Identification of NOX4 as a New Biomarker in Hepatocellular Carcinoma and Its Effect on Sorafenib Therapy
Abstract
:1. Introduction
2. Materials and Methods
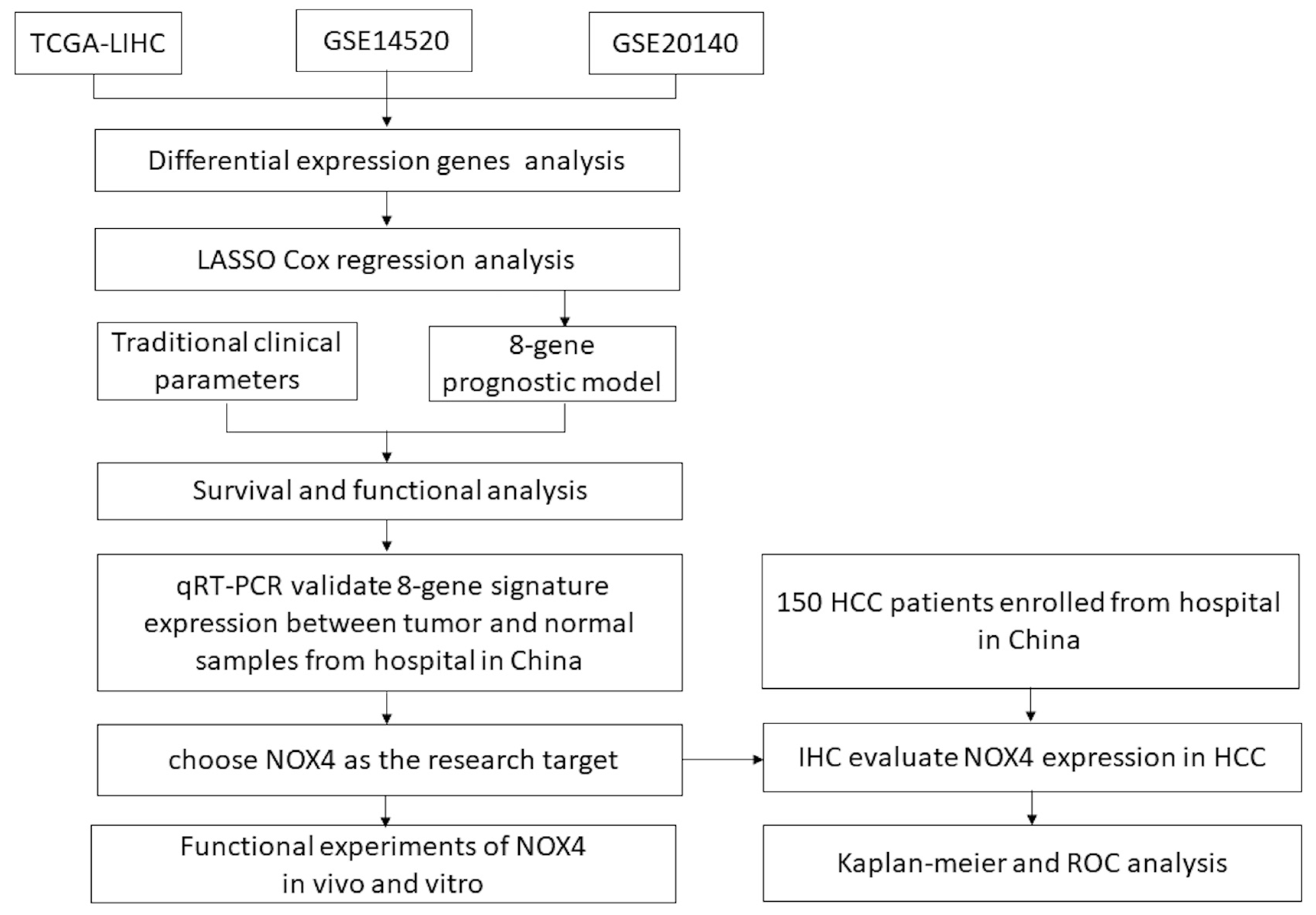
2.1. Data Acquisition from the Cancer Genome Atlas (TCGA) Databases and Gene Expression Omnibus (GEO)
2.2. Differentially Expressed Genes and Functional Annotation and Gene Enrichment Analysis
2.3. Lasso Regression Model Construction and the Nomogram Prediction Model
2.4. Patients and Specimens
2.5. Immunohistochemical Staining and Evaluation
2.6. Cell Culture
2.7. Plasmid Constructions
2.8. Lentivirus Packaging and Stable Cell Lines
2.9. RNA Extraction and Quantitative Real-Time PCR
2.10. Western Blotting
2.11. Cell Proliferation, Colony Formation, Wound Healing, and Intracellular ROS Detection In Vitro
2.12. Nude Mouse Tumor Growth Analysis
2.13. Statistical Analysis
3. Results
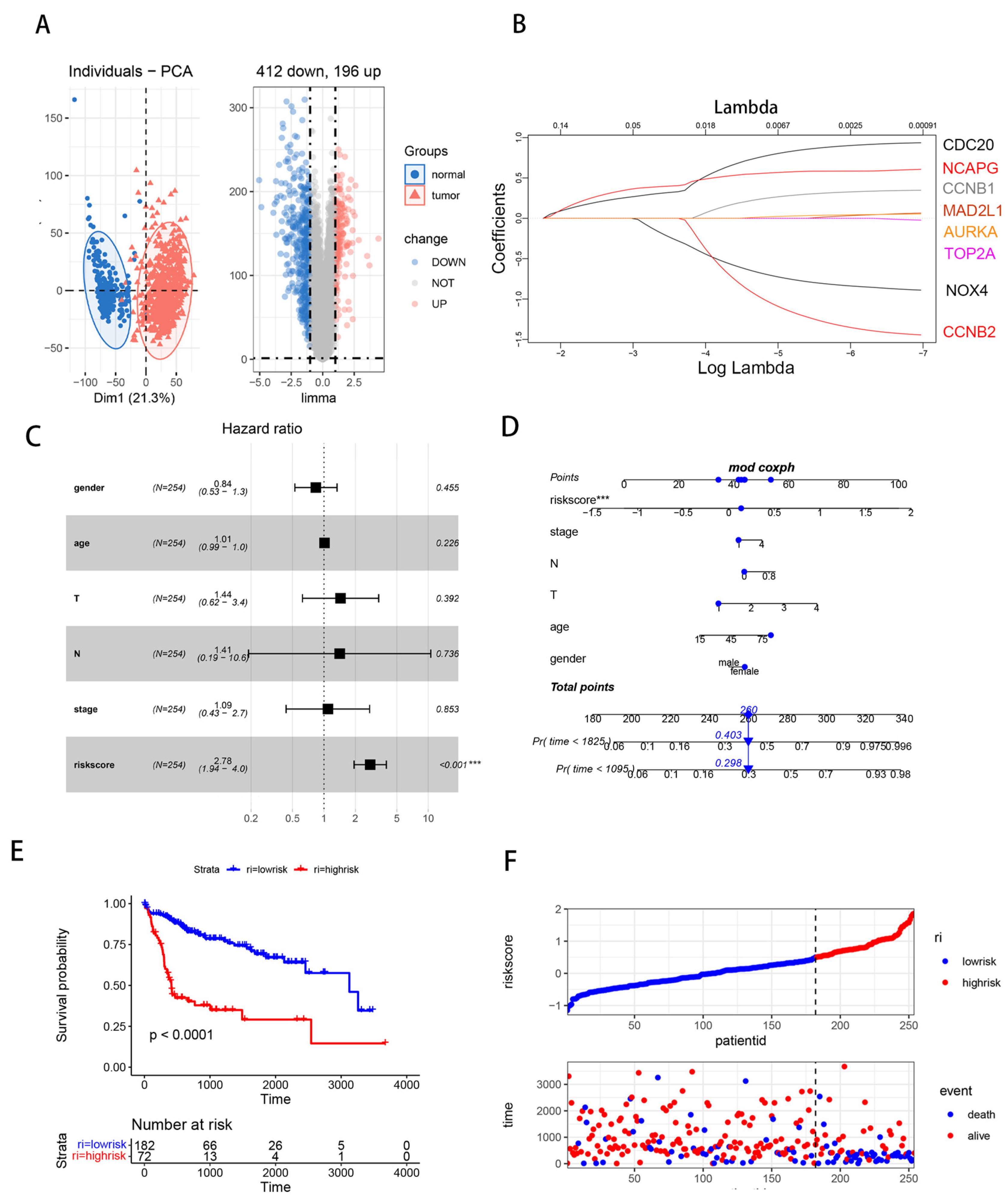
3.1. Differentially Expressed Genes (DEGs) Analysis in HCC
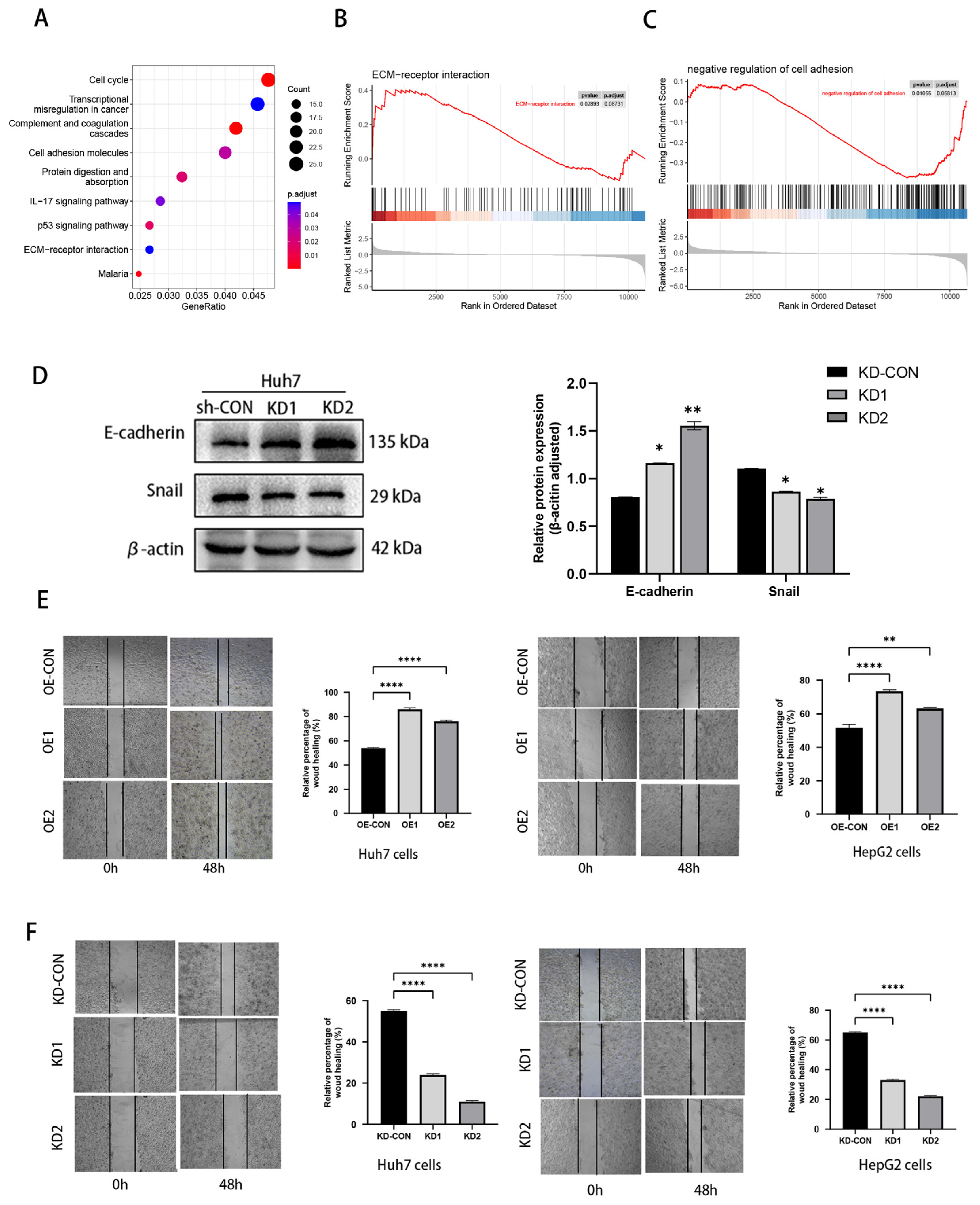
3.2. Hub Gene Identification and Functional Enrichment Analysis
3.3. Association of NOX4 Expression with Clinical Characteristics and Prognosis of HCC Patients
3.4. NOX4 Knockdown Decreased the Proliferation and Migration of HCC Cells In Vitro and In Vivo
3.5. Overexpression of NOX4 Promoted HCC Cell Proliferation In Vitro and Vivo
3.6. NOX4 Influenced HCC Cell Migration: The Epithelial–Mesenchymal Transition (EMT)
3.7. NOX4 Elevation Is Associated with Therapeutic Resistance and Immunotherapy Tolerance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
List of Abbreviations
References
