Machine Learning for COVID-19 Determination Using Surface-Enhanced Raman Spectroscopy
Abstract
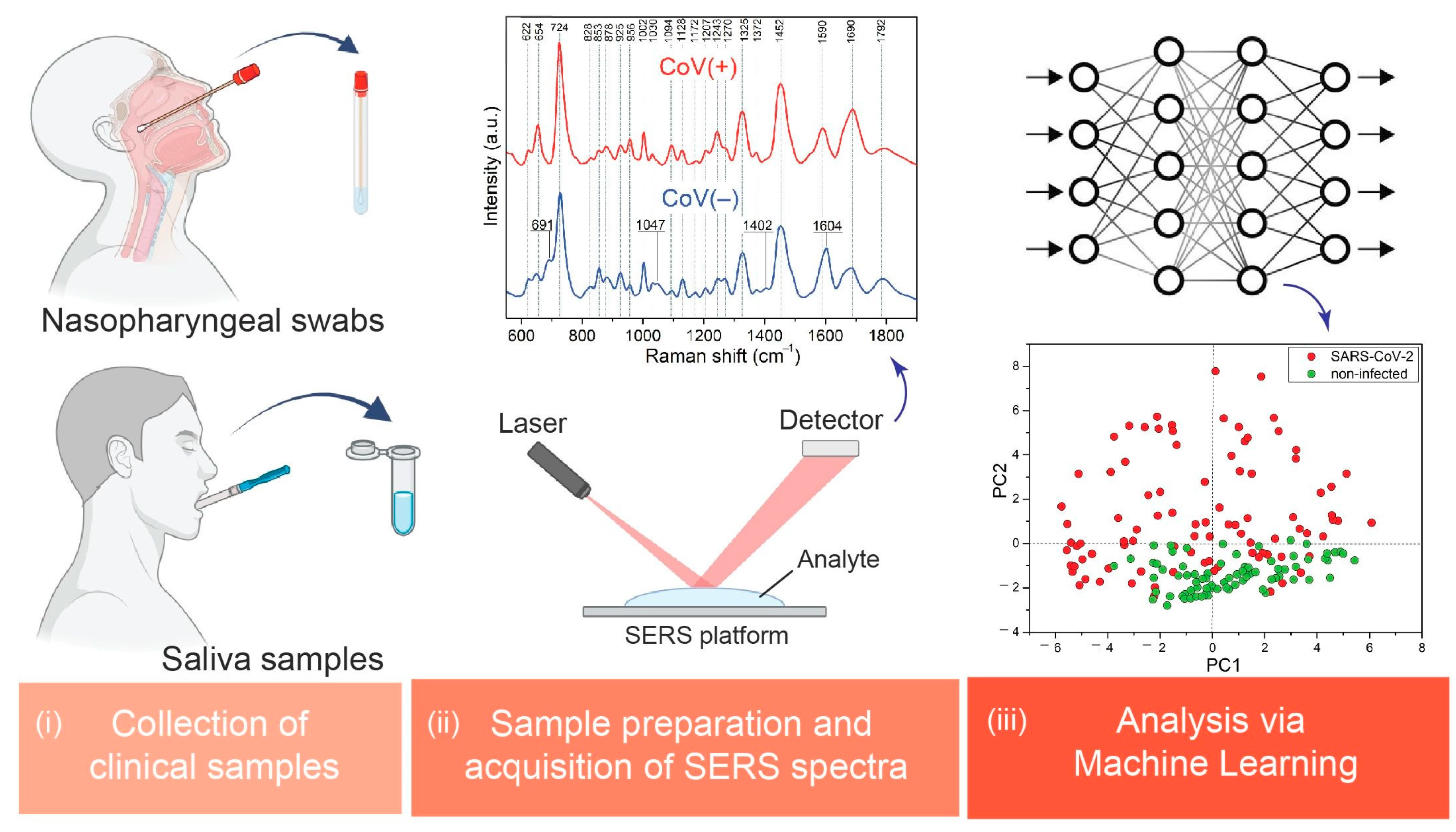
:1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Measurements of the Clinical Samples
2.3. Machine Learning Analysis
2.3.1. Principal Component Analysis (PCA)
- X—Initial matrix of data;
- T—Scores matrix;
- P—Loading matrix;
- E—Error matrix.
