Abstract
Background: Gliomas are neoplasms of the central nervous system that originate in glial cells. The genetic characteristics of this type of neoplasm are the loss of function of tumor suppressor genes such as TP53 and somatic mutations in genes such as IDH1/2. Additionally, in clinical cases, de novo single nucleotide polymorphisms (SNP) are reported, of which their pathogenicity and their effects on the function and stability of the protein are known. Methodology: Non-synonymous SNPs were analyzed for their structural and functional effect on proteins using a set of bioinformatics tools such as SIFT, PolyPhen-2, PhD-SNP, I-Mutant 3.0, MUpro, and mutation3D. A structural comparison between normal and mutated residues for disease-associated coding SNPs was performed using TM-aling and the SWISS MODEL. Results: A total of 13 SNPs were obtained for the TP53 gene, 1 SNP for IDH1, and 1 for IDH2, which would be functionally detrimental and associated with disease. Additionally, these changes compromise the structure and function of the protein; the A161S SNP for TP53 that has not been reported in any databases was classified as detrimental. Conclusions: All non-synonymous SNPs reported for TP53 were in the region of the deoxyribonucleic acid (DNA) binding domain and had a great impact on the function and stability of the protein. In addition, the two polymorphisms detected in IDH1 and IDH2 genes compromise the structure and activity of the protein. Both genes are related to the development of high-grade gliomas. All the data obtained in this study must be validated through experimental approaches.
Keywords:
neoplasia of the central nervous system; computational analysis; protein; variant; TP53; IDH1; IDH2 1. Introduction
High-grade gliomas are primary tumors of the central nervous system (CNS), which, according to the fifth classification of the World Health Organization (WHO) 2021, are classified according to their histological and molecular characteristics into astrocytomas, oligodendrogliomas, and glioblastomas [1]. Patients diagnosed with this neoplasia have a poor prognosis, with only 20% of them surviving around 5 years. More specifically, the two patients diagnosed with glioblastomas were expected to survive about 2 years (14.8% patients) and 10 years (2.6%), Amut-IDH, sin-codel-1p/19q, G3-4, and these patients were expected to survive about 2 years (43.3%) and 10 years (19% patients), Omut-IDH, codel-1p/19q, G3, and these patients are expected to survive about 2 years (68.6% patients) and 10 years (39.3% patients). Despite aggressive treatment with surgery, chemotherapy, and radiotherapy [2,3], oncological diseases arise because of genetic alterations in genes that control critical biological processes. The accumulation of mutations over time allows the survival and progressive transformation of abnormal cell populations that eventually lead to tumor formation. In the case of gliomas, this process is known as gliomagenesis, which is characterized by several biological events, such as activated growth factor, damage to receptor signaling pathways, negative regulation of many apoptotic mechanisms, and imbalance between pro-angiogenic and antiangiogenic factors [4].
Establishing the process of gliomagenesis is of vital importance to identify the underlying molecular mechanisms involved in tumor progression. Several central signaling pathways have been identified in this process: one of these is the RTK/RAS/PI3K, TP53, and RB1 pathway, where TP53 mutations are critical for gli-oma progression [5]. TP53 plays a key role in maintaining cellular homeostasis and is frequently deregulated in oncological pathologies such as gliomas. The protein is positioned at the center of a regulatory network that controls cell proliferation, survival, genome integrity, and other functions. As a transcriptional regulator, TP53 integrates stress signals and promotes cell cycle arrest, senescence, and apoptosis to prevent damaged cells from spreading [6]. The TP53 mutational status is associated with glioblastoma progression [7] and TP53 inactivation is correlated with a more invasive [8], less apoptotic [9], more proliferative, and stem cell-like phenotype [10]. Glioblastoma cell lines that possess inactive mutated TP53 are more resistant to DNA-damaging therapeutic drugs, such as cisplatin.
Within the process of gliomagenesis, IDH1/2 mutations have been identified as frequently occurring in the early stages of human glioma development. IDH mutation has been shown to promote glioma development through DNA and histone methylation [11]. In a study of 1010 diffuse gliomas, IDH1 mutations were detected in 70.9% of tumors, while IDH2 mutations were observed in 3.1% of tumors [11]. These two mutations give rise to neomorphic enzymatic activity, leading to distinct patterns in cancer metabolism, epigenetic alterations, and resistance to therapy. The mutant enzymes, in particular, the IDH1 R132H and IDH2 R172K variants, have been widely studied in low- and high-grade gliomas. These variants are characterized by conferring a specific enzymatic activity that converts alpha-ketoglutarate to 2-hydroxyglutarate; the accumulation of this oncometabolite in gliomas with IDH mutations profoundly affects several cellular processes [12], including metabolic reprogramming, because the Krebs cycle adjusts to compensate for fluctuations in the pathways. An analysis of metabolic flux suggested that cells with IDH mutations exhibit increased oxidative metabolism in the Krebs cycle, while the reductive metabolism of glutamine is suppressed [13]. The objective of this study was to evaluate the functional and structural effects generated by mutations in the genes that encode the TP53 and IDH1/2 proteins in a cohort of 31 patients with high-grade glioma.
2. Materials and Methods
2.1. Study Population
The sample analyzed was obtained from patients who attended the Cancer Institute of the Clínica Las Américas-AUNA Medellín-Colombia between 2019 and 2021 to receive treatment for high-grade gliomas. Of the total population, 31 patients participated by open invitation with a diagnosis of glioblastoma, oligodendroglioma, or astrocytoma.
2.2. Samples for Sequencing
We collected a total of 26 blocks of paraffin-embedded brain tissue and 5 liquid biopsies. From these samples, DNA was extracted and used for the sequencing of 324 genes involved in tumor processes, thus obtaining the mutations for the 31 patients with high-grade gliomas.
