Human Gut Microbiota in Heart Failure: Trying to Unmask an Emerging Organ
Abstract
:1. Introduction
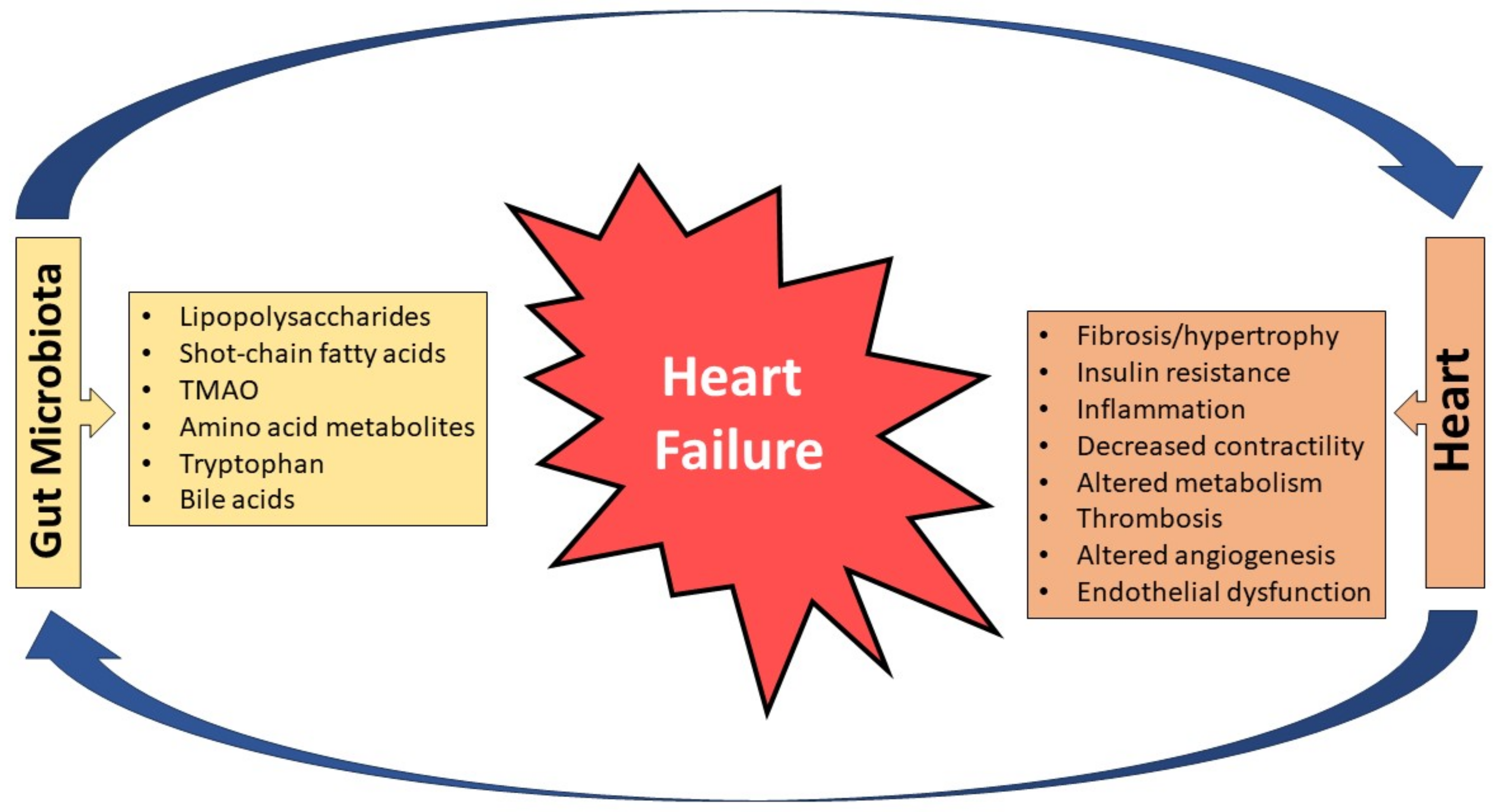
2. Bidirectional Relationship between the Heart and the Gut
3. Understanding the Gut Microbiota
4. Gut Microbiota as a Diagnostic Marker
5. Gut Microbiota and Medications
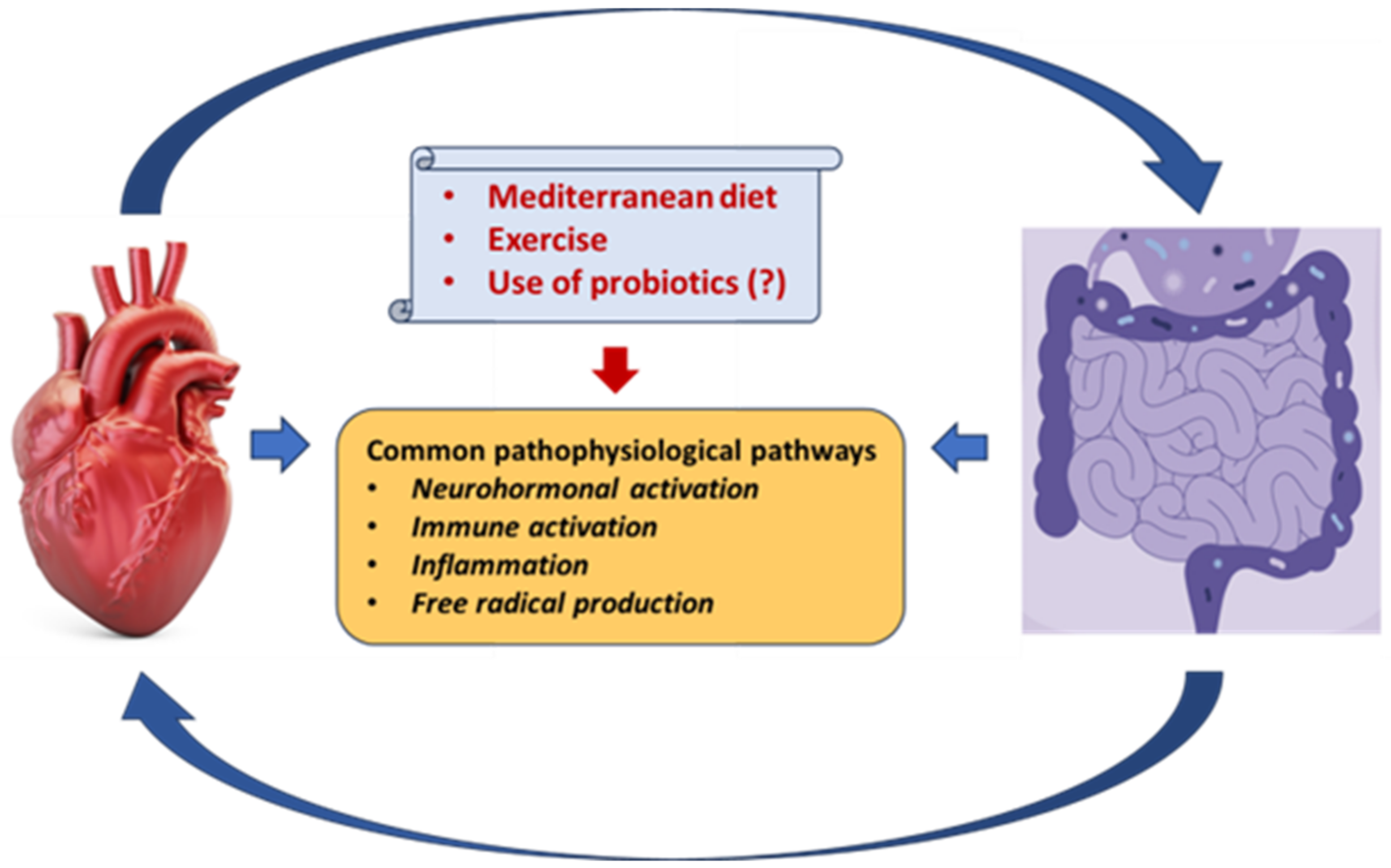
6. Gut Microbiota, Aging, Diet, Exercise Training, and Supplements
7. Future Directions
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Conrad, N.; Judge, A.; Tran, J.; Mohseni, H.; Hedgecott, D.; Crespillo, A.P.; Allison, M.; Hemingway, H.; Cleland, J.G.; McMurray, J.J.V.; et al. Temporal trends and patterns in heart failure incidence: A population-based study of 4 million individuals. Lancet 2018, 391, 572–580. [Google Scholar] [CrossRef]
- Disease, G.B.D.; Injury, I.; Prevalence, C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef]
- Virani, S.S.; Alonso, A.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics-2020 Update: A Report From the American Heart Association. Circulation 2020, 141, e139–e596. [Google Scholar] [CrossRef]
- van Riet, E.E.; Hoes, A.W.; Limburg, A.; Landman, M.A.; van der Hoeven, H.; Rutten, F.H. Prevalence of unrecognized heart failure in older persons with shortness of breath on exertion. Eur. J. Heart Fail. 2014, 16, 772–777. [Google Scholar] [CrossRef]
- Gerber, Y.; Weston, S.A.; Redfield, M.M.; Chamberlain, A.M.; Manemann, S.M.; Jiang, R.; Killian, J.M.; Roger, V.L. A contemporary appraisal of the heart failure epidemic in Olmsted County, Minnesota, 2000 to 2010. JAMA Intern. Med. 2015, 175, 996–1004. [Google Scholar] [CrossRef]
- Tsao, C.W.; Lyass, A.; Enserro, D.; Larson, M.G.; Ho, J.E.; Kizer, J.R.; Gottdiener, J.S.; Psaty, B.M.; Vasan, R.S. Temporal Trends in the Incidence of and Mortality Associated With Heart Failure With Preserved and Reduced Ejection Fraction. JACC Heart Fail. 2018, 6, 678–685. [Google Scholar] [CrossRef]
- Savarese, G.; Lund, L.H. Global Public Health Burden of Heart Failure. Card. Fail. Rev. 2017, 3, 7–11. [Google Scholar] [CrossRef]
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Bohm, M.; Burri, H.; Butler, J.; Celutkiene, J.; Chioncel, O.; et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 2021, 42, 3599–3726. [Google Scholar] [CrossRef]
- Paraskevaidis, I.; Farmakis, D.; Papingiotis, G.; Tsougos, E. Inflammation and Heart Failure: Searching for the Enemy-Reaching the Entelechy. J. Cardiovasc. Dev. Dis. 2023, 10, 19. [Google Scholar] [CrossRef]
- Yancy, C.W.; Jessup, M.; Bozkurt, B.; Butler, J.; Casey, D.E., Jr.; Colvin, M.M.; Drazner, M.H.; Filippatos, G.S.; Fonarow, G.C.; Givertz, M.M.; et al. 2017 ACC/AHA/HFSA Focused Update of the 2013 ACCF/AHA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Failure Society of America. Circulation 2017, 136, e137–e161. [Google Scholar] [CrossRef]
- Anker, S.D.; Egerer, K.R.; Volk, H.D.; Kox, W.J.; Poole-Wilson, P.A.; Coats, A.J. Elevated soluble CD14 receptors and altered cytokines in chronic heart failure. Am. J. Cardiol. 1997, 79, 1426–1430. [Google Scholar] [CrossRef]
- Krack, A.; Sharma, R.; Figulla, H.R.; Anker, S.D. The importance of the gastrointestinal system in the pathogenesis of heart failure. Eur. Heart J. 2005, 26, 2368–2374. [Google Scholar] [CrossRef]
- Mamic, P.; Snyder, M.; Tang, W.H.W. Gut Microbiome-Based Management of Patients With Heart Failure: JACC Review Topic of the Week. J. Am. Coll. Cardiol. 2023, 81, 1729–1739. [Google Scholar] [CrossRef] [PubMed]
- Rogler, G.; Rosano, G. The heart and the gut. Eur. Heart J. 2014, 35, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Jiang, Y.; Xu, H.; Zhou, Y. The Interaction of Gut Microbiota and Heart Failure with Preserved Ejection Fraction: From Mechanism to Potential Therapies. Biomedicines 2023, 11, 442. [Google Scholar] [CrossRef] [PubMed]
- Witkowski, M.; Weeks, T.L.; Hazen, S.L. Gut Microbiota and Cardiovascular Disease. Circ. Res. 2020, 127, 553–570. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, S.; Jin, X.; Li, D.; Lu, J.; Wang, X.; Wu, M. Mitochondria as novel mediators linking gut microbiota to atherosclerosis that is ameliorated by herbal medicine: A review. Front. Pharmacol. 2023, 14, 1082817. [Google Scholar] [CrossRef]
- Gregory, J.C.; Buffa, J.A.; Org, E.; Wang, Z.; Levison, B.S.; Zhu, W.; Wagner, M.A.; Bennett, B.J.; Li, L.; DiDonato, J.A.; et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J. Biol. Chem. 2015, 290, 5647–5660. [Google Scholar] [CrossRef]
- Lian, W.S.; Wang, F.S.; Chen, Y.S.; Tsai, M.H.; Chao, H.R.; Jahr, H.; Wu, R.W.; Ko, J.Y. Gut Microbiota Ecosystem Governance of Host Inflammation, Mitochondrial Respiration and Skeletal Homeostasis. Biomedicines 2022, 10, 860. [Google Scholar] [CrossRef]
- Tang, W.H.W.; Li, D.Y.; Hazen, S.L. Dietary metabolism, the gut microbiome, and heart failure. Nat. Rev. Cardiol. 2019, 16, 137–154. [Google Scholar] [CrossRef]
- Vezza, T.; Abad-Jimenez, Z.; Marti-Cabrera, M.; Rocha, M.; Victor, V.M. Microbiota-Mitochondria Inter-Talk: A Potential Therapeutic Strategy in Obesity and Type 2 Diabetes. Antioxidants 2020, 9, 848. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, T.; Moens de Hase, E.; Van Hul, M.; Paquot, A.; Pelicaen, R.; Regnier, M.; Depommier, C.; Druart, C.; Everard, A.; Maiter, D.; et al. Dysosmobacter welbionis is a newly isolated human commensal bacterium preventing diet-induced obesity and metabolic disorders in mice. Gut 2022, 71, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Barrington, W.T.; Lusis, A.J. Atherosclerosis: Association between the gut microbiome and atherosclerosis. Nat. Rev. Cardiol. 2017, 14, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, A.L.; Backhed, F. Role of gut microbiota in atherosclerosis. Nat. Rev. Cardiol. 2017, 14, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Basak, S.; Banerjee, A.; Pathak, S.; Duttaroy, A.K. Dietary Fats and the Gut Microbiota: Their impacts on lipid-induced metabolic syndrome. J. Funct. Foods 2022, 91, 105026. [Google Scholar] [CrossRef]
- Salvatore, T.; Galiero, R.; Caturano, A.; Vetrano, E.; Rinaldi, L.; Coviello, F.; Di Martino, A.; Albanese, G.; Colantuoni, S.; Medicamento, G.; et al. Dysregulated Epicardial Adipose Tissue as a Risk Factor and Potential Therapeutic Target of Heart Failure with Preserved Ejection Fraction in Diabetes. Biomolecules 2022, 12, 176. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef]