2.3.2. Machine Learning Classification
3. Results
3.1. SERS Measurements and Band Assignments
- (i)
- An origin of 724 cm−1 corresponds to O–O stretching vibration in oxygenated proteins, glycoproteins (e.g., mucin), and to the ring breathing mode of tryptophan.
- (ii)
- An origin of 1325 cm−1 corresponds to amide III band in proteins and DNA.
- (iii)
- An origin of 1452 cm−1 corresponds to C–H stretching of glycoproteins, including mucin.
- (iv)
- An origin of 1585 cm−1 corresponds to ring and C=C vibrations in tyrosine and phenylalanine.
3.2. Principal Component Analysis (PCA)
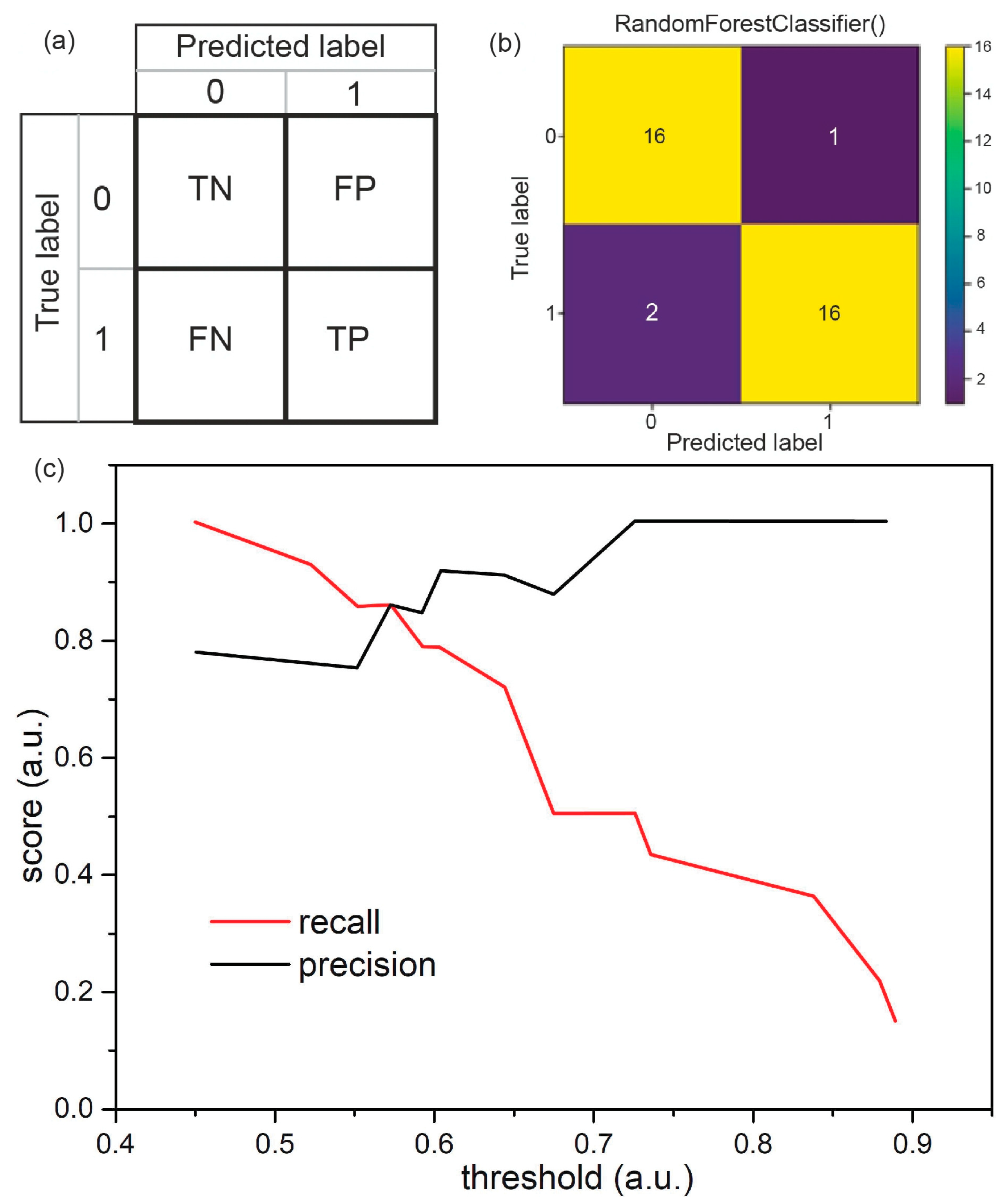
3.3. Classification of the Samples Using Machine Learning Algorithms
3.4. Classification of Saliva Samples via Random Forest
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Oliveira, M.C.; Scharan, K.O.; Thomés, B.I.; Bernardelli, R.S.; Reese, F.B.; Kozesinski-Nakatani, A.C.; Martins, C.C.; Lobo, S.M.A.; Réa-Neto, Á. Diagnostic accuracy of a set of clinical and radiological criteria for screening of COVID-19 using RT-PCR as the reference standard. BMC Pulm. Med. 2023, 23, 81. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Ardebili, A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020, 20, 453–454. [Google Scholar] [CrossRef]
- van Kasteren, P.B.; van der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.B.E.M.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Li, Y.; Lin, C.; Peng, Y.; Zhao, S.; Yang, X.; Yang, Y. Recent Advances of Representative Optical Biosensors for Rapid and Sensitive Diagnostics of SARS-CoV-2. Biosensors 2022, 12, 862. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.A.; Hanora, A.; Solyman, S.M.; Hassan, Y.A. Designing and fabrication of electrochemical nano-biosensor for the fast detection of SARS-CoV-2-RNA. Sci. Rep. 2023, 13, 5139. [Google Scholar] [CrossRef]
- Yakoh, A.; Pimpitak, U.; Rengpipat, S.; Hirankarn, N.; Chailapakul, O.; Chaiyo, S. Paper-based electrochemical biosensor for diagnosing COVID-19: Detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021, 176, 112912. [Google Scholar] [CrossRef]
- Li, Z.; Luo, Y.; Song, Y.; Zhu, Q.; Xu, T.; Zhang, X. One-click investigation of shape influence of silver nanostructures on SERS performance for sensitive detection of COVID-19. Anal. Chim. Acta 2022, 1234, 340523. [Google Scholar] [CrossRef]
- Alnaji, N.; Wasfi, A.; Awwad, F. The design of a point of care FET biosensor to detect and screen COVID-19. Sci. Rep. 2023, 13, 4485. [Google Scholar] [CrossRef]
- Vafabakhsh, M.; Dadmehr, M.; Kazemi Noureini, S.; Es’haghi, Z.; Malekkiani, M.; Hosseini, M. Paper-based colorimetric detection of COVID-19 using aptasenor based on biomimetic peroxidase like activity of ChF/ZnO/CNT nano-hybrid. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 301, 122980. [Google Scholar] [CrossRef]