2.3. Molecular Mutation Panel
Genomic sequencing was performed through the Foundation one® CDx (F1CDx) panel, which is a diagnostic methodology based on next-generation sequencing for the detection of mutations such as substitutions, insertions, copy number alterations (CNA) in 324 selected genes, and gene rearrangements, as well as genomic fingerprints including microsatellite instability, tumor mutational burden, and loss of heterozygosity. They used the results of the sequencing process to perform univariate, bivariate, and multivariate analyses to identify mutated genes that were related to the gliomagenesis process. We identified significant differences in the group of 324 sequenced genes. The genes (PIK3C2B, ERBB3, KIT, and MLH1) were found to have significant differences. However, the genes TP53 and IDH1/2 were chosen for this research due to their importance in the gliomagenesis process, biological plausibility, and the criteria of the investigators, despite not having significant differences between the group of 324 sequenced genes.
2.4. Prediction of Pathogenic SNPs
To predict the changes in the structure and function of the protein produced by the SNPs, we used the SIFT (Sorting Intolerant From Tolerant) software (https://sift.bii.a-star.edu.sg/, accessed on 21 June 2024) [14] and PolyPhen-2 (prediction of functional effects of human nsSNPs (http://genetics.bwh.harvard.edu/pph2/, accessed on 21 June 2024) [15]. PolyPhen-2 stratifies the SNPs into likely damaging, probably damaging, or benign, and generates a score between 0 and 1 where ≤0.85 means a greater probability of damage. As for SIFT, it generates two categories (tolerated/neutral) where values ≤ 0.05 are classified as damaging.
2.5. Prediction of Deleterious SNPs for the Protein
We used the servers for the PhD-SNP (Predictor of human Deleterious Single Nucleotide Polymorphisms) (https://snps.biofold.org/phd-snp/phd-snp.html, accessed on 21 June 2024) and SNPs and GO (predicting disease-associated variations using GO terms) (https://snps.biofold.org/snps-and-go/snps-and-go.html, accessed on 21 June 2024). Both predictors determine whether SNPs can cause disease. For example, PhD-SNP classifies nsSNPs of a gene into neutral or human disease-causing mutations [16], while SNPs and GO predict disease-associated variations using GO terms [17]; the harmful nonsynonymous SNPs predicted were analyzed with these two tools to determine their association with high-grade gliomas, and these were classified as diseases or neutral according to their potential to cause disease. SNPs labeled as a disease were retained for further analysis. The reliability index (RI) in PhD-SNP, SNPs, and GO ranges from 0 to 10, where 10 means the highest reliability.
2.6. Predicting Protein Stability for Functionally Deleterious Glioma-Associated SNPs
The SNPs involved in the gliomagenesis process were further analyzed to determine their effects on the protein stability of the bioinformatics programs I-Mutant 3.0 (https://folding.biofold.org/i-mutant/i-mutant2.0.html, accessed on 21 June 2024) and MUpro (https://mupro.proteomics.ics.uci.edu/, accessed on 21 June 2024); the first one is an algorithm based on an SVM (Support Vector Machine), which automatically predicts the change in the stability of the proteins after a single nucleotide mutation, while the I-Mutant 3.0 algorithm was trained with a data set derived from Protherm [18] that provides an estimate of the change in the Gibbs free energy (Delta Delta G or DDG), calculated by subtracting the DDG value of the non-mutated protein from the DDG value of the mutated protein (unit kcal/moles). Having a DDG > 0 means higher protein stability, while a DDG < 0 refers to lower protein stability. The MUpro server not only calculates using an SVM but also uses a neural network. It gives a prediction value between −1 and 1, where a value < 0 indicates a decrease in stability [19].
2.7. Prediction of Protein Structural Alteration and Loss of Activity
To predict structural changes in different proteins, including alteration in activity and binding, the MutPred2 web application (http://mutpred.mutdb.org/, accessed on 21 June 2024) was used, which is a method and software package based on machine learning that integrates genetic and molecular data to probabilistically reason about the pathogenicity of amino acid substitutions. It is trained on a set of 53,180 pathogenic and 206,946 unlabeled variants obtained from the Human Gene Mutation Database (HGMD), SwissVar, dbSNP, and pairwise alignment between species that can predict more than 50 protein features [20]. The application provides different probability percentages for each of the characteristics; this probability varies between 0 and 1, where the score closest to 1 indicates that it is more likely that the mutation alters the property of the protein, generating a gain or loss of the function [20]. We applied this tool to explore aspects of the activity of the TP53 and IDH proteins and how these are affected by SNPs.
2.8. Observation of Amino Acid Change
The Project-HOPE database (https://www3.cmbi.umcn.nl/hope/, accessed on 21 June 2024) provides information on amino acid changes in protein structures along with changes in different physiological and chemical activities that occur after mutation [21]. From this interface, it is possible to report whether the residue is in a conserved site, interpret mutations that are most likely to be harmful, and therefore significantly affect the function of the protein. We analyzed the SNPs of the TP53 and IDH1/2 genes in this database to obtain information on the produced changes.
2.9. Structural Comparison between Normal and Mutated Residues
The TM-Align application (https://zhanggroup.org/TM-align/, accessed on 21 June 2024) is an algorithm for sequence-independent protein structure comparisons. It gives a score between 0 and 1, where 1 indicates a perfect match between the mutated and wild-type protein structures, while a score ≥ 0.5 indicates that the same fold/topology exists between the structures located in the SCOP/CATH databases. which divides the protein into domains and classifies them at the hierarchical level. For example, SCOP classifies domains into classes, folds, superfamilies, and families. For its part, the four levels in CATH are class, architecture, topology, and homologous superfamily, thus generating a comparison between the structures [22]. Scores of 0.2 correspond to unrelated proteins chosen at random [23]. This tool was used to perform an analysis to establish a structural difference and thus conclude that SNPs affect the protein. Root means square deviation or RMSD scores were also provided, which measures the average distance between atoms of overlapping molecules, where higher scores indicate more structural differences between the two compared molecules and scores closer to 0 indicate greater geometric similarity between the two residues. The SWISS-MODEL program (https://swissmodel.expasy.org/, accessed on 21 June 2024) models the three-dimensional structure of proteins and provides the Ramachandran plot score between two structures [24]. The two values, both the TM and the Ramachandran graph score, helped determine the structural differences caused by the alterations.