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S.; et al. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Kim, H.; Lee, D.H.; Hur, B.Y.; Hwang, Y.J.; Suh, K.S.; Han, J.K. Gadoxetate-enhanced MRI Features of Proliferative Hepatocellular Carcinoma Are Prognostic after Surgery. Radiology. 2021, 300, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Foerster, F.; Galle, P.R. Comparison of the current international guidelines on the management of HCC. JHEP Rep. 2019, 1, 114–119. [Google Scholar] [CrossRef] [Green Version]
- Bai, X.M.; Cui, M.; Yang, W.; Wang, H.; Wang, S.; Zhang, Z.Y.; Wu, W.; Chen, M.H.; Yan, K.; Goldberg, S.N. The 10-year Survival Analysis of Radiofrequency Ablation for Solitary Hepatocellular Carcinoma 5 cm or Smaller: Primary versus Recurrent HCC. Radiology 2021, 300, 458–469. [Google Scholar] [CrossRef]
- Borde, T.; Nezami, N.; Gaupp, F.L.; Savic, L.J.; Taddei, T.; Jaffe, A.; Strazzabosco, M.; Lin, M.D.; Duran, R.; Georgiades, C.; et al. Optimization of the BCLC Staging System for Locoregional Therapy for Hepatocellular Carcinoma by Using Quantitative Tumor Burden Imaging Biomarkers at MRI. Radiology 2022, 304, 228–237. [Google Scholar] [CrossRef]
- Konaté, M.M.; Antony, S.; Doroshow, J.H. Inhibiting the Activity of NADPH Oxidase in Cancer. Antioxid. Redox Signal. 2020, 33, 435–454. [Google Scholar] [CrossRef]
- Koh, H.; Jang, B.; Hyun, C.; Kim, D. Prognostic Value of NOX4 Expression in Cancer Patients: A Systematic Review and Meta-analysis. Dis. Mrk. 2022, 2022, 8567642. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Wang, S.; Shao, M. NADPH Oxidase 4: A Potential Therapeutic Target of Malignancy. Front. Cell Dev. Biol. 2022, 10, 884412. [Google Scholar] [CrossRef] [PubMed]
- Moldogazieva, N.T.; Zavadskiy, S.P.; Sologova, S.S.; Mokhosoev, I.M.; Terentiev, A.A. Predictive biomarkers for systemic therapy of hepatocellular carcinoma. Expert. Rev. Mol. Diagn. 2021, 21, 1147–1164. [Google Scholar] [CrossRef]