- Zheng, P.; Zeng, B.; Zhou, C.; Liu, M.; Fang, Z.; Xu, X.; Zeng, L.; Chen, J.; Fan, S.; Du, X.; et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol. Psychiatry 2016, 21, 786–796. [Google Scholar] [CrossRef]
- Fromentin, S.; Forslund, S.K.; Chechi, K.; Aron-Wisnewsky, J.; Chakaroun, R.; Nielsen, T.; Tremaroli, V.; Ji, B.; Prifti, E.; Myridakis, A.; et al. Microbiome and metabolome features of the cardiometabolic disease spectrum. Nat. Med. 2022, 28, 303–314. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, V.; Duran, P.; Rojas, E.; Diaz, M.P.; Rivas, J.; Nava, M.; Chacin, M.; Cabrera de Bravo, M.; Carrasquero, R.; Ponce, C.C.; et al. The Sick Adipose Tissue: New Insights Into Defective Signaling and Crosstalk With the Myocardium. Front. Endocrinol. 2021, 12, 735070. [Google Scholar] [CrossRef] [PubMed]
- Zhan, S.; Li, N.; Liu, C.; Mao, R.; Wu, D.; Li, T.; Chen, M.; Zhuang, X.; Zeng, Z. Intestinal Fibrosis and Gut Microbiota: Clues From Other Organs. Front. Microbiol. 2021, 12, 694967. [Google Scholar] [CrossRef] [PubMed]
- Nemet, I.; Saha, P.P.; Gupta, N.; Zhu, W.; Romano, K.A.; Skye, S.M.; Cajka, T.; Mohan, M.L.; Li, L.; Wu, Y.; et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell 2020, 180, 862–877.e822. [Google Scholar] [CrossRef] [PubMed]
- Kummen, M.; Mayerhofer, C.C.K.; Vestad, B.; Broch, K.; Awoyemi, A.; Storm-Larsen, C.; Ueland, T.; Yndestad, A.; Hov, J.R.; Troseid, M. Gut Microbiota Signature in Heart Failure Defined From Profiling of 2 Independent Cohorts. J. Am. Coll. Cardiol. 2018, 71, 1184–1186. [Google Scholar] [CrossRef] [PubMed]
- Hietbrink, F.; Besselink, M.G.; Renooij, W.; de Smet, M.B.; Draisma, A.; van der Hoeven, H.; Pickkers, P. Systemic inflammation increases intestinal permeability during experimental human endotoxemia. Shock 2009, 32, 374–378. [Google Scholar] [CrossRef]
- Mu, F.; Tang, M.; Guan, Y.; Lin, R.; Zhao, M.; Zhao, J.; Huang, S.; Zhang, H.; Wang, J.; Tang, H. Knowledge Mapping of the Links Between the Gut Microbiota and Heart Failure: A Scientometric Investigation (2006–2021). Front. Cardiovasc. Med. 2022, 9, 882660. [Google Scholar] [CrossRef]
- Lupu, V.V.; Adam Raileanu, A.; Mihai, C.M.; Morariu, I.D.; Lupu, A.; Starcea, I.M.; Frasinariu, O.E.; Mocanu, A.; Dragan, F.; Fotea, S. The Implication of the Gut Microbiome in Heart Failure. Cells 2023, 12, 1158. [Google Scholar] [CrossRef]
- Ferranti, E.P.; Dunbar, S.B.; Dunlop, A.L.; Corwin, E.J. 20 things you didn’t know about the human gut microbiome. J. Cardiovasc. Nurs. 2014, 29, 479–481. [Google Scholar] [CrossRef]
- Beam, A.; Clinger, E.; Hao, L. Effect of Diet and Dietary Components on the Composition of the Gut Microbiota. Nutrients 2021, 13, 2795. [Google Scholar] [CrossRef]
- Hall, A.B.; Tolonen, A.C.; Xavier, R.J. Human genetic variation and the gut microbiome in disease. Nat. Rev. Genet. 2017, 18, 690–699. [Google Scholar] [CrossRef] [PubMed]
- Koppel, N.; Maini Rekdal, V.; Balskus, E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356, eaag2770. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, J.L.; Backhed, F. Diet-microbiota interactions as moderators of human metabolism. Nature 2016, 535, 56–64. [Google Scholar] [CrossRef]
- Brown, J.M.; Hazen, S.L. Targeting of microbe-derived metabolites to improve human health: The next frontier for drug discovery. J. Biol. Chem. 2017, 292, 8560–8568. [Google Scholar] [CrossRef] [PubMed]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.Z.; Nelson, E.; Chu, P.Y.; Horlock, D.; Fiedler, A.; Ziemann, M.; Tan, J.K.; Kuruppu, S.; Rajapakse, N.W.; El-Osta, A.; et al. High-Fiber Diet and Acetate Supplementation Change the Gut Microbiota and Prevent the Development of Hypertension and Heart Failure in Hypertensive Mice. Circulation 2017, 135, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Vatanen, T.; Kostic, A.D.; d’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hamalainen, A.M.; et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016, 165, 1551. [Google Scholar] [CrossRef]
- Mayerhofer, C.C.K.; Ueland, T.; Broch, K.; Vincent, R.P.; Cross, G.F.; Dahl, C.P.; Aukrust, P.; Gullestad, L.; Hov, J.R.; Troseid, M. Increased Secondary/Primary Bile Acid Ratio in Chronic Heart Failure. J. Card. Fail. 2017, 23, 666–671. [Google Scholar] [CrossRef]
- Binah, O.; Rubinstein, I.; Bomzon, A.; Better, O.S. Effects of bile acids on ventricular muscle contraction and electrophysiological properties: Studies in rat papillary muscle and isolated ventricular myocytes. Naunyn Schmiedebergs Arch. Pharmacol. 1987, 335, 160–165. [Google Scholar] [CrossRef]
- Joubert, P. An in vivo investigation of the negative chronotropic effect of cholic acid in the rat. Clin. Exp. Pharmacol. Physiol. 1978, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gazawi, H.; Ljubuncic, P.; Cogan, U.; Hochgraff, E.; Ben-Shachar, D.; Bomzon, A. The effects of bile acids on beta-adrenoceptors, fluidity, and the extent of lipid peroxidation in rat cardiac membranes. Biochem. Pharmacol. 2000, 59, 1623–1628. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.D.; Chen, W.D.; Wang, M.; Yu, D.; Forman, B.M.; Huang, W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology 2008, 48, 1632–1643. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.T.; Swales, K.E.; Thomas, G.J.; Warner, T.D.; Bishop-Bailey, D. Farnesoid x receptor ligands inhibit vascular smooth muscle cell inflammation and migration. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 2606–2611. [Google Scholar] [CrossRef]
- Purcell, N.H.; Tang, G.; Yu, C.; Mercurio, F.; DiDonato, J.A.; Lin, A. Activation of NF-kappa B is required for hypertrophic growth of primary rat neonatal ventricular cardiomyocytes. Proc. Natl. Acad. Sci. USA 2001, 98, 6668–6673. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.W.; Shaw, J.A.; Kirshenbaum, L.A. Multiple facets of NF-kappaB in the heart: To be or not to NF-kappaB. Circ. Res. 2011, 108, 1122–1132. [Google Scholar] [CrossRef]
- Pu, J.; Yuan, A.; Shan, P.; Gao, E.; Wang, X.; Wang, Y.; Lau, W.B.; Koch, W.; Ma, X.L.; He, B. Cardiomyocyte-expressed farnesoid-X-receptor is a novel apoptosis mediator and contributes to myocardial ischaemia/reperfusion injury. Eur. Heart J. 2013, 34, 1834–1845. [Google Scholar] [CrossRef]
- Calkin, A.C.; Tontonoz, P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 2012, 13, 213–224. [Google Scholar] [CrossRef]
