The Causal Relationship between Inflammatory Cytokines and Liver Cirrhosis in European Descent: A Bidirectional Two-Sample Mendelian Randomization Study and the First Conclusions
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Selection of IVs
2.3. Statistical Analyses
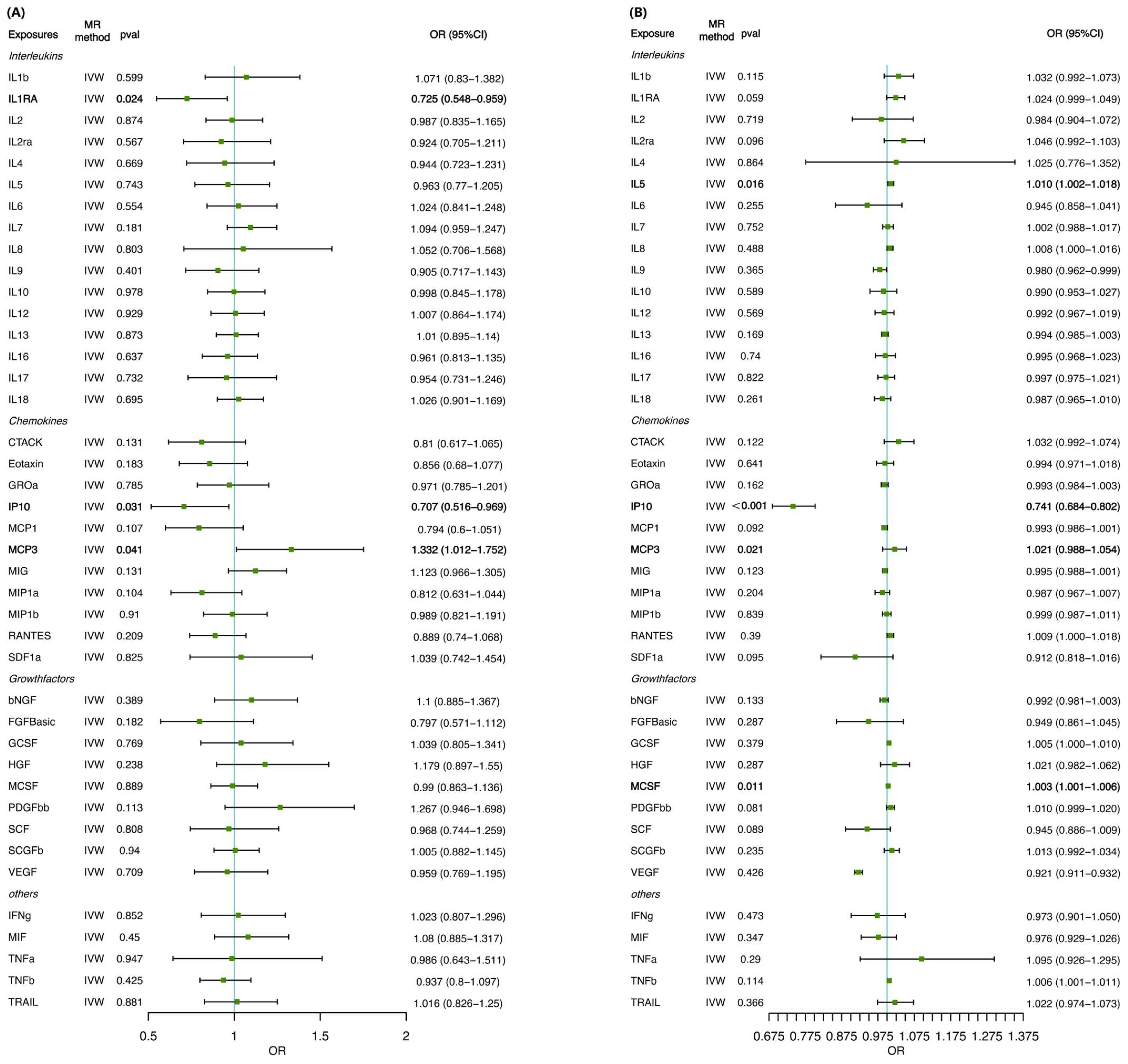
3. Results
3.1. MR Analysis of Inflammatory Cytokines on Cirrhosis Risk
3.2. MR Analysis of the Influence of Cirrhosis on Inflammatory Cytokines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Ginès, P.; Krag, A.; Abraldes, J.G.; Solà, E.; Fabrellas, N.; Kamath, P.S. Liver cirrhosis. Lancet 2021, 398, 1359–1376. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.Q.; Terrault, N.A.; Tacke, F.; Gluud, L.L.; Arrese, M.; Bugianesi, E.; Loomba, R. Global epidemiology of cirrhosis—Aetiology, trends and predictions. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 388–398. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, G.; Garcia-Tsao, G.; Pagliaro, L. Natural history and prognostic indicators of survival in cirrhosis: A systematic review of 118 studies. J. Hepatol. 2006, 44, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Fabrellas, N.; Moreira, R.; Carol, M.; Cervera, M.; de Prada, G.; Perez, M.; Vazquez, E.; Sola, M.; Sancho, R.; Juanola, A.; et al. Psychological Burden of Hepatic Encephalopathy on Patients and Caregivers. Clin. Transl. Gastroenterol. 2020, 11, e00159. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.M.; Kim, W.R.; Moriarty, J.P.; Shah, N.D.; Larson, J.J.; Kamath, P.S. Time trends in the health care burden and mortality of acute on chronic liver failure in the United States. Hepatology 2016, 64, 2165–2172. [Google Scholar] [CrossRef]
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef]
- Arroyo, V.; Angeli, P.; Moreau, R.; Jalan, R.; Clària, J.; Trebicka, J.; Fernández, J.; Gustot, T.; Caraceni, P.; Bernardi, M. The systemic inflammation hypothesis: Towards a new paradigm of acute decompensation and multiorgan failure in cirrhosis. J. Hepatol. 2021, 74, 670–685. [Google Scholar] [CrossRef]
- Friedman, S.L.; Pinzani, M. Hepatic fibrosis 2022: Unmet needs and a blueprint for the future. Hepatology 2022, 75, 473–488. [Google Scholar] [CrossRef]
- Engelmann, C.; Clària, J.; Szabo, G.; Bosch, J.; Bernardi, M. Pathophysiology of decompensated cirrhosis: Portal hypertension, circulatory dysfunction, inflammation, metabolism and mitochondrial dysfunction. J. Hepatol. 2021, 75 (Suppl. S1), S49–S66. [Google Scholar] [CrossRef]
- Rey, I.; Effendi-Ys, R. Association Between Serum IL-6, IL-10, IL-12, and IL-23 Levels and Severity of Liver Cirrhosis. Med. Arch. 2021, 75, 199–203. [Google Scholar] [CrossRef]