2.10. Group of Mutations
SNPs for TP53 that were classified as deleterious by the tools described above were analyzed using the Mutation3D software (http://mutation3d.org/, accessed on 21 June 2024), which predicts and visualizes the spatial arrangement of mutated amino acids in the protein structure, showing whether they are found in an important domain. It can also describe clusters of mutations to discover some more significant SNPs [25].
2.11. Protein–Protein Interaction
The interaction of TP53 and IDH1/2 proteins with other proteins was studied using the STRING web server (https://string-db.org, accessed on 21 June 2024), which predicts the main proteins that show interactions with the query gene. STRING predicts such interactions based on gene fusion, co-expression, function, and experimental data. It shows combined scores for each interacting protein, ranging from 0 to 1, where 0 shows the lowest interaction and 1 indicates the highest interaction [26]; in addition, the KEEG database (https://www.genome.jp/kegg/, accessed on 21 June 2024) was used to identify the metabolic pathways associated with the TP53 and IDH1/2 genes.
3. Results
3.1. Distribution of SNPs
From 31 patients, we selected the genes with the highest mutation frequency from the Foundation one ® panel; therefore, 13 SNPs were cataloged as non-synonymous for the TP53 gene and 1 for the IDH1 gene. Polymorphism for IDH2 was selected for its biological importance in the development of gliomas (Table 1).

Table 1.
List of 15 non-synonymous single nucleotide polymorphisms for the TP53 and IDH1/2 genes. * Mutation not reported in databases such as ClinVar.
3.2. Unique Non-Synonymous Polymorphisms Predicted Using SIFT and PolyPhen-2
The PolyPhen-2 and SIFT software (https://sift.bii.a-star.edu.sg/, accessed on 21 June 2024) were run and thresholds were determined using the MSC (mutation significance cutoff) program available at (https://www.hgid.org/computational-tools/, accessed on 21 June 2024). The MSC for TP53 was (PolyPhen2 = 0.033 and SIFT = 0.133), IDH1 (PolyPhen2 = 0.823 and SIFT = 0.995), and IDH2 (PolyPhen2 = 1.000 and SIFT = 0.995). This score was used for later analysis. We determined, with the PolyPhen-2 tool, that 13 non-synonymous SNPs for the TP53 gene were damaging, while for the SIFT software, only 11 were classified as harmful. The non-synonymous SNPs (A161S, C124T) showed scores greater than 0.00 and are reported as tolerable mutations (Table 1). The same happened with the IDH1 SNP, which is tolerated in SIFT and damaging for PolyPhen-2. For the IDH2 polymorphism, this was harmful for both predictors.
3.3. Disease-Related Single Nucleotide Polymorphisms PhD-SNP SNPs and GO
The two programs identified the 13 SNPs for TP53 as damaging for the protein, as well as the SNPs that had conflicting interpretations (C124T and A161S) (Table 1). Therefore, they were considered harmful and were further analyzed bioinformatically. In the case of IDH1/2, both SNPs were predicted to cause protein alteration; therefore, no SNP was excluded from the analysis.
3.4. Prediction of Protein Stability Alteration Using I-Mutant 3.0 and MUpro Software
Of the 13 single nucleotide polymorphisms for TP53, 12 had a DDG value < 0, showing less protein stability because of the mutation. In the case of the non-synonymous SNPs of the IDH1/2 genes, in both cases, the software predicted that the mutations decreased the protein stability. Reliability index scores are provided along with the DDG value to demonstrate the decrease in structural stability of the proteins. Changes classified as harmful not only cause damage to the phenotype but also reduce the overall stability of the protein (Table 2).

Table 2.
Analysis performed with I-Mutant 3.0 and MUpro that indicate the DDG and predict whether it increases or decreases the stability of the protein; the reliability index ranges from 1 to 10 and ensures that the value or prediction made is within the estimated interval, in this case, the G279E polymorphism has a conflict of interpretation between the two programs.
3.5. Mutations Group
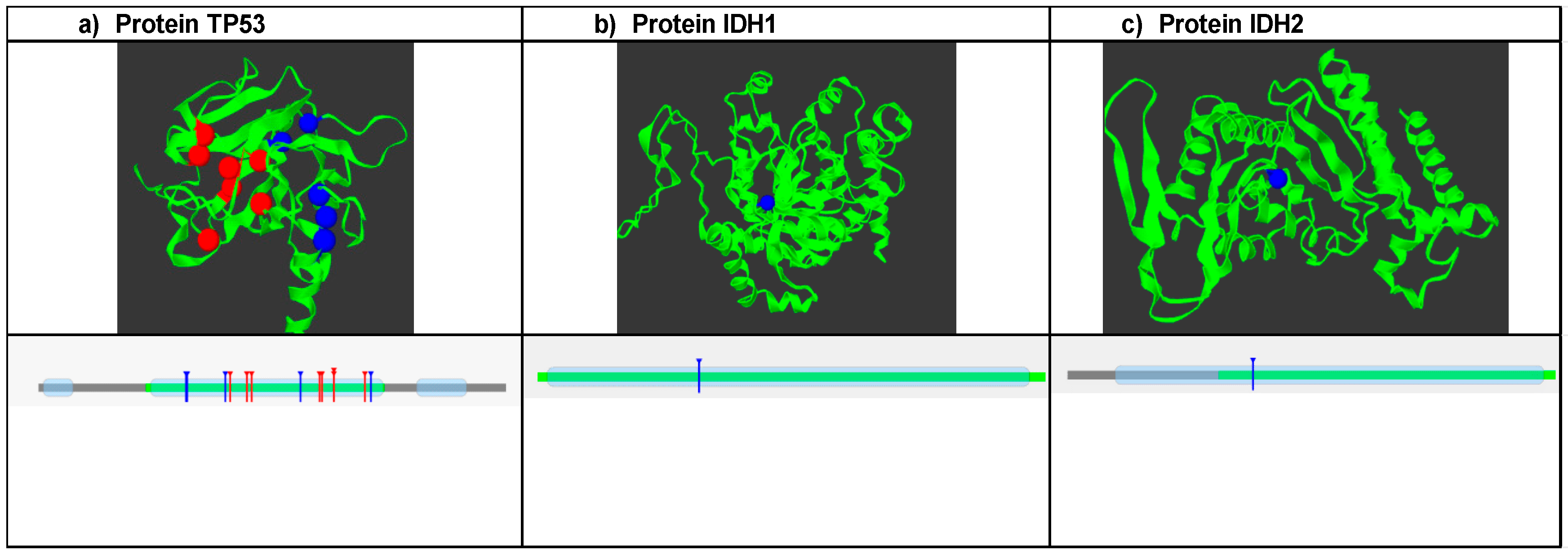
SNPs that were predicted to be damaging, disease-associated, and causing decreased protein stability were further analyzed with mutation3D software (https://www.mutation3d.org/, accessed on 21 June 2024) to predict clusters of mutations. A total of 13 SNPs were analyzed for TP53. The analysis showed that the clustered SNP positions are marked in red and occupy the DNA binding domain of the protein (Figure 1a); conversely, positions 124, 125, and 279 are not included in these frequent mutation sites. Therefore, they are marked in blue. Additionally, it is shown how the groups of mutations are in the DNA binding domain. In the case of IDH1, the R132H mutation is not found in the sites of high mutation frequency, but it is in one of the protein domains (Figure 1b). For the IDH2 protein, the R172K polymorphism was in sites of low mutational frequency, which is why it is marked in blue. Additionally, it is also located in one of the protein domains.