- Gurney, M.A.; Laubitz, D.; Ghishan, F.K.; Kiela, P.R. Pathophysiology of Intestinal Na(+)/H(+) exchange. Cell Mol. Gastroenterol. Hepatol. 2017, 3, 27–40. [Google Scholar] [CrossRef]
- Bernardazzi, C.; Sheikh, I.A.; Xu, H.; Ghishan, F.K. The Physiological Function and Potential Role of the Ubiquitous Na(+)/H(+) Exchanger Isoform 8 (NHE8): An Overview Data. Int. J. Mol. Sci. 2022, 23, 10857. [Google Scholar] [CrossRef]
- Nikolovska, K.; Seidler, U.E.; Stock, C. The Role of Plasma Membrane Sodium/Hydrogen Exchangers in Gastrointestinal Functions: Proliferation and Differentiation, Fluid/Electrolyte Transport and Barrier Integrity. Front. Physiol. 2022, 13, 899286. [Google Scholar] [CrossRef] [PubMed]
- Cook, M.D.; Allen, J.M.; Pence, B.D.; Wallig, M.A.; Gaskins, H.R.; White, B.A.; Woods, J.A. Exercise and gut immune function: Evidence of alterations in colon immune cell homeostasis and microbiome characteristics with exercise training. Immunol. Cell Biol. 2016, 94, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Benedict, C.; Vogel, H.; Jonas, W.; Woting, A.; Blaut, M.; Schurmann, A.; Cedernaes, J. Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals. Mol. Metab. 2016, 5, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Zeng, D.N.; Chi, L.; Tan, Y.; Galzote, C.; Cardona, C.; Lax, S.; Gilbert, J.; Quan, Z.X. The Influence of Age and Gender on Skin-Associated Microbial Communities in Urban and Rural Human Populations. PLoS ONE 2015, 10, e0141842. [Google Scholar] [CrossRef] [PubMed]
- Zozaya, M.; Ferris, M.J.; Siren, J.D.; Lillis, R.; Myers, L.; Nsuami, M.J.; Eren, A.M.; Brown, J.; Taylor, C.M.; Martin, D.H. Bacterial communities in penile skin, male urethra, and vaginas of heterosexual couples with and without bacterial vaginosis. Microbiome 2016, 4, 16. [Google Scholar] [CrossRef]
- Lax, S.; Smith, D.P.; Hampton-Marcell, J.; Owens, S.M.; Handley, K.M.; Scott, N.M.; Gibbons, S.M.; Larsen, P.; Shogan, B.D.; Weiss, S.; et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 2014, 345, 1048–1052. [Google Scholar] [CrossRef]
- David, L.A.; Materna, A.C.; Friedman, J.; Campos-Baptista, M.I.; Blackburn, M.C.; Perrotta, A.; Erdman, S.E.; Alm, E.J. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014, 15, R89. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Costello, E.K.; Berg-Lyons, D.; Gonzalez, A.; Stombaugh, J.; Knights, D.; Gajer, P.; Ravel, J.; Fierer, N.; et al. Moving pictures of the human microbiome. Genome Biol. 2011, 12, R50. [Google Scholar] [CrossRef]
- Knight, R.; Jansson, J.; Field, D.; Fierer, N.; Desai, N.; Fuhrman, J.A.; Hugenholtz, P.; van der Lelie, D.; Meyer, F.; Stevens, R.; et al. Unlocking the potential of metagenomics through replicated experimental design. Nat. Biotechnol. 2012, 30, 513–520. [Google Scholar] [CrossRef]
- Geva-Zatorsky, N.; Sefik, E.; Kua, L.; Pasman, L.; Tan, T.G.; Ortiz-Lopez, A.; Yanortsang, T.B.; Yang, L.; Jupp, R.; Mathis, D.; et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 2017, 168, 928–943.e911. [Google Scholar] [CrossRef] [PubMed]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, N.; Gao, J.; Li, H.; Cai, W.; Zhang, X.; Ma, Y.; Niu, X.; Yang, G.; Zhou, X.; et al. The Effect of Different l-Carnitine Administration Routes on the Development of Atherosclerosis in ApoE Knockout Mice. Mol. Nutr. Food Res. 2018, 62, 1700299. [Google Scholar] [CrossRef] [PubMed]
- Organ, C.L.; Otsuka, H.; Bhushan, S.; Wang, Z.; Bradley, J.; Trivedi, R.; Polhemus, D.J.; Tang, W.H.; Wu, Y.; Hazen, S.L.; et al. Choline Diet and Its Gut Microbe-Derived Metabolite, Trimethylamine N-Oxide, Exacerbate Pressure Overload-Induced Heart Failure. Circ. Heart Fail. 2016, 9, e002314. [Google Scholar] [CrossRef]
- Zhu, W.; Gregory, J.C.; Org, E.; Buffa, J.A.; Gupta, N.; Wang, Z.; Li, L.; Fu, X.; Wu, Y.; Mehrabian, M.; et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell 2016, 165, 111–124. [Google Scholar] [CrossRef]
- Bennett, B.J.; de Aguiar Vallim, T.Q.; Wang, Z.; Shih, D.M.; Meng, Y.; Gregory, J.; Allayee, H.; Lee, R.; Graham, M.; Crooke, R.; et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013, 17, 49–60. [Google Scholar] [CrossRef]
- Tang, W.H.; Wang, Z.; Fan, Y.; Levison, B.; Hazen, J.E.; Donahue, L.M.; Wu, Y.; Hazen, S.L. Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-N-oxide in patients with heart failure: Refining the gut hypothesis. J. Am. Coll. Cardiol. 2014, 64, 1908–1914. [Google Scholar] [CrossRef]
- Tang, W.H.; Wang, Z.; Shrestha, K.; Borowski, A.G.; Wu, Y.; Troughton, R.W.; Klein, A.L.; Hazen, S.L. Intestinal microbiota-dependent phosphatidylcholine metabolites, diastolic dysfunction, and adverse clinical outcomes in chronic systolic heart failure. J. Card. Fail. 2015, 21, 91–96. [Google Scholar] [CrossRef]
- Troseid, M.; Ueland, T.; Hov, J.R.; Svardal, A.; Gregersen, I.; Dahl, C.P.; Aakhus, S.; Gude, E.; Bjorndal, B.; Halvorsen, B.; et al. Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J. Intern. Med. 2015, 277, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Schuett, K.; Kleber, M.E.; Scharnagl, H.; Lorkowski, S.; Marz, W.; Niessner, A.; Marx, N.; Meinitzer, A. Trimethylamine-N-oxide and Heart Failure With Reduced Versus Preserved Ejection Fraction. J. Am. Coll. Cardiol. 2017, 70, 3202–3204. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jin, M.; Liu, L.; Yu, Z.; Lu, X.; Zhang, H. Trimethylamine N-oxide and cardiovascular outcomes in patients with chronic heart failure after myocardial infarction. ESC Heart Fail. 2020, 7, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Heaney, L.M.; Bhandari, S.S.; Jones, D.J.; Ng, L.L. Trimethylamine N-oxide and prognosis in acute heart failure. Heart 2016, 102, 841–848. [Google Scholar] [CrossRef]
- Savi, M.; Bocchi, L.; Bresciani, L.; Falco, A.; Quaini, F.; Mena, P.; Brighenti, F.; Crozier, A.; Stilli, D.; Del Rio, D. Trimethylamine-N-Oxide (TMAO)-Induced Impairment of Cardiomyocyte Function and the Protective Role of Urolithin B-Glucuronide. Molecules 2018, 23, 549. [Google Scholar] [CrossRef]
- Li, X.; Fan, Z.; Cui, J.; Li, D.; Lu, J.; Cui, X.; Xie, L.; Wu, Y.; Lin, Q.; Li, Y. Trimethylamine N-Oxide in Heart Failure: A Meta-Analysis of Prognostic Value. Front. Cardiovasc. Med. 2022, 9, 817396. [Google Scholar] [CrossRef]