- Albillos, A.; Lario, M.; Álvarez-Mon, M. Cirrhosis-associated immune dysfunction: Distinctive features and clinical relevance. J. Hepatol. 2014, 61, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Hammerich, L.; Tacke, F. Hepatic inflammatory responses in liver fibrosis. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 633–646. [Google Scholar] [CrossRef] [PubMed]
- Davey Smith, G.; Hemani, G. Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014, 23, R89–R98. [Google Scholar] [CrossRef] [PubMed]
- Skrivankova, V.W.; Richmond, R.C.; Woolf, B.A.R.; Davies, N.M.; Swanson, S.A.; VanderWeele, T.J.; Timpson, N.J.; Higgins, J.P.T.; Dimou, N.; Langenberg, C.; et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): Explanation and elaboration. BMJ 2021, 375, n2233. [Google Scholar] [CrossRef] [PubMed]
- Skrivankova, V.W.; Richmond, R.C.; Woolf, B.A.R.; Yarmolinsky, J.; Davies, N.M.; Swanson, S.A.; VanderWeele, T.J.; Higgins, J.P.T.; Timpson, N.J.; Dimou, N.; et al. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization: The STROBE-MR Statement. JAMA 2021, 326, 1614–1621. [Google Scholar] [CrossRef] [PubMed]
- Sallis, H.M.; Palmer, T.; Tilling, K.; Davey Smith, G.; Munafò, M.R. Using allele scores to identify confounding by reverse causation: Studies of alcohol consumption as an exemplar. Int. J. Epidemiol. 2023, 52, 536–544. [Google Scholar] [CrossRef]
- Emdin, C.A.; Khera, A.V.; Kathiresan, S. Mendelian Randomization. JAMA 2017, 318, 1925–1926. [Google Scholar] [CrossRef] [PubMed]
- Ahola-Olli, A.V.; Würtz, P.; Havulinna, A.S.; Aalto, K.; Pitkänen, N.; Lehtimäki, T.; Kähönen, M.; Lyytikäinen, L.P.; Raitoharju, E.; Seppälä, I.; et al. Genome-wide Association Study Identifies 27 Loci Influencing Concentrations of Circulating Cytokines and Growth Factors. Am. J. Hum. Genet. 2017, 100, 40–50. [Google Scholar] [CrossRef]
- Emdin, C.A.; Haas, M.E.; Khera, A.V.; Aragam, K.; Chaffin, M.; Klarin, D.; Hindy, G.; Jiang, L.; Wei, W.Q.; Feng, Q.; et al. A missense variant in Mitochondrial Amidoxime Reducing Component 1 gene and protection against liver disease. PLoS Genet. 2020, 16, e1008629. [Google Scholar] [CrossRef]
- Buch, S.; Stickel, F.; Trépo, E.; Way, M.; Herrmann, A.; Nischalke, H.D.; Brosch, M.; Rosendahl, J.; Berg, T.; Ridinger, M.; et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat. Genet. 2015, 47, 1443–1448. [Google Scholar] [CrossRef]
- He, H.; Liao, S.; Zeng, Y.; Liang, L.; Chen, J.; Tao, C. Causal relationships between metabolic-associated fatty liver disease and iron status: Two-sample Mendelian randomization. Liver Int. Off. J. Int. Assoc. Study Liver 2022, 42, 2759–2768. [Google Scholar] [CrossRef] [PubMed]