- Huang, P.C.; Zhou, Y.; Porter, E.B.; Saxena, R.G.; Gomez, A.; Ykema, M.; Senehi, N.L.; Lee, D.; Tseng, C.P.; Alvarez, P.J.; et al. Organic Electrochemical Transistors functionalized with Protein Minibinders for Sensitive and Specific Detection of SARS-CoV-2. Adv. Mater. Interfaces 2023, 10, 2202409. [Google Scholar] [CrossRef]
- Kim, H.E.; Schuck, A.; Park, H.; Huh, H.J.; Kang, M.; Kim, Y.-S. Gold nanostructures modified carbon-based electrode enhanced with methylene blue for point-of-care COVID-19 tests using isothermal amplification. Talanta 2023, 265, 124841. [Google Scholar] [CrossRef]
- GhaderiShekhiAbadi, P.; Irani, M.; Noorisepehr, M.; Maleki, A. Magnetic biosensors for identification of SARS-CoV-2, Influenza, HIV, and Ebola viruses: A review. Nanotechnology 2023, 34, 272001. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, B.; Murray, J.; Haverstick, J.; Chen, X.; Tripp, R.A.; Zhao, Y. Rapid and quantitative detection of respiratory viruses using surface-enhanced Raman spectroscopy and machine learning. Biosens. Bioelectron. 2022, 217, 114721. [Google Scholar] [CrossRef]
- Driskell, J.D.; Kwarta, K.M.; Lipert, R.J.; Porter, M.D.; Neill, J.D.; Ridpath, J.F. Low-level detection of viral pathogens by a surface-enhanced Raman scattering based immunoassay. Anal. Chem. 2005, 77, 6147–6154. [Google Scholar] [CrossRef]
- Luo, S.C.; Sivashanmugan, K.; Der Liao, J.; Yao, C.K.; Peng, H.C. Nanofabricated SERS-active substrates for single-molecule to virus detection in vitro: A review. Biosens. Bioelectron. 2014, 61, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Saviñon-Flores, F.; Méndez, E.; López-Castaños, M.; Carabarin-Lima, A.; López-Castaños, K.A.; González-Fuentes, M.A.; Méndez-Albores, A. A Review on SERS-Based Detection of Human Virus Infections: Influenza and Coronavirus. Biosensors 2021, 11, 66. [Google Scholar] [CrossRef]
- Lin, C.; Li, Y.; Peng, Y.; Zhao, S.; Xu, M.; Zhang, L.; Huang, Z.; Shi, J.; Yang, Y. REVIEW Open Access Recent development of surface-enhanced Raman scattering for biosensing. J. Nanobiotechnol. 2023, 21, 149. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, J.-W.; Li, F.; Usman, M.; Wu, C.-Y.; Liu, Q.-H.; Kang, H.-Q.; Liu, W.; Gu, B. Identification of Bacterial Pathogens at Genus and Species Levels through Combination of Raman Spectrometry and Deep-Learning Algorithms. Microbiol. Spectr. 2022, 10, e02580-22. [Google Scholar] [CrossRef]
- Tang, J.W.; Lyu, J.W.; Lai, J.X.; Zhang, X.D.; Du, Y.G.; Zhang, X.Q.; Zhang, Y.D.; Gu, B.; Zhang, X.; Gu, B.; et al. Determination of Shigella spp. via label-free SERS spectra coupled with deep learning. Microchem. J. 2023, 189, 108539. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Z.; Ning, Y.; Miao, P.; Li, Z.; Wang, H. Simultaneous quantitative analysis of Escherichia coli, Staphylococcus aureus and Salmonella typhimurium using surface-enhanced Raman spectroscopy coupled with partial least squares regression and artificial neural networks. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 2023, 293, 122510. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Q.; Dukes, P.V.; Lee, W.; Sarkis, M.; Vo-Dinh, T. Machine learning using convolutional neural networks for SERS analysis of biomarkers in medical diagnostics. J. Raman Spectrosc. 2022, 53, 2044–2057. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Chen, C.; Chen, C.; Wu, L.; Wu, X.; Lu, C.; Zhang, X.; Chao, P.; Lv, X.; Jia, Z.; et al. Coupling annealed silver nanoparticles with a porous silicon Bragg mirror SERS substrate and machine learning for rapid non-invasive disease diagnosis. Anal. Chim. Acta 2023, 1254, 341116. [Google Scholar] [CrossRef] [PubMed]
- Tabarov, A.; Vitkin, V.; Andreeva, O.; Shemanaeva, A.; Popov, E.; Dobroslavin, A.; Kurikova, V.; Kuznetsova, O.; Grigorenko, K.; Tzibizov, I.; et al. Detection of A and B Influenza Viruses by Surface-Enhanced Raman Scattering Spectroscopy and Machine Learning. Biosensors 2022, 12, 1065. [Google Scholar] [CrossRef]
- dos Santos, D.P.; Sena, M.M.; Almeida, M.R.; Mazali, I.O.; Olivieri, A.C.; Villa, J.E.L. Unraveling surface-enhanced Raman spectroscopy results through chemometrics and machine learning: Principles, progress, and trends. Anal. Bioanal. Chem. 2023, 415, 3945–3966. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, Y.; Liu, C.; Jiang, Q.Y.; Chen, F.; Cao, Y. SERS-Based Biosensors Combined with Machine Learning for Medical Application**. ChemistryOpen 2023, 12, e202200192. [Google Scholar] [CrossRef]
- Chen, H.; Park, S.G.; Choi, N.; Kwon, H.J.; Kang, T.; Lee, M.K.; Choo, J. Sensitive Detection of SARS-CoV-2 Using a SERS-Based Aptasensor. ACS Sensors 2021, 6, 2378–2385. [Google Scholar] [CrossRef]
- Peng, Y.; Lin, C.; Long, L.; Masaki, T.; Tang, M.; Yang, L.; Liu, J.; Huang, Z.; Li, Z.; Luo, X.; et al. Charge-Transfer Resonance and Electromagnetic Enhancement Synergistically Enabling MXenes with Excellent SERS Sensitivity for SARS-CoV-2 S Protein Detection. Nano-Micro Lett. 2021, 13, 52. [Google Scholar] [CrossRef]
- Yang, Y.; Peng, Y.; Lin, C.; Long, L.; Hu, J.; He, J.; Zeng, H.; Huang, Z.; Li, Z.Y.; Tanemura, M.; et al. Human ACE2-Functionalized Gold “Virus-Trap” Nanostructures for Accurate Capture of SARS-CoV-2 and Single-Virus SERS Detection. Nano-Micro Lett. 2021, 13, 109. [Google Scholar] [CrossRef]
- Liu, H.; Dai, E.; Xiao, R.; Zhou, Z.; Zhang, M.; Bai, Z.; Shao, Y.; Qi, K.; Tu, J.; Wang, C.; et al. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens. Actuators B Chem. 2021, 329, 129196. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Lee, H.C.; Diao, K.Y.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020, 26, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Ikponmwoba, E.; Ukorigho, O.; Moitra, P.; Pan, D.; Gartia, M.R.; Owoyele, O. A Machine Learning Framework for Detecting COVID-19 Infection Using Surface-Enhanced Raman Scattering. Biosensors 2022, 12, 589. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, V.; Joseph, M.M.; Yadev, I.; Sharma, H.; Shamna, K.; Saurav, S.; Sreejith, R.P.; Anand, V.; Beegum, R.; Regi David, S.; et al. A non-invasive ultrasensitive diagnostic approach for COVID-19 infection using salivary label-free SERS fingerprinting and artificial intelligence. J. Photochem. Photobiol. B Biol. 2022, 234, 112545. [Google Scholar] [CrossRef]