- Cui, X.; Ye, L.; Li, J.; Jin, L.; Wang, W.; Li, S.; Bao, M.; Wu, S.; Li, L.; Geng, B.; et al. Metagenomic and metabolomic analyses unveil dysbiosis of gut microbiota in chronic heart failure patients. Sci. Rep. 2018, 8, 635. [Google Scholar] [CrossRef]
- Li, D.Y.; Tang, W.H.W. Contributory Role of Gut Microbiota and Their Metabolites Toward Cardiovascular Complications in Chronic Kidney Disease. Semin. Nephrol. 2018, 38, 193–205. [Google Scholar] [CrossRef]
- Tang, W.H.; Wang, Z.; Kennedy, D.J.; Wu, Y.; Buffa, J.A.; Agatisa-Boyle, B.; Li, X.S.; Levison, B.S.; Hazen, S.L. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 2015, 116, 448–455. [Google Scholar] [CrossRef]
- Vanholder, R.; Schepers, E.; Pletinck, A.; Nagler, E.V.; Glorieux, G. The uremic toxicity of indoxyl sulfate and p-cresyl sulfate: A systematic review. J. Am. Soc. Nephrol. 2014, 25, 1897–1907. [Google Scholar] [CrossRef]
- Anand, I.S.; Gupta, P. Anemia and Iron Deficiency in Heart Failure: Current Concepts and Emerging Therapies. Circulation 2018, 138, 80–98. [Google Scholar] [CrossRef] [PubMed]
- Iorio, A.; Senni, M.; Barbati, G.; Greene, S.J.; Poli, S.; Zambon, E.; Di Nora, C.; Cioffi, G.; Tarantini, L.; Gavazzi, A.; et al. Prevalence and prognostic impact of non-cardiac co-morbidities in heart failure outpatients with preserved and reduced ejection fraction: A community-based study. Eur. J. Heart Fail. 2018, 20, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, A.J.; Das, N.K.; Ramakrishnan, S.K.; Jain, C.; Jurkovic, M.T.; Wu, J.; Nemeth, E.; Lakhal-Littleton, S.; Colacino, J.A.; Shah, Y.M. Hepatic hepcidin/intestinal HIF-2alpha axis maintains iron absorption during iron deficiency and overload. J. Clin. Invest. 2019, 129, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.M.; Wang, J.; Xie, J.X. Regulation of iron metabolism by hypoxia-inducible factors. Sheng Li Xue Bao 2017, 69, 598–610. [Google Scholar] [PubMed]
- Mastrogiannaki, M.; Matak, P.; Peyssonnaux, C. The gut in iron homeostasis: Role of HIF-2 under normal and pathological conditions. Blood 2013, 122, 885–892. [Google Scholar] [CrossRef]
- Das, N.K.; Schwartz, A.J.; Barthel, G.; Inohara, N.; Liu, Q.; Sankar, A.; Hill, D.R.; Ma, X.; Lamberg, O.; Schnizlein, M.K.; et al. Microbial Metabolite Signaling Is Required for Systemic Iron Homeostasis. Cell Metab. 2020, 31, 115–130.e116. [Google Scholar] [CrossRef]
- Malesza, I.J.; Bartkowiak-Wieczorek, J.; Winkler-Galicki, J.; Nowicka, A.; Dzieciolowska, D.; Blaszczyk, M.; Gajniak, P.; Slowinska, K.; Niepolski, L.; Walkowiak, J.; et al. The Dark Side of Iron: The Relationship between Iron, Inflammation and Gut Microbiota in Selected Diseases Associated with Iron Deficiency Anaemia-A Narrative Review. Nutrients 2022, 14, 3478. [Google Scholar] [CrossRef]
- Al-Sulaiti, H.; Diboun, I.; Agha, M.V.; Mohamed, F.F.S.; Atkin, S.; Domling, A.S.; Elrayess, M.A.; Mazloum, N.A. Metabolic signature of obesity-associated insulin resistance and type 2 diabetes. J. Transl. Med. 2019, 17, 348. [Google Scholar] [CrossRef]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, Y.; Ren, H.; Wang, S.; Zhong, H.; Zhao, X.; Ma, J.; Gu, X.; Xue, Y.; Huang, S.; et al. Gut microbiome-related effects of berberine and probiotics on type 2 diabetes (the PREMOTE study). Nat. Commun. 2020, 11, 5015. [Google Scholar] [CrossRef]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Lesko, L.J.; Atkinson, A.J., Jr. Use of biomarkers and surrogate endpoints in drug development and regulatory decision making: Criteria, validation, strategies. Annu. Rev. Pharmacol. Toxicol. 2001, 41, 347–366. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, F.; Ghosh, T.S.; O’Toole, P.W. The Healthy Microbiome-What Is the Definition of a Healthy Gut Microbiome? Gastroenterology 2021, 160, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Gong, T.; Zhang, J.; Gu, Q.; Gao, X.; Weng, X.; Liu, J.; Sun, J. Gut Microbiome Signatures Are Biomarkers for Cognitive Impairment in Patients With Ischemic Stroke. Front. Aging Neurosci. 2020, 12, 511562. [Google Scholar] [CrossRef]
- Schuijs, M.J.; Willart, M.A.; Vergote, K.; Gras, D.; Deswarte, K.; Ege, M.J.; Madeira, F.B.; Beyaert, R.; van Loo, G.; Bracher, F.; et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science 2015, 349, 1106–1110. [Google Scholar] [CrossRef] [PubMed]
- Pascal, M.; Perez-Gordo, M.; Caballero, T.; Escribese, M.M.; Lopez Longo, M.N.; Luengo, O.; Manso, L.; Matheu, V.; Seoane, E.; Zamorano, M.; et al. Microbiome and Allergic Diseases. Front. Immunol. 2018, 9, 1584. [Google Scholar] [CrossRef] [PubMed]
- Renz, H.; Brandtzaeg, P.; Hornef, M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol. 2011, 12, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Littman, D.R. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 2012, 30, 759–795. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P.; Vouret-Craviari, V. Microbes-induced EMT at the crossroad of inflammation and cancer. Gut Microbes 2012, 3, 176–185. [Google Scholar] [CrossRef]
- Salaspuro, M.P. Acetaldehyde, microbes, and cancer of the digestive tract. Crit. Rev. Clin. Lab. Sci. 2003, 40, 183–208. [Google Scholar] [CrossRef]
- Khan, A.A.; Shrivastava, A.; Khurshid, M. Normal to cancer microbiome transformation and its implication in cancer diagnosis. Biochim. Biophys. Acta 2012, 1826, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Sears, C.L.; Garrett, W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014, 15, 317–328. [Google Scholar] [CrossRef]
- Whisner, C.M.; Athena Aktipis, C. The Role of the Microbiome in Cancer Initiation and Progression: How Microbes and Cancer Cells Utilize Excess Energy and Promote One Another’s Growth. Curr. Nutr. Rep. 2019, 8, 42–51. [Google Scholar] [CrossRef]
- Peng, J.; Xiao, X.; Hu, M.; Zhang, X. Interaction between gut microbiome and cardiovascular disease. Life Sci. 2018, 214, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.; Franzosa, E.A.; Lloyd-Price, J.; McIver, L.J.; Schwager, R.; Poon, T.W.; Ananthakrishnan, A.N.; Andrews, E.; Barron, G.; Lake, K.; et al. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat. Microbiol. 2018, 3, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.Y.; Wu, T.T.; Liu, Z.Q.; Li, A.; Guo, Q.Q.; Ma, Y.Y.; Zhang, Z.L.; Xun, Y.L.; Zhang, J.C.; Wang, W.R.; et al. Gut Microbiome-Based Diagnostic Model to Predict Coronary Artery Disease. J. Agric. Food Chem. 2020, 68, 3548–3557. [Google Scholar] [CrossRef]