- Palmer, T.M.; Lawlor, D.A.; Harbord, R.M.; Sheehan, N.A.; Tobias, J.H.; Timpson, N.J.; Davey Smith, G.; Sterne, J.A. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat. Methods Med. Res. 2012, 21, 223–242. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Verbanck, M.; Chen, C.Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef]
- Pratim Das, P.; Medhi, S. Role of inflammasomes and cytokines in immune dysfunction of liver cirrhosis. Cytokine 2023, 170, 156347. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, P.; Xiao, N.; Liu, Q.; Zhu, X. Interleukin-8 predicts short-term mortality in acute-on-chronic liver failure patients with hepatitis B-related-related cirrhosis background. Ann. Med. 2023, 55, 2287708. [Google Scholar] [CrossRef]
- Salgüero, S.; Medrano, L.M.; González-García, J.; Berenguer, J.; Montes, M.L.; Diéz, C.; Garcia-Broncano, P.; Llop-Herrera, E.; Pérez-Latorre, L.; Bellóno, J.M.; et al. Plasma IP-10 and IL-6 are linked to Child-Pugh B cirrhosis in patients with advanced HCV-related cirrhosis: A cross-sectional study. Sci. Rep. 2020, 10, 10384. [Google Scholar] [CrossRef]
- Casari, M.; Siegl, D.; Deppermann, C.; Schuppan, D. Macrophages and platelets in liver fibrosis and hepatocellular carcinoma. Front. Immunol. 2023, 14, 1277808. [Google Scholar] [CrossRef]
- Sinzel, M.; Tan, T.; Wendling, P.; Kalbacher, H.; Özbalci, C.; Chelius, X.; Westermann, B.; Brügger, B.; Rapaport, D.; Dimmer, K.S. Mcp3 is a novel mitochondrial outer membrane protein that follows a unique IMP-dependent biogenesis pathway. EMBO Rep. 2016, 17, 965–981. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.L.; McVicar, D.W.; Ben-Baruch, A.; Kuhns, D.B.; Johnston, J.; Oppenheim, J.J.; Wang, J.M. Monocyte chemotactic protein-3 (MCP3) interacts with multiple leukocyte receptors: Binding and signaling of MCP3 through shared as well as unique receptors on monocytes and neutrophils. Eur. J. Immunol. 1995, 25, 2612–2617. [Google Scholar] [CrossRef]
- Seki, E.; de Minicis, S.; Inokuchi, S.; Taura, K.; Miyai, K.; van Rooijen, N.; Schwabe, R.F.; Brenner, D.A. CCR2 promotes hepatic fibrosis in mice. Hepatology 2009, 50, 185–197. [Google Scholar] [CrossRef]
- Owusu Sekyere, S.; Port, K.; Deterding, K.; Cornberg, M.; Wedemeyer, H. Inflammatory patterns in plasma associate with hepatocellular carcinoma development in cured hepatitis C cirrhotic patients. United Eur. Gastroenterol. J. 2021, 9, 486–496. [Google Scholar] [CrossRef]
