martínez, P., Yubero-Serrano, E. M., Arroyo-López, F. N., & Garrido-Fernández, A. (2023). Application of Compositional Data Analysis to Study the Relationship between Bacterial Diversity in Human Faeces and Sex, Age, and Weight. Biomedicines, 11(8), 2134. https://doi.org/10.3390/biomedicines11082134