- Liu, H.; Chen, X.; Hu, X.; Niu, H.; Tian, R.; Wang, H.; Pang, H.; Jiang, L.; Qiu, B.; Chen, X.; et al. Alterations in the gut microbiome and metabolism with coronary artery disease severity. Microbiome 2019, 7, 68. [Google Scholar] [CrossRef]
- Liu, M.; Wang, M.; Peng, T.; Ma, W.; Wang, Q.; Niu, X.; Hu, L.; Qi, B.; Guo, D.; Ren, G.; et al. Gut-microbiome-based predictive model for ST-elevation myocardial infarction in young male patients. Front. Microbiol. 2022, 13, 1031878. [Google Scholar] [CrossRef]
- Temraz, S.; Nassar, F.; Nasr, R.; Charafeddine, M.; Mukherji, D.; Shamseddine, A. Gut Microbiome: A Promising Biomarker for Immunotherapy in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4155. [Google Scholar] [CrossRef]
- Lin, H.; He, Q.Y.; Shi, L.; Sleeman, M.; Baker, M.S.; Nice, E.C. Proteomics and the microbiome: Pitfalls and potential. Expert. Rev. Proteom. 2019, 16, 501–511. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N. Microbiome-Based Biomarkers for IBD. Inflamm. Bowel Dis. 2020, 26, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Yazaki, Y.; Voors, A.A.; Jones, D.J.L.; Chan, D.C.S.; Anker, S.D.; Cleland, J.G.; Dickstein, K.; Filippatos, G.; Hillege, H.L.; et al. Association with outcomes and response to treatment of trimethylamine N-oxide in heart failure: Results from BIOSTAT-CHF. Eur. J. Heart Fail. 2019, 21, 877–886. [Google Scholar] [CrossRef]
- Savji, N.; Meijers, W.C.; Bartz, T.M.; Bhambhani, V.; Cushman, M.; Nayor, M.; Kizer, J.R.; Sarma, A.; Blaha, M.J.; Gansevoort, R.T.; et al. The Association of Obesity and Cardiometabolic Traits With Incident HFpEF and HFrEF. JACC Heart Fail. 2018, 6, 701–709. [Google Scholar] [CrossRef]
- Salzano, A.; Cassambai, S.; Yazaki, Y.; Israr, M.Z.; Bernieh, D.; Wong, M.; Suzuki, T. The Gut Axis Involvement in Heart Failure: Focus on Trimethylamine N-oxide. Cardiol. Clin. 2022, 40, 161–169. [Google Scholar] [CrossRef]
- Xu, J.; Yang, Y. Gut microbiome and its meta-omics perspectives: Profound implications for cardiovascular diseases. Gut Microbes 2021, 13, 1936379. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Zheng, S.; Shen, Z.; Luo, Y.; Hai, X. Trimethylamine N-Oxide is Associated with Heart Failure Risk in Patients with Preserved Ejection Fraction. Lab. Med. 2021, 52, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid. Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Zhao, T.; Gu, J.; Zhang, H.; Wang, Z.; Zhang, W.; Zhao, Y.; Zheng, Y.; Zhang, W.; Zhou, H.; Zhang, G.; et al. Sodium Butyrate-Modulated Mitochondrial Function in High-Insulin Induced HepG2 Cell Dysfunction. Oxid. Med. Cell Longev. 2020, 2020, 1904609. [Google Scholar] [CrossRef]
- Tang, X.; Ma, S.; Li, Y.; Sun, Y.; Zhang, K.; Zhou, Q.; Yu, R. Evaluating the Activity of Sodium Butyrate to Prevent Osteoporosis in Rats by Promoting Osteal GSK-3beta/Nrf2 Signaling and Mitochondrial Function. J. Agric. Food Chem. 2020, 68, 6588–6603. [Google Scholar] [CrossRef]
- Kaye, D.M.; Shihata, W.A.; Jama, H.A.; Tsyganov, K.; Ziemann, M.; Kiriazis, H.; Horlock, D.; Vijay, A.; Giam, B.; Vinh, A.; et al. Deficiency of Prebiotic Fiber and Insufficient Signaling Through Gut Metabolite-Sensing Receptors Leads to Cardiovascular Disease. Circulation 2020, 141, 1393–1403. [Google Scholar] [CrossRef]
- Pluznick, J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes 2014, 5, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.J.; Zheng, L.; Campbell, E.L.; Saeedi, B.; Scholz, C.C.; Bayless, A.J.; Wilson, K.E.; Glover, L.E.; Kominsky, D.J.; Magnuson, A.; et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 2015, 17, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Van der Hee, B.; Wells, J.M. Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends. Microbiol. 2021, 29, 700–712. [Google Scholar] [CrossRef]
- Challa, A.A.; Lewandowski, E.D. Short-Chain Carbon Sources: Exploiting Pleiotropic Effects for Heart Failure Therapy. JACC Basic Transl. Sci. 2022, 7, 730–742. [Google Scholar] [CrossRef]
- Lymperopoulos, A.; Suster, M.S.; Borges, J.I. Short-Chain Fatty Acid Receptors and Cardiovascular Function. Int. J. Mol. Sci. 2022, 23, 3303. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Inoue, D.; Maeda, T.; Hara, T.; Ichimura, A.; Miyauchi, S.; Kobayashi, M.; Hirasawa, A.; Tsujimoto, G. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc. Natl. Acad. Sci. USA 2011, 108, 8030–8035. [Google Scholar] [CrossRef] [PubMed]
- Niebauer, J.; Volk, H.D.; Kemp, M.; Dominguez, M.; Schumann, R.R.; Rauchhaus, M.; Poole-Wilson, P.A.; Coats, A.J.; Anker, S.D. Endotoxin and immune activation in chronic heart failure: A prospective cohort study. Lancet 1999, 353, 1838–1842. [Google Scholar] [CrossRef]
- Zong, X.; Fan, Q.; Yang, Q.; Pan, R.; Zhuang, L.; Tao, R. Phenylacetylglutamine as a risk factor and prognostic indicator of heart failure. ESC Heart Fail. 2022, 9, 2645–2653. [Google Scholar] [CrossRef]
- Romano, K.A.; Nemet, I.; Prasad Saha, P.; Haghikia, A.; Li, X.S.; Mohan, M.L.; Lovano, B.; Castel, L.; Witkowski, M.; Buffa, J.A.; et al. Gut Microbiota-Generated Phenylacetylglutamine and Heart Failure. Circ. Heart Fail. 2023, 16, e009972. [Google Scholar] [CrossRef]
- Fang, C.; Zuo, K.; Jiao, K.; Zhu, X.; Fu, Y.; Zhong, J.; Xu, L.; Yang, X. PAGln, an Atrial Fibrillation-Linked Gut Microbial Metabolite, Acts as a Promoter of Atrial Myocyte Injury. Biomolecules 2022, 12, 1120. [Google Scholar] [CrossRef]
- Ren, X.; Wang, X.; Yuan, M.; Tian, C.; Li, H.; Yang, X.; Li, X.; Li, Y.; Yang, Y.; Liu, N.; et al. Mechanisms and Treatments of Oxidative Stress in Atrial Fibrillation. Curr. Pharm. Des. 2018, 24, 3062–3071. [Google Scholar] [CrossRef] [PubMed]
- Mesubi, O.O.; Anderson, M.E. Atrial remodelling in atrial fibrillation: CaMKII as a nodal proarrhythmic signal. Cardiovasc. Res. 2016, 109, 542–557. [Google Scholar] [CrossRef]
- Triposkiadis, F.; Xanthopoulos, A.; Parissis, J.; Butler, J.; Farmakis, D. Pathogenesis of chronic heart failure: Cardiovascular aging, risk factors, comorbidities, and disease modifiers. Heart Fail. Rev. 2022, 27, 337–344. [Google Scholar] [CrossRef]
- Dias, C.K.; Starke, R.; Pylro, V.S.; Morais, D.K. Database limitations for studying the human gut microbiome. PeerJ Comput. Sci. 2020, 6, e289. [Google Scholar] [CrossRef] [PubMed]
- Inkpen, S.A.; Douglas, G.M.; Brunet, T.D.P.; Leuschen, K.; Doolittle, W.F.; Langille, M.G.I. The coupling of taxonomy and function in microbiomes. Biol. Philos. 2017, 32, 1225–1243. [Google Scholar] [CrossRef]