- Rampa, D.R.; Feng, H.; Allur-Subramaniyan, S.; Shim, K.; Pekcec, A.; Lee, D.; Doods, H.; Wu, D. Kinin B1 receptor blockade attenuates hepatic fibrosis and portal hypertension in chronic liver diseases in mice. J. Transl. Med. 2022, 20, 590. [Google Scholar] [CrossRef] [PubMed]
- Shahera, U.; Munshi, S.; Jahan, M.; Nessa, A.; Alam, S.; Tabassum, S. IP-10, p53, and Foxp3 Expression in Hepatocytes of Chronic Hepatitis B Patients with Cirrhosis and Hepatocellular Carcinoma. Euroasian J. Hepato-Gastroenterol. 2016, 6, 149–153. [Google Scholar] [CrossRef]
- Talaat, R.M.; Elsharnoby, S.; Abdelkhalek, M.S.; El-Shenawy, S.Z.; Elmasry, S. The Impact of Interferon-γ (IFN-γ) and IFN-γ-Inducible Protein 10 (IP-10) Genes’ Polymorphism on Risk of Hepatitis C Virus-Related Liver Cirrhosis. Immunol. Investig. 2022, 51, 688–704. [Google Scholar] [CrossRef]
- Katoh, R.; Maekawa, S.; Osawa, L.; Komiyama, Y.; Nakakuki, N.; Takada, H.; Matsuda, S.; Muraoka, M.; Suzuki, Y.; Sato, M.; et al. Significance of serum IP-10/CXCL10 measurement in predicting post-direct acting antiviral treatment liver function in patients with HCV-decompensated liver cirrhosis. Hepatol. Res. Off. J. Jpn. Soc. Hepatol. 2023, 53, 280–288. [Google Scholar] [CrossRef]
- Zhao, Y.; Ye, W.; Wang, Y.D.; Chen, W.D. HGF/c-Met: A Key Promoter in Liver Regeneration. Front. Pharmacol. 2022, 13, 808855. [Google Scholar] [CrossRef]
- Dong, X.; Luo, Y.; Lu, S.; Ma, H.; Zhang, W.; Zhu, Y.; Sun, G.; Sun, X. Ursodesoxycholic acid alleviates liver fibrosis via proregeneration by activation of the ID1-WNT2/HGF signaling pathway. Clin. Transl. Med. 2021, 11, e296. [Google Scholar] [CrossRef]
- Liang, F.; Xu, X.; Tu, Y. Resveratrol inhibited hepatocyte apoptosis and alleviated liver fibrosis through miR-190a-5p /HGF axis. Bioorganic Med. Chem. 2022, 57, 116593. [Google Scholar] [CrossRef] [PubMed]
- Stöß, C.; Laschinger, M.; Wang, B.; Lu, M.; Altmayr, F.; Hartmann, D.; Hüser, N.; Holzmann, B. TLR3 promotes hepatocyte proliferation after partial hepatectomy by stimulating uPA expression and the release of tissue-bound HGF. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 10387–10397. [Google Scholar] [CrossRef]
- Shivaramu, S.; Maiti, S.K.; Banu, S.A.; Kalaiselvan, E.; Sharun, K.; Mishra, M.; Mohan, D.; Palakkara, S.; Kumar, S.; Sahoo, M.; et al. Synergistic Hepatoprotective Effects of Mesenchymal Stem Cells and Platelet-Rich Plasma in a Rat Model of Bile Duct Ligation-Induced Liver Cirrhosis. Cells 2024, 13, 404. [Google Scholar] [CrossRef] [PubMed]