- Feng, Z.C.; Li, H.L.; Zhao, H.F.; Jiang, Y.; Liu, Q.; Chen, Q.; Wang, W.; Rong, P.F. Preoperative CT for Characterization of Aggressive Macrotrabecular-Massive Subtype and Vessels That Encapsulate Tumor Clusters Pattern in Hepatocellular Carcinoma. Radiology. 2021, 300, 219–229. [Google Scholar] [CrossRef]
- YuFeng, Z.; Ming, Q. Expression and prognostic roles of PABPC1 in hepatocellular carcinoma. Int. J. Surg. 2020, 84, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, J. The Prognostic and Immunotherapeutic Significance of AHSA1 in Pan-Cancer, and Its Relationship with the Proliferation and Metastasis of Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 845585. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.; Cao, T.; Qian, Y.; Wang, H.; Ju, S.; Chen, Y.; Chen, T.; Yang, J.; Liang, B.; Hou, S. CDC20 is a novel biomarker for improved clinical predictions in epithelial ovarian cancer. Am. J. Cancer Res. 2022, 12, 3303–3317. [Google Scholar]
- Greil, C.; Engelhardt, M.; Wäsch, R. The Role of the APC/C and Its Coactivators Cdh1 and Cdc20 in Cancer Development and Therapy. Front. Genet. 2022, 13, 941565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Ai, J.; Wang, J.; Sun, C.; Lu, H.; He, A.; Li, M.; Liao, Y.; Lei, J.; Zhou, F.; et al. NCAPG Promotes the Prolif-eration of Hepatocellular Carcinoma through the CKII-Dependent Regulation of PTEN. J. Transl. Med. 2022, 20, 325. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, S.; Zhang, Y.; Li, B.; Xu, H.; Zuo, S. Identification of MAD2L1 as a Potential Biomarker in Hepatocellular Carcinoma via Comprehensive Bioinformatics Analysis. BioMed Res. Int. 2022, 2022, 9868022. [Google Scholar] [CrossRef]
- Jiali, M.; Wei, Y.; Deng, Q.; Li, L.; Li, X. Study on the Expression of TOP2A in Hepatocellular Car-cinoma and Its Relationship with Patient Prognosis. Cancer Cell Int. 2022, 22, 29. [Google Scholar]