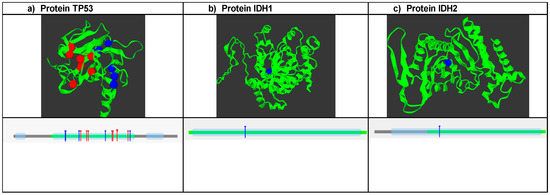
Figure 1.
Analysis of mutation clusters using Mutation 3D software (https://www.mutation3d.org/, accessed on 21 June 2024). (a) Shows the structure of the protein and the location of frequent mutation sites marked in red, and the blue dots are mutations that are outside of these; the lower graph shows the position of the mutation clusters in the domains of the TP53 protein. (b) Structure of the IDH1 protein: where the R132H mutation is represented, it is located outside of the frequent mutation sites, and the lower graph shows the position of the polymorphism in the protein domain. (c) IDH2 protein and the position of the R172K mutation are shown, which are located within a domain of this.
3.6. Impact of Single Nucleotide Polymorphisms on Protein Structural Activity
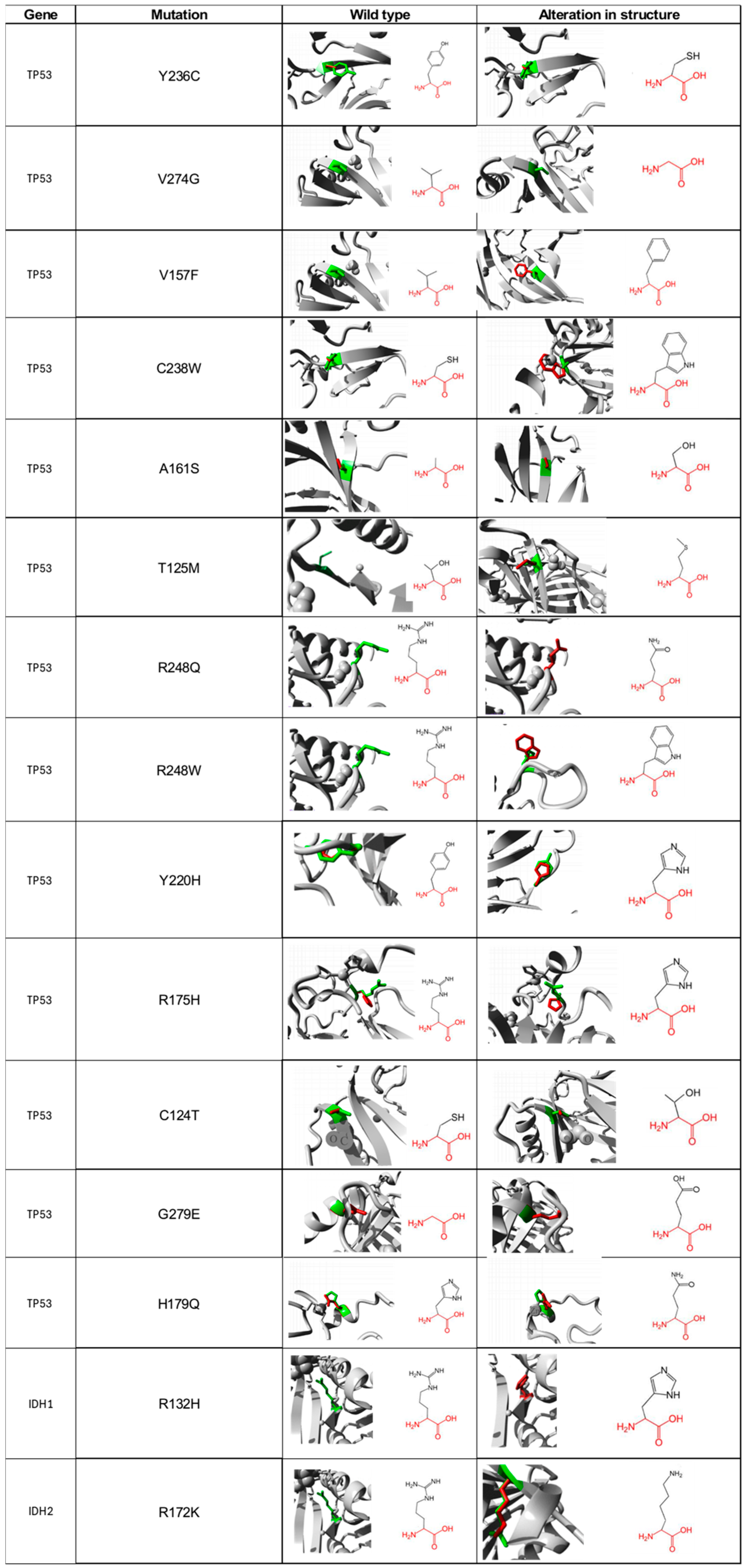
For the 13 non-synonymous SNP of TP53 and the 2 of IDH1/2 that were suggested as deleterious, we carried out a functional characterization of each of the amino acids, in both the wild type and mutated form using the Project HOPE database. For the Y236C mutation of the TP53 gene, the wild-type amino acid differs in size with respect to the mutant, which will cause an empty space in the body of the protein. The mutated residue is more hydrophobic; therefore, the change will produce the loss of hydrogen bonds in the body of the protein, resulting in altered folding. As for the V274G mutation, it will cause a loss of hydrophobic interactions in the body of the protein; in addition, this mutation will generate an empty space because the mutated residue is smaller (Figure 2). The main difference in the V157F change is the size between the mutated residue and the wild type since there is a probability that the mutated amino acid does not fit at a spatial level. For the C238W change, the normal residue is 100% conserved for this position, so a change in this position would result in damage to the protein. For the A161S mutation, the residues differ in size; additionally, there is a loss of hydrophobic interactions in the body of the protein. The T125M mutation causes the loss of hydrophobicity; therefore, there are fewer hydrogen bonds in the body of the protein, which could alter protein folding. The R248Q and R248W changes are characterized by having a difference in charge between the wild type and the mutated amino acid, so interactions with other molecules could be lost. The wild-type amino acid is not conserved in the case of the Y220H mutation; however, the mutation will cause an empty space