- Kamo, T.; Akazawa, H.; Suzuki, J.I.; Komuro, I. Novel Concept of a Heart-Gut Axis in the Pathophysiology of Heart Failure. Korean Circ. J. 2017, 47, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Joice, R.; Yasuda, K.; Shafquat, A.; Morgan, X.C.; Huttenhower, C. Determining microbial products and identifying molecular targets in the human microbiome. Cell Metab. 2014, 20, 731–741. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, Z.; Tang, W.H.W.; Hazen, S.L. Gut Microbe-Generated Trimethylamine N-Oxide From Dietary Choline Is Prothrombotic in Subjects. Circulation 2017, 135, 1671–1673. [Google Scholar] [CrossRef]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef]
- Jackson, M.A.; Goodrich, J.K.; Maxan, M.E.; Freedberg, D.E.; Abrams, J.A.; Poole, A.C.; Sutter, J.L.; Welter, D.; Ley, R.E.; Bell, J.T.; et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut 2016, 65, 749–756. [Google Scholar] [CrossRef]
- Imhann, F.; Bonder, M.J.; Vich Vila, A.; Fu, J.; Mujagic, Z.; Vork, L.; Tigchelaar, E.F.; Jankipersadsing, S.A.; Cenit, M.C.; Harmsen, H.J.; et al. Proton pump inhibitors affect the gut microbiome. Gut 2016, 65, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Manneras-Holm, L.; Stahlman, M.; Olsson, L.M.; Serino, M.; Planas-Felix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Pedersen, H.K.; et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015, 528, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.; Pruteanu, M.; Kuhn, M.; Zeller, G.; Telzerow, A.; Anderson, E.E.; Brochado, A.R.; Fernandez, K.C.; Dose, H.; Mori, H.; et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018, 555, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Hata, S.; Okamura, T.; Kobayashi, A.; Bamba, R.; Miyoshi, T.; Nakajima, H.; Kitagawa, N.; Hashimoto, Y.; Majima, S.; Senmaru, T.; et al. Gut Microbiota Changes by an SGLT2 Inhibitor, Luseogliflozin, Alters Metabolites Compared with Those in a Low Carbohydrate Diet in db/db Mice. Nutrients 2022, 14, 3531. [Google Scholar] [CrossRef]
- Tuteja, S.; Ferguson, J.F. Gut Microbiome and Response to Cardiovascular Drugs. Circ. Genom. Precis Med. 2019, 12, 421–429. [Google Scholar] [CrossRef]
- Alhajri, N.; Khursheed, R.; Ali, M.T.; Abu Izneid, T.; Al-Kabbani, O.; Al-Haidar, M.B.; Al-Hemeiri, F.; Alhashmi, M.; Pottoo, F.H. Cardiovascular Health and The Intestinal Microbial Ecosystem: The Impact of Cardiovascular Therapies on the Gut Microbiota. Microorganisms 2021, 9, 2013. [Google Scholar] [CrossRef]
- Vich Vila, A.; Collij, V.; Sanna, S.; Sinha, T.; Imhann, F.; Bourgonje, A.R.; Mujagic, Z.; Jonkers, D.; Masclee, A.A.M.; Fu, J.; et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun. 2020, 11, 362. [Google Scholar] [CrossRef]
- Sanches Machado d’Almeida, K.; Ronchi Spillere, S.; Zuchinali, P.; Correa Souza, G. Mediterranean Diet and Other Dietary Patterns in Primary Prevention of Heart Failure and Changes in Cardiac Function Markers: A Systematic Review. Nutrients 2018, 10, 58. [Google Scholar] [CrossRef]
- Wang, Z.; Bergeron, N.; Levison, B.S.; Li, X.S.; Chiu, S.; Jia, X.; Koeth, R.A.; Li, L.; Wu, Y.; Tang, W.H.W.; et al. Impact of chronic dietary red meat, white meat, or non-meat protein on trimethylamine N-oxide metabolism and renal excretion in healthy men and women. Eur. Heart J. 2019, 40, 583–594. [Google Scholar] [CrossRef]
- Mayerhofer, C.C.K.; Kummen, M.; Holm, K.; Broch, K.; Awoyemi, A.; Vestad, B.; Storm-Larsen, C.; Seljeflot, I.; Ueland, T.; Bohov, P.; et al. Low fibre intake is associated with gut microbiota alterations in chronic heart failure. ESC Heart Fail. 2020, 7, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Mousa, S.; Sarfraz, M.; Mousa, W.K. The Interplay between Gut Microbiota and Oral Medications and Its Impact on Advancing Precision Medicine. Metabolites 2023, 13, 674. [Google Scholar] [CrossRef] [PubMed]
- McCoubrey, L.E.; Elbadawi, M.; Orlu, M.; Gaisford, S.; Basit, A.W. Machine Learning Uncovers Adverse Drug Effects on Intestinal Bacteria. Pharmaceutics 2021, 13, 1026. [Google Scholar] [CrossRef]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017, 5, 14. [Google Scholar] [CrossRef]
- Jie, Z.; Xia, H.; Zhong, S.L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, H.Y.; Hu, X.M.; Zhang, Y.; Zhang, S.Y. Current understanding of gut microbiota alterations and related therapeutic intervention strategies in heart failure. Chin. Med. J. 2019, 132, 1843–1855. [Google Scholar] [CrossRef]
- Ros, M.; Carrascosa, J.M. Current nutritional and pharmacological anti-aging interventions. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165612. [Google Scholar] [CrossRef]
- McPhee, J.S.; French, D.P.; Jackson, D.; Nazroo, J.; Pendleton, N.; Degens, H. Physical activity in older age: Perspectives for healthy ageing and frailty. Biogerontology 2016, 17, 567–580. [Google Scholar] [CrossRef]
- Godos, J.; Grosso, G.; Ferri, R.; Caraci, F.; Lanza, G.; Al-Qahtani, W.H.; Caruso, G.; Castellano, S. Mediterranean diet, mental health, cognitive status, quality of life, and successful aging in southern Italian older adults. Exp. Gerontol. 2023, 175, 112143. [Google Scholar] [CrossRef]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C.; et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016, 65, 1812–1821. [Google Scholar] [CrossRef]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Kawano, Y.; Edwards, M.; Huang, Y.; Bilate, A.M.; Araujo, L.P.; Tanoue, T.; Atarashi, K.; Ladinsky, M.S.; Reiner, S.L.; Wang, H.H.; et al. Microbiota imbalance induced by dietary sugar disrupts immune-mediated protection from metabolic syndrome. Cell 2022, 185, 3501–3519 e3520. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Liu, Y.; Wu, Z.; Wang, J.; Zhang, X. Effects of Diet and Exercise on Circadian Rhythm: Role of Gut Microbiota in Immune and Metabolic Systems. Nutrients 2023, 15, 2743. [Google Scholar] [CrossRef] [PubMed]
- Frampton, J.; Murphy, K.G.; Frost, G.; Chambers, E.S. Short-chain fatty acids as potential regulators of skeletal muscle metabolism and function. Nat. Metab. 2020, 2, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Bermon, S.; Petriz, B.; Kajeniene, A.; Prestes, J.; Castell, L.; Franco, O.L. The microbiota: An exercise immunology perspective. Exerc. Immunol. Rev. 2015, 21, 70–79. [Google Scholar]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 2018, 67, 625–633. [Google Scholar] [CrossRef]