- Yang, Y.; Li, H.; Jones, L.; Murray, J.; Haverstick, J.; Naikare, H.K.; Mosley, Y.Y.C.; Tripp, R.A.; Ai, B.; Zhao, Y. Rapid Detection of SARS-CoV-2 RNA in Human Nasopharyngeal Specimens Using Surface-Enhanced Raman Spectroscopy and Deep Learning Algorithms. ACS Sensors 2023, 8, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.S.H.; Lee, S.; Lee, S.; Kim, H.; Kang, T.; Lee, D.; Jeong, K.H. Highly Adsorptive Au-TiO2Nanocomposites for the SERS Face Mask Allow the Machine-Learning-Based Quantitative Assay of SARS-CoV-2 in Artificial Breath Aerosols. ACS Appl. Mater. Interfaces 2022, 14, 54550–54557. [Google Scholar] [CrossRef] [PubMed]
- Ansah, I.B.; Leming, M.; Lee, S.H.; Yang, J.-Y.; Mun, C.; Noh, K.; An, T.; Lee, S.; Kim, D.-H.; Kim, M.; et al. Label-free detection and discrimination of respiratory pathogens based on electrochemical synthesis of biomaterials-mediated plasmonic composites and machine learning analysis. Biosens. Bioelectron. 2023, 227, 115178. [Google Scholar] [CrossRef] [PubMed]
- Berus, S.M.; Nowicka, A.B.; Wieruszewska, J.; Niciński, K.; Kowalska, A.A.; Szymborski, T.R.; Dróżdż, I.; Borowiec, M.; Waluk, J.; Kamińska, A. SERS Signature of SARS-CoV-2 in Saliva and Nasopharyngeal Swabs: Towards Perspective COVID-19 Point-of-Care Diagnostics. Int. J. Mol. Sci. 2023, 24, 9706. [Google Scholar] [CrossRef]
- Szymborski, T.; Stepanenko, Y.; Niciński, K.; Piecyk, P.; Berus, S.M.; Adamczyk-Popławska, M.; Kamińska, A. Ultrasensitive SERS platform made via femtosecond laser micromachining for biomedical applications. J. Mater. Res. Technol. 2021, 12, 1496–1507. [Google Scholar] [CrossRef]
- Ralbovsky, N.M.; Lednev, I.K. Towards development of a novel universal medical diagnostic method: Raman spectroscopy and machine learning. Chem. Soc. Rev. 2020, 49, 7428–7453. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.Y.; Jordan, M.I. On Discriminative vs. Generative Classifiers: A comparison of logistic regression and naive Bayes. Adv. Neural Inf. Process. Syst. 2001, 14, 1–8. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Suykens, J.A.K.; Vandewalle, J. Least squares support vector machine classifiers. Neural Process. Lett. 1999, 9, 293–300. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Isho, B.; Abe, K.T.; Zuo, M.; Jamal, A.J.; Rathod, B.; Wang, J.H.; Li, Z.; Chao, G.; Rojas, O.L.; Bang, Y.M.; et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020, 5, eabe5511. [Google Scholar] [CrossRef] [PubMed]
- Baghizadeh Fini, M. Oral saliva and COVID-19. Oral Oncol. 2020, 108, 104821. [Google Scholar] [CrossRef] [PubMed]