in the body of the protein, and a difference in hydrophobicity is also present. The R175H alteration is identified by generating changes in charge between residues [27]. Both C124T and H179Q changes differ in size between amino acids; therefore, an empty space will be generated that may result in the loss of external interactions with other molecules. Regarding the G279E change, the wild-type amino acid has a neutral charge, unlike the mutated, which has a negative charge, which can cause problems in the proper folding of the protein.
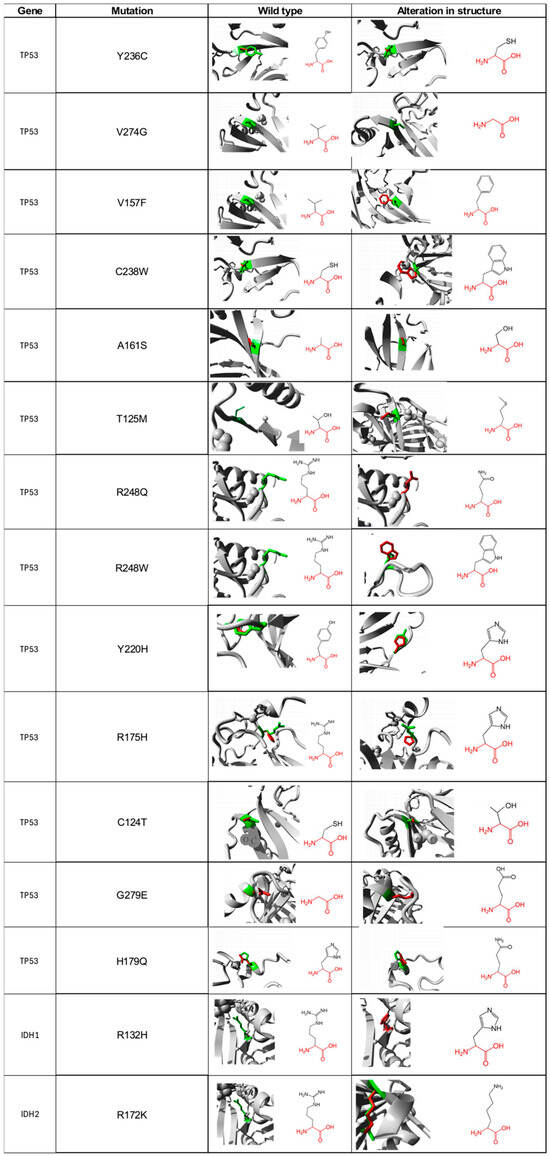
Figure 2.
Visual representation of amino acid structural changes caused by SNPs in the TP53 and IDH1/2 genes. Data taken from the Project-HOPE web server, a total of 13 deleterious SNPs were identified for TP53, 1 for IDH1, and 1 for IDH2.
For its part, the R132H mutation found in the IDH1 protein predicts that the mutated residue is smaller, and its charge is neutral, unlike the wild type, which has a positive charge, which will prevent its interaction with other molecules that, in this specific case, could not interact with isocitrate (Figure 2). For the R172K mutation located in the IDH2 protein, the wild-type amino acid is 100% conserved, so changes in it could be harmful to the protein. Additionally, the mutated residue is smaller, which produces an empty space in the body of the protein.
3.7. Alteration and Loss of Activity
We further analyzed SNPs using the MutPred2 web server to calculate alterations in protein activity (Table 3). According to the analysis, we found that Y220H, R175H, and H179Q polymorphisms of the TP53 protein mainly affected the metal binding property, while changes in the Y236C, V274G, and V157F altered the stability of the protein (Table 3). For mutations A161S, T125M, R248Q, and R248W no results were obtained. On the other hand, for the IDH1 protein, we found that this R132H mutation, which presented changes in metal binding, also impacted the allosteric site and loss of relative accessibility to the solvent (Table 3). Regarding the R172K mutation for the IDH2 protein, it presents changes in activity such as loss of the catalytic site and metal and DNA binding sites (Table 3).

Table 3.
Protein activity data, structural alteration, and loss of binding from MutPred2, altered properties for TP53 SNPs, altered properties for non-synonymous SNP of the IDH1 protein, and altered properties for the IDH2 protein polymorphism, where probabilities close to 1 have a greater possibility that the mutation is affecting the protein property.