- Gharbia, S.; Nazarie, S.R.; Dinescu, S.; Balta, C.; Herman, H.; Peteu, V.E.; Gherghiceanu, M.; Hermenean, A.; Costache, M. Adipose-Derived Stem Cells (ADSCs) Supplemented with Hepatocyte Growth Factor (HGF) Attenuate Hepatic Stellate Cell Activation and Liver Fibrosis by Inhibiting the TGF-β/Smad Signaling Pathway in Chemical-Induced Liver Fibrosis Associated with Diabetes. Cells 2022, 11, 3338. [Google Scholar] [CrossRef] [PubMed]




| Characteristic | Abbreviation | GWAS ID | Sample Size | Number of SNPs | Cases Definition | Exclusion Criteria | Quality Control | |
|---|---|---|---|---|---|---|---|---|
| Interleukins | Interleukin-1-beta | IL-1β | ebi-a-GCST004448 | 3309 | 983,642 | The GWAS meta-analysis included up to 8293 Finnish individuals from three independent population cohorts: the Cardiovascular Risk in Young Finns Study (YFS), FINRISK1997, and FINRISK2002. The study cohort characteristics are reported in Table S1 in the original study. On average, the YFS participants are younger than the C23FINRISK1997 and FINRISK2002 participants (37 versus 60 years). | - | Linear regression with a probe as a dependent variable was used to test associations between cytokine-associated variants and transcripts. Age and sex were used as covariates. Genotype dosage was calculated for each included variant with Qctool (version 2) software. More details referred to the original study. |
| Interleukin-1-receptor antagonist | IL-1RA | ebi-a-GCST004447 | 3638 | 9,564,741 | ||||
| Interleukin-2 | IL-2 | ebi-a-GCST004455 | 3475 | 9,512,914 | ||||
| Interleukin-2 receptor antagonist | IL-2RA | ebi-a-GCST004454 | 3677 | 9,583,519 | ||||
| Interleukin-4 | IL-4 | ebi-a-GCST004453 | 8124 | 9,786,064 | ||||
| Interleukin-5 | IL-5 | ebi-a-GCST004452 | 3364 | 9,450,731 | ||||
| Interleukin-6 | IL-6 | ebi-a-GCST004446 | 8189 | 9,790,590 | ||||
| Interleukin-7 | IL-7 | ebi-a-GCST004451 | 3409 | 9,692,306 | ||||
| Interleukin-8 | IL-8 | ebi-a-GCST004445 | 3526 | 9,517,348 | ||||
| Interleukin-9 | IL-9 | ebi-a-GCST004450 | 3634 | 9,567,876 | ||||
| Interleukin-10 | IL-10 | ebi-a-GCST004444 | 7681 | 9,793,415 | ||||
| Interleukin-12p70 | IL-12p70 | ebi-a-GCST004439 | 8270 | 9,799,886 | ||||
| Interleukin-13 | IL-13 | ebi-a-GCST004443 | 3557 | 9,539,073 | ||||
| Interleukin-16 | IL-16 | ebi-a-GCST004430 | 3483 | 9,551,485 | ||||
| Interleukin-17 | IL-17 | ebi-a-GCST004442 | 7760 | 9,786,653 | ||||
| Interleukin-18 | IL-18 | ebi-a-GCST004441 | 3636 | 9,785,222 | ||||
| Chemokines | Cutaneous T cell attracting | CTACK | ebi-a-GCST004420 | 3631 | 9,568,408 | |||
| Eotaxin | Eotaxin | ebi-a-GCST004460 | 8153 | 9,793,404 | ||||
| Growth-regulated protein alpha | GRPa | ebi-a-GCST004457 | 3505 | 9,528,505 | ||||
| Interferon gamma-induced protein 10 | IP-10 | ebi-a-GCST004440 | 3685 | 9,576,881 | ||||