- Torreggiani, A.; Barata-Vallejo, S.; Chatgilialoglu, C. Combined Raman and IR spectroscopic study on the radical-based modifications of methionine. Anal. Bioanal. Chem. 2011, 401, 1231–1239. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Han, Q. Oral methioninase for Covid-19 methionine-restriction therapy. In Vivo 2020, 34, 1593–1596. [Google Scholar] [CrossRef] [PubMed]
- Franco-Martínez, L.; Cerón, J.J.; Vicente-Romero, M.R.; Bernal, E.; Cantero, A.T.; Tecles, F.; Resalt, C.S.; Martínez, M.; Tvarijonaviciute, A.; Martínez-Subiela, S. Salivary Ferritin Changes in Patients with COVID-19. Int. J. Environ. Res. Public Health 2021, 19, 41. [Google Scholar] [CrossRef] [PubMed]
- Hailemichael, W.; Kiros, M.; Akelew, Y.; Getu, S.; Andualem, H. Neopterin: A Promising Candidate Biomarker for Severe COVID-19; Dove Press: Macclesfield, UK, 2021; Volume 14, p. 245. [Google Scholar] [CrossRef]
- Kamińska, A.; Witkowska, E.; Kowalska, A.; Skoczyńska, A.; Gawryszewska, I.; Guziewicz, E.; Snigurenko, D.; Waluk, J. Highly efficient SERS-based detection of cerebrospinal fluid neopterin as a diagnostic marker of bacterial infection. Anal. Bioanal. Chem. 2016, 408, 4319–4327. [Google Scholar] [CrossRef]
- Lin, X.; Lin, D.; Ge, X.; Qiu, S.; Feng, S.; Chen, R. Noninvasive Detection of Nasopharyngeal Carcinoma Based on Saliva Proteins Using Surface-Enhanced Raman Spectroscopy. J. Biomed. Opt. 2017, 22, 105004. [Google Scholar] [CrossRef]
- Li, X.; Yang, T.; Lin, J. Spectral Analysis of Human Saliva for Detection of Lung Cancer Using Surface-Enhanced Raman Spectroscopy. J. Biomed. Opt. 2012, 17, 037003. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2015, 50, 46–111. [Google Scholar] [CrossRef]
- Austin, L.A.; Osseiran, S.; Evans, C.L. Raman Technologies in Cancer Diagnostics. Analyst 2016, 141, 476–503. [Google Scholar] [CrossRef]
- Cao, G.; Chen, M.; Chen, Y.; Huang, Z.; Lin, J.; Lin, J.; Xu, Z.; Wu, S.; Huang, W.; Weng, G.; et al. A Potential Method for Non-Invasive Acute Myocardial Infarction Detection Based on Saliva Raman Spectroscopy and Multivariate Analysis. Laser Phys. Lett. 2015, 12, 125702. [Google Scholar] [CrossRef]
- Oliveira, E.M.; Rogero, M.; Ferreira, E.C.; Gomes Neto, J.A. Simultaneous Determination of Phosphite and Phosphate in Fertilizers by Raman Spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 246, 119025. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Zheng, X.S.; Zong, C.; Li, M.H.; Zhang, L.Y.; Li, W.; Ren, B. Drop-Coating Deposition and Surface-Enhanced Raman Spectroscopies (DCDRS and SERS) Provide Complementary Information of Whole Human Tears. J. Raman Spectrosc. 2014, 45, 565–573. [Google Scholar] [CrossRef]
- Virkler, K.; Lednev, I.K. Forensic Body Fluid Identification: The Raman Spectroscopic Signature of Saliva. Analyst 2010, 135, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Carlomagno, C.; Bertazioli, D.; Gualerzi, A.; Picciolini, S.; Banfi, P.I.; Lax, A.; Messina, E.; Navarro, J.; Bianchi, L.; Caronni, A.; et al. COVID-19 salivary Raman fingerprint: Innovative approach for the detection of current and past SARS-CoV-2 infections. Sci. Rep. 2021, 11, 4943. [Google Scholar] [CrossRef]