- Guo, S.; Chen, X. The human Nox4: Gene, structure, physiological function and pathological significance. J. Drug. Target. 2015, 23, 888–896. [Google Scholar] [CrossRef]
- Perillo, B.; Di Donato, M.; Pezone, A.; Di Zazzo, E.; Giovannelli, P.; Galasso, G.; Castoria, G.; Migliaccio, A. ROS in cancer therapy: The bright side of the moon. Exp. Mol. Med. 2020, 52, 192–203. [Google Scholar] [CrossRef]
- Cheung, E.C.; Vousden, K.H. The role of ROS in tumour development and progression. Nat. Rev. Cancer 2022, 22, 280–297. [Google Scholar] [CrossRef]
- Szczepaniak, P.; Siedlinski, M.; Hodorowicz-Zaniewska, D.; Nosalski, R.; Mikolajczyk, T.P.; Dobosz, A.M.; Dikalova, A.; Dikalov, S.; Streb, J.; Gara, K.; et al. Breast cancer chemotherapy induces vascular dysfunction and hypertension through a NOX4-dependent mechanism. J. Clin. Investig. 2022, 132, e149117. [Google Scholar] [CrossRef]
- Bi, Y.; Lei, X.; Chai, N.; Linghu, E. NOX4: A potential therapeutic target for pancreatic cancer and its mechanism. J. Transl. Med. 2021, 19, 515. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.-H.; Wu, C.-P.; Lee, H.-T.; Liang, J.-A.; Yu, C.-Y.; Lin, Y.-J. NADPH oxidase subunit 4 mediates cycling hypoxia-promoted radiation resistance in glioblastoma multiforme. Free Radic. Biol. Med. 2012, 53, 649–658. [Google Scholar] [CrossRef]
- Kumar, B.; Koul, S.; Khandrika, L.; Meacham, R.B.; Koul, H.K. Oxidative Stress Is Inherent in Prostate Cancer Cells and Is Required for Aggressive Phenotype. Cancer Res. 2008, 68, 1777–1785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, M.-S.; Wang, H.; Ansari, K.H.; Li, X.-P.; Sun, W.; Fan, Y.-Z. Gallbladder cancer-associated fibroblasts promote vasculogenic mimicry formation and tumor growth in gallbladder cancer via upregulating the expression of NOX4, a poor prognosis factor, through IL-6-JAK-STAT3 signal pathway. J. Exp. Clin. Cancer Res. 2020, 39, 234. [Google Scholar] [CrossRef]