| Monocyte chemoattractant protein-1 | MCP1 | ebi-a-GCST004438 | 8293 | 9,801,908 | ||||
| Monocyte chemoattractant protein-3 | MCP3 | ebi-a-GCST004437 | 843 | 7,630,881 | ||||
| Monokine induced by gamma interferon | MIG | ebi-a-GCST004435 | 3685 | 9,579,894 | ||||
| Macrophage inflammatory protein 1a | MIP1α | ebi-a-GCST004434 | 3522 | 9,519,267 | ||||
| Macrophage inflammatory protein 1b | MIP1β | ebi-a-GCST004433 | 8243 | 9,802,973 | ||||
| Regulated on activation, normal T cell expressed and secreted | RANTES | ebi-a-GCST004431 | 3421 | 9,523,827 | ||||
| Stromal-cell-derived factor 1 alpha | SDF-1α | ebi-a-GCST004427 | 5998 | 9,736,366 | ||||
| Growth factors | Beta-nerve growth factor | βNGF | ebi-a-GCST004421 | 3531 | 9,537,863 | |||
| Granulocyte-colony stimulating factor | GCSF | ebi-a-GCST004458 | 7904 | 9,788,961 | ||||
| Fibroblast growth factor basic | FGFBasic | ebi-a-GCST004459 | 7565 | 9,790,946 | ||||
| Hepatocyte growth factor | HGF | ebi-a-GCST004449 | 8292 | 9,802,538 | ||||
| Macrophage colony stimulating factor | MCSF | ebi-a-GCST004436 | 840 | 9,184,521 | ||||
| Platelet-derived growth factor BB | PDGFbb | ebi-a-GCST004432 | 8293 | 9,800,009 | ||||
| Stem cell factor | SCF | ebi-a-GCST004429 | 8290 | 9,796,683 | ||||
| Stem cell growth factor beta | SCGFβ | ebi-a-GCST004428 | 3682 | 9,574,890 | ||||
| Vascular endothelial growth factor | VEGF | ebi-a-GCST004422 | 7118 | 9,784,803 | ||||
| Others | Interferon gamma | IFN -γ | ebi-a-GCST004456 | 7701 | 9,785,363 | |||
| Macrophage Migration Inhibitory Factor | MIF | ebi-a-GCST004423 | 3494 | 9,537,573 | ||||
| Tumor necrosis factor alpha | TGFα | ebi-a-GCST004426 | 3454 | 9,500,449 | ||||
| Tumor necrosis factor beta | TGFβ | ebi-a-GCST004425 | 1559 | 6,304,298 | ||||
| TNF-related apoptosis inducing ligand | TRAIL | ebi-a-GCST004424 | 8186 | 9,698,525 | ||||
| CIRRHOSIS | Cirrhosis | - | finn-b-CIRRHOSIS_BROAD | 1931 cases and 216,861 controls | 16,380,466 | Cirrhosis ascertained through ICD-10 code K74, ICD-9 code 571 (hepatic fibrosis and cirrhosis) | The study excluded cases of cirrhosis secondary to primary biliary cholangitis and primary sclerosis cholangitis, as these autoimmune disorders are directed against the biliary (and not hepatic) parenchyma < 30 g/day for men, chronic viral hepatitis (hepatitis B and hepatitis C), autoimmune liver diseases, hereditary hemochromatosis, α1-antitrypsin deficiency, Wilson’s disease, and drug-induced liver injury | A genome-wide association study in each cohort was performed using logistic regression with adjustment for age, sex, and ten principal