- Gan, X.T.; Ettinger, G.; Huang, C.X.; Burton, J.P.; Haist, J.V.; Rajapurohitam, V.; Sidaway, J.E.; Martin, G.; Gloor, G.B.; Swann, J.R.; et al. Probiotic administration attenuates myocardial hypertrophy and heart failure after myocardial infarction in the rat. Circ. Heart Fail. 2014, 7, 491–499. [Google Scholar] [CrossRef]
- Awoyemi, A.; Mayerhofer, C.; Felix, A.S.; Hov, J.R.; Moscavitch, S.D.; Lappegard, K.T.; Hovland, A.; Halvorsen, S.; Halvorsen, B.; Gregersen, I.; et al. Rifaximin or Saccharomyces boulardii in heart failure with reduced ejection fraction: Results from the randomized GutHeart trial. EBioMedicine 2021, 70, 103511. [Google Scholar] [CrossRef]
- Roger, A.J.; Munoz-Gomez, S.A.; Kamikawa, R. The Origin and Diversification of Mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef]
- Ni Lochlainn, M.; Nessa, A.; Sheedy, A.; Horsfall, R.; Garcia, M.P.; Hart, D.; Akdag, G.; Yarand, D.; Wadge, S.; Baleanu, A.F.; et al. The PROMOTe study: Targeting the gut microbiome with prebiotics to overcome age-related anabolic resistance: Protocol for a double-blinded, randomised, placebo-controlled trial. BMC Geriatr. 2021, 21, 407. [Google Scholar] [CrossRef]
- Elias, A.J.; Barna, V.; Patoni, C.; Demeter, D.; Veres, D.S.; Bunduc, S.; Eross, B.; Hegyi, P.; Foldvari-Nagy, L.; Lenti, K. Probiotic supplementation during antibiotic treatment is unjustified in maintaining the gut microbiome diversity: A systematic review and meta-analysis. BMC Med. 2023, 21, 262. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, P.A.; Garces, D.; Prado-Vivar, B.; Flores, N.; Fornasini, M.; Cohen, H.; Salvador, I.; Cargua, O.; Baldeon, M.E. Effect of Saccharomyces boulardii CNCM I-745 as complementary treatment of Helicobacter pylori infection on gut microbiome. Eur J. Clin. Microbiol. Infect. Dis. 2020, 39, 1365–1372. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, W.; Lee, A.; He, J.; Huang, B.; Zheng, W.; Su, T.; Lai, S.; Long, Y.; Chu, H.; et al. The impact of Helicobacter pylori infection, eradication therapy and probiotic supplementation on gut microenvironment homeostasis: An open-label, randomized clinical trial. EBioMedicine 2018, 35, 87–96. [Google Scholar] [CrossRef] [PubMed]
- De Wolfe, T.J.; Eggers, S.; Barker, A.K.; Kates, A.E.; Dill-McFarland, K.A.; Suen, G.; Safdar, N. Oral probiotic combination of Lactobacillus and Bifidobacterium alters the gastrointestinal microbiota during antibiotic treatment for Clostridium difficile infection. PLoS ONE 2018, 13, e0204253. [Google Scholar] [CrossRef] [PubMed]
- Kabbani, T.A.; Pallav, K.; Dowd, S.E.; Villafuerte-Galvez, J.; Vanga, R.R.; Castillo, N.E.; Hansen, J.; Dennis, M.; Leffler, D.A.; Kelly, C.P. Prospective randomized controlled study on the effects of Saccharomyces boulardii CNCM I-745 and amoxicillin-clavulanate or the combination on the gut microbiota of healthy volunteers. Gut Microbes 2017, 8, 17–32. [Google Scholar] [CrossRef]
- Kakiuchi, T.; Mizoe, A.; Yamamoto, K.; Imamura, I.; Hashiguchi, K.; Kawakubo, H.; Yamaguchi, D.; Fujioka, Y.; Nakayama, A.; Okuda, M.; et al. Effect of probiotics during vonoprazan-containing triple therapy on gut microbiota in Helicobacter pylori infection: A randomized controlled trial. Helicobacter 2020, 25, e12690. [Google Scholar] [CrossRef]
- MacPherson, C.W.; Mathieu, O.; Tremblay, J.; Champagne, J.; Nantel, A.; Girard, S.A.; Tompkins, T.A. Gut Bacterial Microbiota and its Resistome Rapidly Recover to Basal State Levels after Short-term Amoxicillin-Clavulanic Acid Treatment in Healthy Adults. Sci. Rep. 2018, 8, 11192. [Google Scholar] [CrossRef]
- Oh, B.; Kim, B.S.; Kim, J.W.; Kim, J.S.; Koh, S.J.; Kim, B.G.; Lee, K.L.; Chun, J. The Effect of Probiotics on Gut Microbiota during the Helicobacter pylori Eradication: Randomized Controlled Trial. Helicobacter 2016, 21, 165–174. [Google Scholar] [CrossRef]
- Tang, B.; Tang, L.; Huang, C.; Tian, C.; Chen, L.; He, Z.; Yang, G.; Zuo, L.; Zhao, G.; Liu, E.; et al. The Effect of Probiotics Supplementation on Gut Microbiota After Helicobacter pylori Eradication: A Multicenter Randomized Controlled Trial. Infect Dis. Ther. 2021, 10, 317–333. [Google Scholar] [CrossRef]
- Zhong, H.; Wang, X.G.; Wang, J.; Chen, Y.J.; Qin, H.L.; Yang, R. Impact of probiotics supplement on the gut microbiota in neonates with antibiotic exposure: An open-label single-center randomized parallel controlled study. World J. Pediatr. 2021, 17, 385–393. [Google Scholar] [CrossRef]
- Engelbrektson, A.; Korzenik, J.R.; Pittler, A.; Sanders, M.E.; Klaenhammer, T.R.; Leyer, G.; Kitts, C.L. Probiotics to minimize the disruption of faecal microbiota in healthy subjects undergoing antibiotic therapy. J. Med. Microbiol. 2009, 58, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Forssten, S.; Evans, M.; Wilson, D.; Ouwehand, A.C. Influence of a probiotic mixture on antibiotic induced microbiota disturbances. World J. Gastroenterol. 2014, 20, 11878–11885. [Google Scholar] [CrossRef] [PubMed]
- Madden, J.A.; Plummer, S.F.; Tang, J.; Garaiova, I.; Plummer, N.T.; Herbison, M.; Hunter, J.O.; Shimada, T.; Cheng, L.; Shirakawa, T. Effect of probiotics on preventing disruption of the intestinal microflora following antibiotic therapy: A double-blind, placebo-controlled pilot study. Int. Immunopharmacol. 2005, 5, 1091–1097. [Google Scholar] [CrossRef] [PubMed]
- Plummer, S.F.; Garaiova, I.; Sarvotham, T.; Cottrell, S.L.; Le Scouiller, S.; Weaver, M.A.; Tang, J.; Dee, P.; Hunter, J. Effects of probiotics on the composition of the intestinal microbiota following antibiotic therapy. Int. J. Antimicrob. Agents 2005, 26, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.J.; Chen, X.F.; Zhang, Z.X.; Li, Y.C.; Deng, J.; Tu, J.; Song, Z.Q.; Zou, Q.H. Effects of anti-Helicobacter pylori concomitant therapy and probiotic supplementation on the throat and gut microbiota in humans. Microb. Pathog. 2017, 109, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Amarri, S.; Morelli, L. Evaluation of the Effects of Enterogermina, 2 Billion Bacillus clausii Spores, on the Intestinal Flora of Children Antibiotic Treated for Bacterial Upper Respiratory Tract Infections: Open, Pilot Study.-DIAMANTE. 2008. EUCTR2006-002482-39-IT. Available online: https://www.cochranelibrary.com/central/doi/10.1002/central/CN-01818519/full (accessed on 17 September 2023).