- Jain, P.; Dvorkin-Gheva, A.; Mollen, E.; Malbeteau, L.; Xie, M.; Jessa, F.; Dhavarasa, P.; Chung, S.; Brown, K.R.; Jang, G.H.; et al. NOX4 links metabolic regulation in pancreatic cancer to endoplasmic reticulum redox vulnerability and dependence on PRDX4. Sci. Adv. 2021, 7, abf7114. [Google Scholar] [CrossRef]
- Eun, H.S.; Chun, K.; Song, I.-S.; Oh, C.-H.; Seong, I.-O.; Yeo, M.-K.; Kim, K.-H. High nuclear NADPH oxidase 4 expression levels are correlated with cancer development and poor prognosis in hepatocellular carcinoma. Pathology 2019, 51, 579–585. [Google Scholar] [CrossRef]
- Crosas-Molist, E.; Bertran, E.; Sancho, P.; López-Luque, J.; Fernando, J.; Sánchez, A.; Fernández, M.; Navarro, E.; Fabregat, I. The NADPH oxidase NOX4 inhibits hepatocyte proliferation and liver cancer progression. Free. Radic. Biol. Med. 2014, 69, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, D.-H.; Shah, S.R.; Kim, H.-N.; Kshitiz; Kim, P.; Quiñones-Hinojosa, A.; Levchenko, A. Switch-like enhancement of epithelial-mesenchymal transition by YAP through feedback regulation of WT1 and Rho-family GTPases. Nat. Commun. 2019, 10, 2797. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Steed, A.; Co, M.; Chen, X. Cancer stem cells, epithelial-mesenchymal transition, ATP and their roles in drug resistance in cancer. Cancer Drug. Resist. 2021, 4, 684–709. [Google Scholar] [CrossRef]
- Zhang, P.-F.; Li, K.-S.; Shen, Y.-H.; Gao, P.-T.; Dong, Z.-R.; Cai, J.-B.; Zhang, C.; Huang, X.-Y.; Tian, M.-X.; Hu, Z.-Q.; et al. Galectin-1 induces hepatocellular carcinoma EMT and sorafenib resistance by activating FAK/PI3K/AKT signaling. Cell Death Dis. 2016, 7, e2201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.Y.; Ke, A.W.; Shi, G.M.; Zhang, X.; Zhang, C.; Shi, Y.H.; Wang, X.-Y.; Ding, Z.