components of ancestry. We tested the association of fourteen million variants with a minor allele frequency of greater than 0.1% with cirrhosis in each cohort. PLINK (2015-1-25) was used for all analyses. To combine estimates across cohorts, inverse variance fixed-effects meta-analysis, as implemented by METAL (2010-9-1), was used. Quantile–quantile analysis was used to examine the presence of population stratification. No evidence of inflation was observed (lambda 1.02; Supplementary Figure S2 in the original study). Both additive and recessive analyses were performed. |
| - | UK Biobank | 2701 cases and 16,206 controls | - | Hospitalization or death due to physician-diagnosed cirrhosis: K70.2 (alcoholic fibrosis and sclerosis of the liver), K70.3 (alcoholic cirrhosis of the liver), K70.4 (alcoholic hepatic failure), K74.0 (hepatic fibrosis), K74.1 (hepatic sclerosis), K74.2 (hepatic fibrosis with hepatic sclerosis), K74.6 (other and unspecified cirrhosis of liver), K76.6 (portal hypertension), or I85 (esophageal varices). Controls were free of liver disease. | - | - | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, S.; Zhou, Y.; Zhang, H.; Zhu, Y.; Jiang, P.; Xie, C.; Feng, T.; Zeng, Y.; He, H.; Luo, Y.; et al. The Causal Relationship between Inflammatory Cytokines and Liver Cirrhosis in European Descent: A Bidirectional Two-Sample Mendelian Randomization Study and the First Conclusions. Biomedicines 2024, 12, 2264. https://doi.org/10.3390/biomedicines12102264
Shi S, Zhou Y, Zhang H, Zhu Y, Jiang P, Xie C, Feng T, Zeng Y, He H, Luo Y, et al. The Causal Relationship between Inflammatory Cytokines and Liver Cirrhosis in European Descent: A Bidirectional Two-Sample Mendelian Randomization Study and the First Conclusions. Biomedicines. 2024; 12(10):2264. https://doi.org/10.3390/biomedicines12102264
Chicago/Turabian StyleShi, Shiya, Yanjie Zhou, He Zhang, Yalan Zhu, Pengjun Jiang, Chengxia Xie, Tianyu Feng, Yuping Zeng, He He, Yao Luo, and et al. 2024. "The Causal Relationship between Inflammatory Cytokines and Liver Cirrhosis in European Descent: A Bidirectional Two-Sample Mendelian Randomization Study and the First Conclusions" Biomedicines 12, no. 10: 2264. https://doi.org/10.3390/biomedicines12102264
APA StyleShi, S., Zhou, Y., Zhang, H., Zhu, Y., Jiang, P., Xie, C., Feng, T., Zeng, Y., He, H., Luo, Y., & Chen, J. (2024). The Causal Relationship between Inflammatory Cytokines and Liver Cirrhosis in European Descent: A Bidirectional Two-Sample Mendelian Randomization Study and the First Conclusions. Biomedicines, 12(10), 2264. https://doi.org/10.3390/biomedicines12102264