- Ceccon, D.M.; Amaral, P.H.R.; Andrade, L.M.; da Silva, M.I.; Andrade, L.A.; Moraes, T.F.; Bagno, F.F.; Rocha, R.P.; de Almeida Marques, D.P.; Ferreira, G.M.; et al. New, fast, and precise method of COVID-19 detection in nasopharyngeal and tracheal aspirate samples combining optical spectroscopy and machine learning. Braz. J. Microbiol. 2023, 54, 769–777. [Google Scholar] [CrossRef]
- Goulart, A.C.C.; Zângaro, R.A.; Carvalho, H.C.; Lednev, I.K.; Silveira, L., Jr. Diagnosing COVID-19 in nasopharyngeal secretion through Raman spectroscopy: A feasibility study. Lasers Med. Sci. 2023, 38, 210. [Google Scholar] [CrossRef]



| Type | Total Number of Samples | CoV(+) | CoV(−) |
|---|---|---|---|
| saliva | 175 | 81 | 94 |
| nasopharyngeal swab | 114 | 51 | 63 |
| total: | 289 | 132 | 157 |
| Number of Dimensions | Variance Perseverance Ratio (%) | |
|---|---|---|
| Saliva | Nasopharyngeal Swab | |
| 1 | 24.5 | 30.0 |
| 2 | 41.3 | 44.5 |
| 5 | 70.0 | 70.0 |
| 10 | 86.8 | 86.3 |
| 20 | 96.3 | 96.3 |
| 30 | 98.6 | 98.8 |
| 40 | 99.4 | 99.5 |
| Classifier Type | Saliva | Nasopharyngeal Swabs | ||||
|---|---|---|---|---|---|---|
| Precision (%) | Recall (%) | Adjusted * Balanced Accuracy (%) | Precision (%) | Recall (%) | Adjusted ** Balanced Accuracy (%) | |
| gaussian naive Bayes (GNB) | 71.4 | 68.7 | 79.1 | 50 | 63.6 | 58.1 |
| 85.7 | 100 | 92.1 | 71.4 | 100 | 84.6 | |
| 81.4 | 83 | 83 | 62.6 | 82.7 | 71.5 | |
| random forest (RF) | 85.7 | 62.5 | 78.6 | 38.4 | 45.4 | 39.4 |
| 93.7 | 93.7 | 94.2 | 90 | 90 | 90.8 | |
| 90.4 | 81.6 | 87.1 | 65.7 | 61.1 | 66.1 | |
| support vector classifier (SVC) | 75 | 68.7 | 74.3 | 46.1 | 60 | 53.1 |
| 93.7 | 100 | 91.1 | 81.8 | 90.9 | 86.7 | |
| 85 | 87.7 | 85.8 | 69 | 78.2 | 74 | |
| logistic regression (LR) | 80 | 62.5 | 78.6 | 41.2 | 50 | 40.1 |
| 92.8 | 100 | 89.5 | 77.8 | 70 | 77.3 | |
| 87.1 | 85.1 | 86.7 | 61.2 | 62.7 | 62.8 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szymborski, T.R.; Berus, S.M.; Nowicka, A.B.; Słowiński, G.; Kamińska, A. Machine Learning for COVID-19 Determination Using Surface-Enhanced Raman Spectroscopy. Biomedicines 2024, 12, 167. https://doi.org/10.3390/biomedicines12010167
Szymborski TR, Berus SM, Nowicka AB, Słowiński G, Kamińska A. Machine Learning for COVID-19 Determination Using Surface-Enhanced Raman Spectroscopy. Biomedicines. 2024; 12(1):167. https://doi.org/10.3390/biomedicines12010167
Chicago/Turabian StyleSzymborski, Tomasz R., Sylwia M. Berus, Ariadna B. Nowicka, Grzegorz Słowiński, and Agnieszka Kamińska. 2024. "Machine Learning for COVID-19 Determination Using Surface-Enhanced Raman Spectroscopy" Biomedicines 12, no. 1: 167. https://doi.org/10.3390/biomedicines12010167
APA StyleSzymborski, T. R., Berus, S. M., Nowicka, A. B., Słowiński, G., & Kamińska, A. (2024). Machine Learning for COVID-19 Determination Using Surface-Enhanced Raman Spectroscopy. Biomedicines, 12(1), 167. https://doi.org/10.3390/biomedicines12010167