-B.; Xiao, Y.-S.; Yan, J.; et al. αB-crystallin complexes with 14-3-3ζ to induce epitheli-al-mesenchymal transition and resistance to sorafenib in hepatocellular carcinoma. Hepatology 2013, 57, 2235–2247. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Cheng, X.; Yu, J.; Li, Y. Stabilization of snail maintains the sorafenib resistance of hepatocellular carcinoma cells. Arch. Biochem. Biophys. 2021, 699, 108754. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Chen, Z.; Zhang, W.; Cheng, Y.; Zhang, B.; Wu, F.; Wang, Q.; Wang, S.; Rong, D.; Reiter, F.P.; et al. The mechanisms of sorafenib resistance in hepatocellular carcinoma: Theoretical basis and therapeutic aspects. Signal Transduct. Target. Ther. 2020, 5, 87. [Google Scholar] [CrossRef] [PubMed]
- Granito, A.; Forgione, A.; Marinelli, S.; Renzulli, M.; Ielasi, L.; Sansone, V.; Benevento, F.; Piscaglia, F.; Tovoli, F. Experience with regorafenib in the treatment of hepatocellular carcinoma. Ther. Adv. Gastroenterol. 2021, 14, 17562848211016959. [Google Scholar] [CrossRef]
- Trevisani, F.; Brandi, G.; Garuti, F.; Barbera, M.A.; Tortora, R.; Gardini, A.C.; Granito, A.; Tovoli, F.; De Lorenzo, S.; Inghilesi, A.L.; et al. Metronomic capecitabine as second-line treatment for hepatocellular carcinoma after sorafenib discontinuation. J. Cancer Res. Clin. Oncol. 2017, 144, 403–414. [Google Scholar] [CrossRef] [Green Version]
- Stefanini, B.; Ielasi, L.; Chen, R.; Abbati, C.; Tonnini, M.; Tovoli, F.; Granito, A. TKIs in combination with immunotherapy for hepatocellular carcinoma. Expert. Rev. Anticancer Ther. 2023, 23, 279–291. [Google Scholar] [CrossRef]

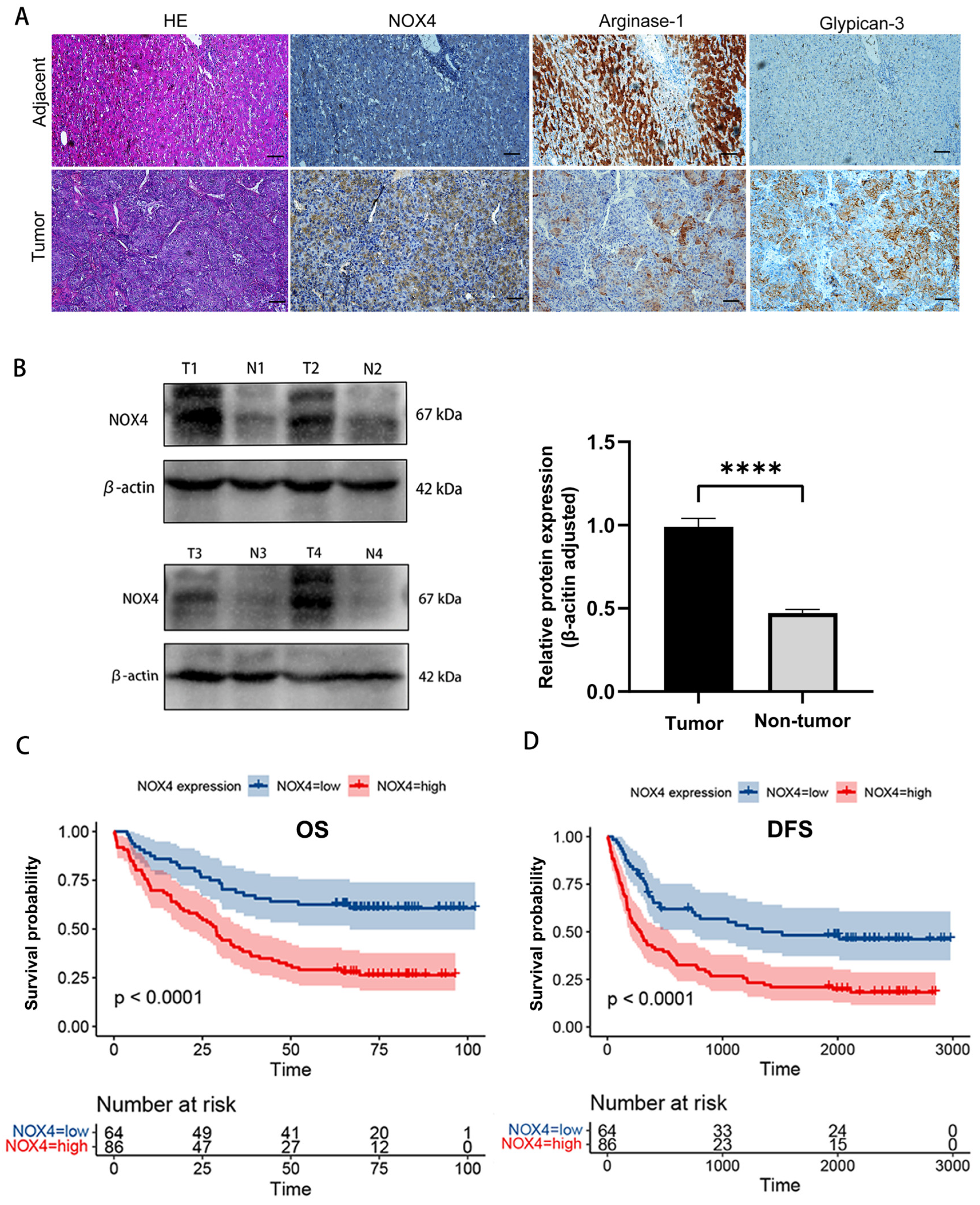



| Characteristics | NOX4 Expression | |||
|---|---|---|---|---|
| Overall (n = 150) | Low (n = 64) | High (n = 86) | p-Value | |