3.8. Structural Differences between Normal and Mutated Residues Using TM-Aling and Ramachandran Plots
We ran the TM-aling and SWISS-MODEL programs to further analyze the changes between the structures of the wild-type proteins and the homology-modeled mutated proteins. The lower the TM score and the higher the RMSD value, the greater the difference between the structures. On the contrary, it may also be the case that the TM scores may be high as well as the RMSD value. Furthermore, the higher the score of the Ramachandran graph, the more the structure will be favored (Table 4). It was shown that the C238W polymorphism for TP53 affects the structure of the protein, while for the IDH1 protein, it was found that R132H was the most deleterious SNP, having a low TM score and higher RMSD, with a significant score in favor of the Ramachandran plot. This scenario is different from the non-synonymous SNP of IDH2; it has a high TM score and a very low RMSD score.

Table 4.
TM-matching score, RMSD value, and Ramachandran plot of identified SNP in TP53, IDH1, and IDH2. a a score of 1 indicates a perfect match between mutated and wild-type protein structures; b a higher RMSD score means more structural differences between the two compared proteins; c the Ramachandran plot score is calculated in percentages and provides an idea of how well two structures seem to compare to each other.
3.9. Protein–Protein Interaction
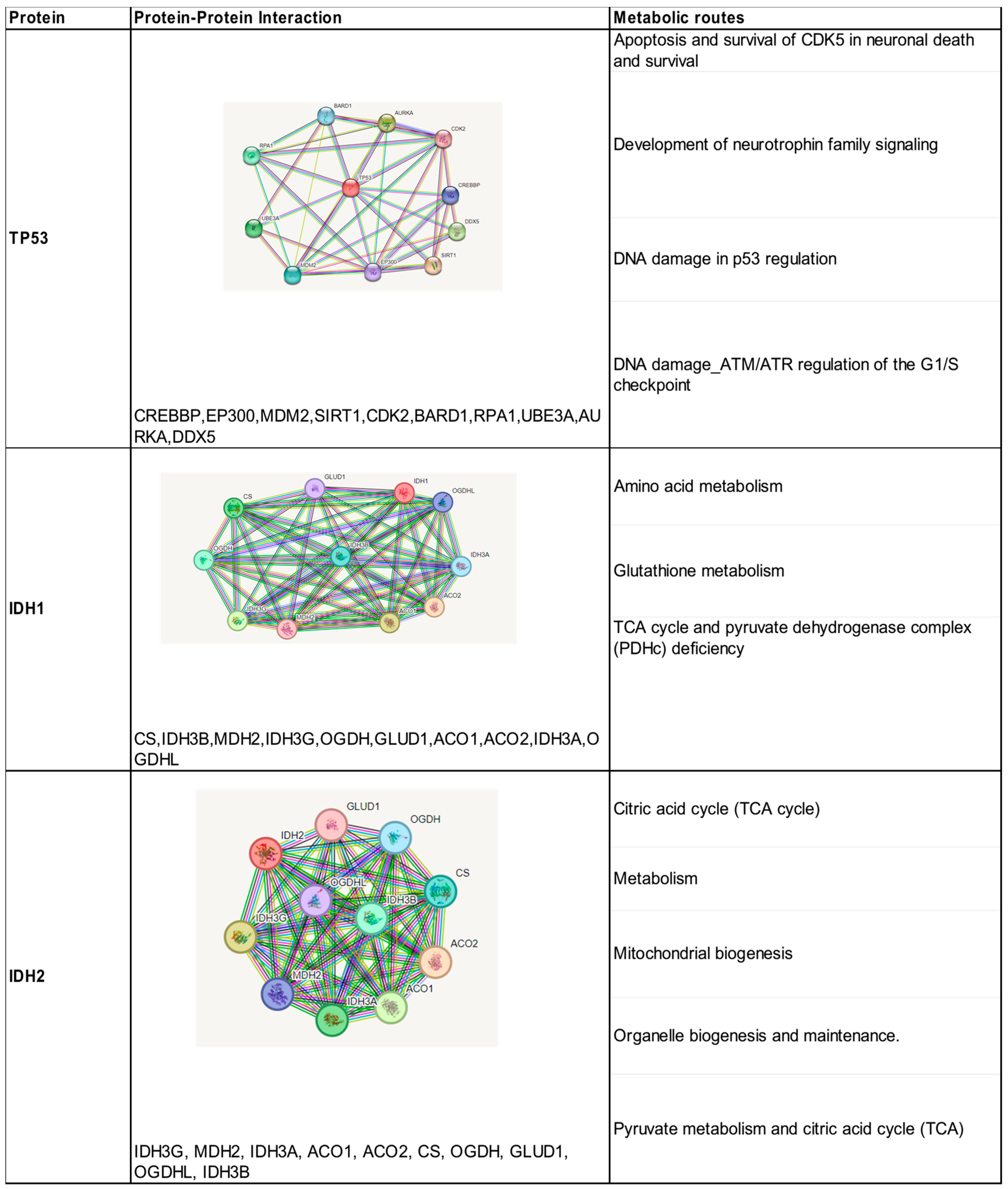
The search in the STRING database resulted in the direct interaction of TP53 with 10 proteins, with confidence scores (>0.95). These results were obtained according to our experiments, data sets, and text mining (Figure 3). All these proteins predicted strong functional associations with TP53 and were shown to be involved in the activation of different pathways such as cell cycle regulation, DNA damage response, metabolism, apoptosis, and autophagy. The analysis of the interaction network shows that CREBBP, MDM2, and EP300 proteins contribute to the co-activation of TP53 as a transcription factor. The protein–protein interaction analyzed for the IDH1 and IDH2 isoenzymes showed the presence of 10 proteins with confidence scores between 0.99 and 0.97, all of them were involved in the oxidative decarboxylation process of isocitrate (Figure 3). They are involved in the metabolic pathway of tricarboxylic acids generated in the mitochondria.