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef]
- Liang, J.Q.; Li, T.; Nakatsu, G.; Chen, Y.X.; Yau, T.O.; Chu, E.; Wong, S.; Szeto, C.H.; Ng, S.C.; Chan, F.K.L.; et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut 2020, 69, 1248–1257. [Google Scholar] [CrossRef]
- Van der Meulen, T.A.; Harmsen, H.J.M.; Vila, A.V.; Kurilshikov, A.; Liefers, S.C.; Zhernakova, A.; Fu, J.; Wijmenga, C.; Weersma, R.K.; de Leeuw, K.; et al. Shared gut, but distinct oral microbiota composition in primary Sjogren’s syndrome and systemic lupus erythematosus. J. Autoimmun. 2019, 97, 77–87. [Google Scholar] [CrossRef]
- Shaukat, A.; Levin, T.R. Current and future colorectal cancer screening strategies. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 521–531. [Google Scholar] [CrossRef]
- Pasolli, E.; Truong, D.T.; Malik, F.; Waldron, L.; Segata, N. Machine Learning Meta-analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLoS Comput. Biol. 2016, 12, e1004977. [Google Scholar] [CrossRef]
- Gacesa, R.; Kurilshikov, A.; Vich Vila, A.; Sinha, T.; Klaassen, M.A.Y.; Bolte, L.A.; Andreu-Sanchez, S.; Chen, L.; Collij, V.; Hu, S.; et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature 2022, 604, 732–739. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Kelly, L. Multiclass Disease Classification from Microbial Whole-Community Metagenomes. In Pacific Symposium on Biocomputing 2020; World Scientific: Singapore, 2019; Volume 25, pp. 55–66. [Google Scholar]
- Ghannam, R.B.; Techtmann, S.M. Machine learning applications in microbial ecology, human microbiome studies, and environmental monitoring. Comput. Struct. Biotechnol. J. 2021, 19, 1092–1107. [Google Scholar] [CrossRef] [PubMed]
- Goodswen, S.J.; Kennedy, P.J.; Ellis, J.T. Applying Machine Learning to Predict the Exportome of Bovine and Canine Babesia Species That Cause Babesiosis. Pathogens 2021, 10, 660. [Google Scholar] [CrossRef]
- Marcos-Zambrano, L.J.; Karaduzovic-Hadziabdic, K.; Loncar Turukalo, T.; Przymus, P.; Trajkovik, V.; Aasmets, O.; Berland, M.; Gruca, A.; Hasic, J.; Hron, K.; et al. Applications of Machine Learning in Human Microbiome Studies: A Review on Feature Selection, Biomarker Identification, Disease Prediction and Treatment. Front. Microbiol. 2021, 12, 634511. [Google Scholar] [CrossRef]
- Li, M.; Liu, J.; Zhu, J.; Wang, H.; Sun, C.; Gao, N.L.; Zhao, X.M.; Chen, W.H. Performance of Gut Microbiome as an Independent Diagnostic Tool for 20 Diseases: Cross-Cohort Validation of Machine-Learning Classifiers. Gut Microbes 2023, 15, 2205386. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, M.R.; Aggarwal, P.; Costa, R.G.F.; Cole, A.M.; Trinchieri, G. Targeting the gut microbiota for cancer therapy. Nat. Rev. Cancer 2022, 22, 703–722. [Google Scholar] [CrossRef] [PubMed]
- Cammarota, G.; Ianiro, G.; Ahern, A.; Carbone, C.; Temko, A.; Claesson, M.J.; Gasbarrini, A.; Tortora, G. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 635–648. [Google Scholar] [CrossRef] [PubMed]
| Study | Country | Study Design * | Population | Antibiotics (and Additional) Treatment | Probiotic Supplementation | ||||
|---|---|---|---|---|---|---|---|---|---|
| Number of Randomized Patients (Female %) | Age (Years—Mean ± SD) in the Intervention (and Control) Groups | Indication | Type | Duration (Days) | Type | Duration (Days) | |||
| Cárdenas et al. (2020) [182] | Ecuador | Single-blinded RCT | 38 (60.5) | 37.9 ± 7.2 (39.5 ± 10.7) | Helicobacter pylori infection | Amoxicillin, tinidazole, and omeprazole | 14 | Saccharomyces boulardii | 14 |
| Chen et al. (2018) [183] | China | Open-label RCT | 70 (78.5) | 43.89 ± 12.50 (43.20 ± 12.45) | Helicobacter pylori infection | Pantoprazole, amoxycillin, furazolidone, and colloidal bismuthpectin | 14 | Clostridium butyricum | 14 |
| De Wolfe et al. (2018) [184] | USA | Double-blinded, placebo-controlled RCT | 31 (N.D.) | N.D. (N.D.) | Clostridioides difficile infection | Vancomycin, metronidazole, or fidaxomicin | 28 | Lactobacillus acidophilus, Lactobacillus paracasei, Bifidobacterium lactis Bi-07, and Bifidobacterium lactis Bi-04 | 28 |
| Kabbani et al. (2017) [185] | USA | Open-label RCT | 24 (59) | N.D. (N.D.) | Healthy volunteers—no indication | Amoxycillin-clavulanate | 7 | Saccharomyces boulardii | 14 |
| Kakiuchi et al. (2020) [186] | Japan | Open-label RCT | 65 (44.6) | 15.31 ± 0.32 (15.08 ± 0.28) | Helicobacter pylori infection | Vonoprazan, amoxycillin, and clarithromycin | 7 | Enterococcus faecium | 7 |
| MacPherson et al. (2018) [187] | Canada | Double-blinded, placebo-controlled RCT | 70 (N.D.) | N.D. (N.D.) | Healthy volunteers—no indication | Amoxycillin trihydrate, potassium clavulanate | 7 | Lactobacillus rhamnosus and Lactobacillus helveticus | 14 |
| Oh et al. (2016) [188] | Korea | RCT | 20 (30) | 51.7 ± 0.79 (49.3 ± 3.56) | Helicobacter pylori infection | Clarithromycin, Amoxycillin, Lansoprazole | 14 | Streptococcus faecium and Bacillus subtilis | 14 |
| Tang et al. (2021) [189] | China | Placebo-controlled multicenter RCT | 151 (34.4) | 43.29 ± 11.30 (45.32 ± 10.98) | Helicobacter pylori infection | Esomeprazole, amoxycillin, furazolidone, and bismuth potassium citrate | 14 | Enterococcus faecium and Bacillus subtilis | 28 |
| Zhong et al. (2021) [190] | China | Open-label parallel RCT | 42 (52.4) | All neonates (All neonates) | 15 neonates with neonatal pneumonia, 5 neonates with urinary tract infection, and 35 neonates with non-specific infection | Piperacillin-tazobactam | 7 | Bifidobacterium longum, Lactobacillus acidophilus and Enterococcus faecalis | 7 |
| Engelbrektson et al. (2009) [191] | USA | Placebo-controlled RCT | 40 (77.5) | 36.5 ± N.D. (39.5 ± N.D.) | Healthy volunteers—no indication | Amoxicillin and clavulanic acid | 7 | Bifidobacterium lactis BI-04, Bifidobacterium lactis Bi-07, Lactobacillus acidophilus, Lactobacillus paracasei and Bifidobacterium bifidum | 21 |
| Forssten et al. (2014) [192] | Finland | Double-blinded, parallel RCT | 80 (50) | 33.7 ± 9.4 (30.9 ± 10.3) | Healthy volunteers—no indication | Amoxicillin and clavulanate | 7 | Lactobacillus acidophilus, Bifidobacterium animalis ssp. Lactis | 14 |
| Madden et al. (2005) [193] | UK | Pilot-scale, double-blinded RCT | 13 (53.8) | 60 ± N.D. (49 ± N.D.) | Helicobacter pylori infection | Amoxicillin, metronidazole, and lansoprazole | 8 | Lactobacillus acidophilus and 2 strains of Bifidobacterium bifidum | 14 |
| Plummer et al. (2005) [194] | UK | Double-blinded RCT | 155 (N.D.) | N.D. N.D. | Helicobacter pylori infection | Amoxicillin, clarithromycin, and lansoprazole | 7 | Lactobacillus acidophilus and 2 strains of Bifidobacterium spp. | 21 |
| Wang et al. (2017) [195] | China | Double-blinded RCT | 20 (45) | 37.1 ± 12.3 (42.8 ± 13.8) | Helicobacter pylori infection | Esomeprazole, amoxicillin, clarithromycin, and tinidazole | 14 | Saccharomyces boulardii | 14 |
| Amarri et al. (2008) [196] | Italy | Open-label, national, parallel RCT | 58 (50) | 40 ± 18.9 months (42.1 ± 18.9 months) | Bacterial upper respiratory tract infections | Amoxicillin | 5–10 | Antibiotic-resistant Bacillus clausii | 12–17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paraskevaidis, I.; Xanthopoulos, A.; Tsougos, E.; Triposkiadis, F. Human Gut Microbiota in Heart Failure: Trying to Unmask an Emerging Organ. Biomedicines 2023, 11, 2574. https://doi.org/10.3390/biomedicines11092574
Paraskevaidis I, Xanthopoulos A, Tsougos E, Triposkiadis F. Human Gut Microbiota in Heart Failure: Trying to Unmask an Emerging Organ. Biomedicines. 2023; 11(9):2574. https://doi.org/10.3390/biomedicines11092574
Chicago/Turabian StyleParaskevaidis, Ioannis, Andrew Xanthopoulos, Elias Tsougos, and Filippos Triposkiadis. 2023. "Human Gut Microbiota in Heart Failure: Trying to Unmask an Emerging Organ" Biomedicines 11, no. 9: 2574. https://doi.org/10.3390/biomedicines11092574
APA StyleParaskevaidis, I., Xanthopoulos, A., Tsougos, E., & Triposkiadis, F. (2023). Human Gut Microbiota in Heart Failure: Trying to Unmask an Emerging Organ. Biomedicines, 11(9), 2574. https://doi.org/10.3390/biomedicines11092574