| Age (years, mean ± SD) | 51.2 ± 12.1 | 48.9 ± 13.8 | 52.9 ± 10.3 | 0.057 |
| Sex (n, %) | ||||
| male | 130 (86.7) | 51 (79.7) | 79 (91.9) | |
| female | 20 (13.3) | 13 (20.3) | 7 (8.1) | 0.030 |
| Number of tumors (n, %) | ||||
| single | 131 (87.3) | 58 (90.6) | 73 (84.9) | |
| multiple | 19 (12.7) | 6 (9.4) | 13 (15.1) | 0.296 |
| Child-Pugh class (n, %) | ||||
| A | 134 (89.3) | 58 (90.6) | 76 (88.4) | |
| B | 10 (6.7) | 4 (6.3) | 6 (6.9) | |
| C | 6 (4.0) | 2 (3.1) | 4 (4.7) | 0.877 |
| Etiologies of hepatitis (n, %) | ||||
| none | 20 (13.3) | 6 (9.4) | 14 (16.3) | |
| HBV | 114 (76.0) | 53 (82.8) | 61 (70.9) | |
| HCV | 5 (3.3) | 1(1.6) | 4 (4.7) | |
| alcohol | 3 (2.0) | 2 (3.1) | 1 (1.2) | |
| others | 8 (5.4) | 2 (3.1) | 6 (6.9) | 0.312 |
| Cirrhosis (n, %) | ||||
| absence | 69 (46.0) | 35 (54.7) | 34 (39.5) | |
| presence | 81 (54.0) | 29 (45.3) | 52 (60.5) | 0.066 |
| AFP (n, %) | ||||
| ≤400 ng/mL | 120 (80.0) | 51 (79.7) | 69 (80.2) | |
| >400 ng/mL | 30(20.0) | 13 (20.3) | 17 (19.8) | 0.934 |
| Vascular invasion | ||||
| absence | 130 (86.7) | 59 (92.2) | 71 (82.6) | |
| presence | 20 (13.3) | 5 (7.8) | 15 (17.4) | 0.086 |
| Differentiation grade (n, %) | ||||
| high | 18 (12.0) | 9 (14.1) | 9 (10.5) | |
| median | 69 (46.0) | 32 (50.0) | 37 (43.0) | |
| low | 63 (42.0) | 23 (35.9) | 40 (46.5) | 0.415 |
| Prognostic Variables | Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|---|
| No. of Patients | HR (95% CI) | p | HR (95% CI) | p | |
| Expression of NOX4 | |||||
| low | 64 | [Reference] | [Reference] | ||
| high | 86 | 2.522 (1.588–4.005) | <0.001 | 2.397 (1.486–3.867) | <0.001 |
| Age | |||||
| ≤55 years | 93 | [Reference] | [Reference] | ||
| >55 years | 57 | 1.616 (1.059–2.465) | 0.026 | 1.577 (1.024–2.429) | 0.039 |
| Sex | |||||
| female | 20 | [Reference] | [Reference] | ||
| male | 130 | 2.177 (1.050–4.510) | 0.036 | 1.601 (0.758–3.382) | 0.217 |