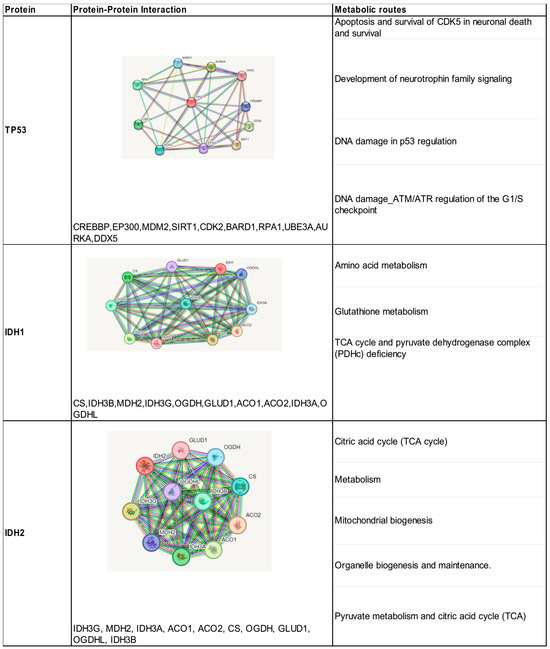
Figure 3.
Identification of protein–protein interactions and metabolic pathways of the proteins encoded by the three mutated genes associated with high-grade gliomas.
4. Discussion
In the development of gliomas, a significant number of biological processes are compromised in which genes such as TP53 and IDH1/2 are involved. The comprehensive characterization of the non-synonymous SNPs that may be present in the coding regions allows us to elucidate the relationship between the genetic variants and their functional and structural impact on a protein, which can lead to discovering possible markers for the diagnosis and prognosis of this type of neoplasms.
The bioinformatics analysis found that the 13 polymorphisms reported for the TP53 gene were detrimental since they modify the structure and function of the protein. Each of the variants participates in the p53-ARF-MDM2 metabolic pathway, which is mutated in 84% of glioblastomas. This pathway is related to processes of cell invasion, migration, and proliferation, as well as to evasion of apoptosis [6]. All SNPs were in the DNA binding site, which is considered a site of high mutation frequency, given that molecular profiling of various tumors has shown that most of the 190 amino acids found in this protein domain are mutated in one or more cancers, and, in many cases, are homozygous in the tumor. Therefore, they are believed to contribute to the development of neoplasia [28]. These mutations are found at different frequencies and in various oncological diseases; for example, in the case of the SNP rs730882026 (Y236C), different databases such as ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, accessed on 21 June 2024) and Varsome (https://varsome.com/, accessed on 21 June 2024) report this variant with conflicting interpretation, unlike the results obtained where it is shown that it is detrimental. In the literature, it has been associated with lung cancer, has not been associated with high-grade gliomas [29,30], and has been related to resistance to the chemotherapeutic agent Crizotinib® [31]. For its part, the variant rs1057520006 (V274G) generates a loss of protein function; despite this, it has a conflicting interpretation in the databases. This mutation was classified as detrimental in this study.
Somatic mutations in TP53 are typically single-nucleotide missense changes that allow the production of a full-length protein. Some of these mutated proteins not only lose tumor suppressor function but can also acquire oncogenic properties, including increased invasion, proliferation, and chemo-resistance. This is the case for the rs121912654 (V157F) variant associated with lung cancer and hepatocellular carcinoma, where the mutation predicted a worse outcome for the patient [32,33]. According to ClinVar, its interpretation is not clear; however, the variant in this study was detrimental. For the SNP rs193920789 (C238W), its interpretation appears to be detrimental, as do our obtained results, where it is predicted that it is one of the variants that modify the structure and function of the protein the most. There are no reports in the literature indicating a detrimental association with this polymorphism or its clinical significance.
In our study, we report on a polymorphism (A161S) not yet classified in the databases. This represents an SNP with conflicting interpretation since there are predictors, such as SIFT that classify it as tolerable. Therefore, experimental validation is required, e.g., an in-vitro functional assay, to determine whether this SNP impacts protein function. For its part, the rs786201057 (T125M) variant has shown that the mutated protein has deficiencies in transactivation activity [34] and has also been related to diseases such as Li-Fraumeni syndrome [35]. This variant can be classified as detrimental because predictors and other programs associate it with a loss of protein structure and function. The polymorphisms (R248Q and R248W) are considered sites of high mutation frequency; several studies have shown that these changes promote cell migration and proliferation, through AKT signaling [36]. The R248 is mutated in three amino acids, R248Q, R248W, and R248L, and it has been shown that the first change induces more aggressive tumors in mice compared with other mutation hotspots [37]. The second change acquires neomorphic tumorigenic functions, particularly in invasion and the metastasis of solid tumors [38]. The R248Q mutation has mainly been associated with ovarian cancer and myeloid leukemia, while R248W is associated with pancreatic and lung cancer [39]. The SNP rs530941076, classified as detrimental, is related to different types of cancer such as breast, lung, and liver [40]. This change produces a slight alteration of intramolecular interactions which causes a loss of DNA binding [41].
One of the TP53 gene variants reported with high frequency in oncological diseases is R175H. It is at a critical point for the structure, characterized by lower thermodynamic stability [42]. This SNP is classified as detrimental according to our results. This mutation has been associated with colorectal, breast, lung, pancreas, and myeloid leukemia [43]. In the specific case of gliomas, this has been related to a worse prognosis for the patient and rapid cancer progression. In high-grade glioma, recurrent TP53 point mutations may be the key to tumor progression, thus emphasizing its importance in gliomagenesis [44]. The C124T polymorphism is located deep in the DNA binding domain; however, it does not play a significant role in modulating the DNA binding affinity of TP53 [45]. In this study, this SNP provided conflicting interpretations because the predictors were unable to conclude whether it was detrimental, and the rest of the web servers did not conclude it modified the structure and function of the protein.
In the different databases, the G279E variant is reported with conflicting interpretations. In this study, this SNP was detrimental; however, the stability of the protein was increased, which could be related to the gliomagenesis process. Finally, the H179Q SNP is classified as detrimental; cells that express this mutated protein are immortal and show attenuation of the G1 checkpoint function, loss of expression of the p21 protein [46], and it is related to osteosarcomas and melanomas [47].