| Number of tumors | |||||
| single | 131 | [Reference] | [Reference] | ||
| multiple | 19 | 2.542 (1.450–4.459) | 0.001 | 2.533 (1.406–4.561) | 0.002 |
| HBV infection | |||||
| negative | 36 | [Reference] | |||
| positive | 114 | 1.332 (0.081–2.215) | 0.270 | ||
| Cirrhosis | |||||
| absence | 69 | [Reference] | |||
| presence | 81 | 1.452 (0.945–2.231) | 0.089 | ||
| AFP | |||||
| ≤400 ng/mL | 120 | [Reference] | |||
| >400 ng/mL | 30 | 1.217 (0.724–2.0460 | 0.458 | ||
| Vascular invasion | |||||
| absence | 130 | [Reference] | [Reference] | ||
| presence | 20 | 2.103 (1.237–3.576) | 0.006 | 1.155 (0.662–2.016) | 0.611 |
| Differentiation grade | |||||
| high | 18 | [Reference] | [Reference] | ||
| median | 69 | 2.994 (1.065–8.418) | 0.038 | 2.485 (0.875–7.053) | 0.087 |
| low | 63 | 6.150 (2.211–17.108) | 0.001 | 5.310 (1.881–14.989) | 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.-Z.; Liu, Q.-Q.; Chang, D.-H.; Li, S.-X.; Yang, L.-T.; Zhou, P.; Deng, J.-B.; Huang, C.-H.; Xiao, Y.-D. Identification of NOX4 as a New Biomarker in Hepatocellular Carcinoma and Its Effect on Sorafenib Therapy. Biomedicines 2023, 11, 2196. https://doi.org/10.3390/biomedicines11082196
Li H-Z, Liu Q-Q, Chang D-H, Li S-X, Yang L-T, Zhou P, Deng J-B, Huang C-H, Xiao Y-D. Identification of NOX4 as a New Biomarker in Hepatocellular Carcinoma and Its Effect on Sorafenib Therapy. Biomedicines. 2023; 11(8):2196. https://doi.org/10.3390/biomedicines11082196
Chicago/Turabian StyleLi, Hui-Zhou, Qing-Qing Liu, De-Hua Chang, Shu-Xian Li, Long-Tao Yang, Peng Zhou, Jiang-Bei Deng, Chang-Hao Huang, and Yu-Dong Xiao. 2023. "Identification of NOX4 as a New Biomarker in Hepatocellular Carcinoma and Its Effect on Sorafenib Therapy" Biomedicines 11, no. 8: 2196. https://doi.org/10.3390/biomedicines11082196