The TP53 gene has a key role in several cellular processes. It often undergoes mutations, which facilitate the appearance of oncological disease. Its role in gliomas is related to the onset of neoplasia, for example, in 30% to 40% of astrocytoma cases, a loss of TP53 function can be observed at an early stage [48]. Additionally, mutations in this gene are related to low-grade gliomas, unlike this study where all patients were adults and were diagnosed with high-grade gliomas, which confirms that these mutations may be an early-onset event.
The R132H mutation in IDH1 is considered an early event in gliomagenesis. At a clinical level, glioma patients carrying mutant IDH1 respond better to antitumor therapies. However, the mechanism by which mutations in this gene contribute to gliomagenesis and therapeutic response has remained difficult to elucidate [49]. The presence of R132H in glial cells significantly enhanced cell senescence in response to temozolomide and radiation through a DNA damage-mediated mechanism. The R132H change occurs heterozygously in the tumor since the homozygous form produces an inactive protein. The biological impact of the mutated protein in oncogenesis is related to the state of homeostasis in the Krebs cycle. Glutamine and/or Glutamate serve as key substrates to offset the metabolic impact by promoting lipid and glutathione synthetic pathways. Gliomas with IDH1 mutation show a different metabolic pattern compared to other solid tumors [13]. Regarding treatment, many studies have shown that gliomas with IDH mutations respond better to standard therapeutic methods such as temozolomide and radiotherapy [50]. Bioinformatic characterization studies of the R132H SNP have revealed that it is generated at a critical point in the protein [51]. This mutation resulted in conflicting interpretations for the pathogenicity predictors, but at the same time, this mutation was the one that most modified the structure of the protein (see Table 4). It is found in more than 95% of secondary glioblastomas and in less than 10% of primary glioblastomas, being classified as a definitive marker for this type of neoplasm [52].
The R172K mutation in the IDH2 enzyme is related to different malignancies, including gliomas, chondrosarcomas, and myeloid leukemia, among others. This mutation confers neoactivity to the enzyme and confers epigenetic alterations to the DNA [53]. The implications of this mutation in the structure of the enzyme are related to its conformational state since its natural state is to be in an open conformation, but when it binds to isocitrate, it adopts a closed conformation and properly assembles the active site for the normal reaction to take place [54]. Therefore, R172K is expected to affect enzyme activity; the IDH2 variants are less common, with R172K observed in 3% of glioma patients, and detection of the R172K variant has implications for glioma diagnosis, prognosis, and potential treatment [55]. In this study, the variant was pathogenic, which agrees with the databases and affects the function and structure of the protein.
Regarding mutations that lead to the loss of binding of P53 and IDH1/2 proteins to metal ions, the following have been described: the P53 protein depends on certain endogenous metals for its function, especially on Zinc (Zn) ions [56]. Structural studies have demonstrated the presence of Zn ions in its active site [57] and the importance of these metal ions in the following functions has been verified: binding of the DNA-binding domain (DBD) to DNA occurs only in the presence of Zn; the absence of Zn decreases the activity of the protein; the DNA binding domain is stabilized by Zn; the interaction between P53–DNA is altered by mutations in the DNA binding domain; mutations in the DBD domain lead to misfolding of the protein and poor binding to Zn ions; mutations in the DBD domain lead to the poor transcriptional activity of the P53 protein; and in the absence of Zn, binding to DNA is considered less stable. Additionally, it has been discovered that Zn can help reactivate mutated proteins and that Zn sequestration leads to the inactivation of the P53 protein [58,59,60,61,62,63,64,65,66,67,68,69,70]. This highlights the importance of P53 mutations (Y220H = 27%, R175H = 67%, and H179Q = 72%), which alter, to a certain extent, the binding of metals to the P53 protein and could lead to all these alterations in the function of the protein.
On the other hand, it has been observed that divalent metal ions such as magnesium and manganese (Mg2 and Mn2), when binding to the substrate (isocitrate) of the IDH1 protein, enhance the catalytic activity of the protein [71]. The R132H mutation represents about 80% of the mutations found in this gene and reduces the activity of the wild-type IDH1. But it confers a gain of function in the mutated proteins to produce oncometabolites such as (2-hydroxyglutarate) [x16]. The IDH1 mutation (R132H = 55%), by reducing the metal binding of the protein, could affect the catalytic activity of the wild-type IDH1 by converting isocitrate to α-ketoglutarate in the cytoplasm.
The association between IDH2 gene mutations and metal ions has been little explored. However, due to the similarity in amino acid sequences, shared domains, and three-dimensional structure with respect to IDH1, it could be suggested that their metabolic activity would be affected in a similar way. The IDH2 mutation (R172K = 40%), by reducing the metal binding of the protein, could affect the catalytic activity of the wild-type IDH2 by converting isocitrate to α-ketoglutarate in the mitochondria.
All the single nucleotide polymorphisms for the TP53 and IDH1/2 proteins discussed in this manuscript are associated with promoting the development of glioma tumorigenesis since it has been shown that the proteins have some effect both in their level of activity and in their structure. The combination of these mutations has not been associated with patient survival results [72], so more correlation studies are needed between the described mutations and their outcome. The combination of new bioinformatic methods with next-generation sequencing is part of daily clinical care and an essential tool for the diagnosis of low- and high-grade gliomas, providing more precise information, which can help reduce the need for additional surgeries, and providing important information for additional radiotherapy or chemotherapy treatments depending on the diagnosis. Despite these advances, it is important to note that there is still much to be done in this area, particularly in the development of innovative and improved bioinformatics models for so-called translational research.
5. Conclusions
Computational analysis and identification of deleterious polymorphisms of TP53 and IDH1/2 in patients with high-grade gliomas are essential to distinguish differences in the properties of mutated and wild-type proteins. The collection of this data is applicable to the diagnosis and prognosis of this type of patient and may additionally indicate treatments based on the genetics of the tumor. More clinical research is needed where the association of these polymorphisms with the severity of the neoplasia can be studied.
Author Contributions
Conceptualization, S.V.G. and R.G.P.S.; methodology, L.D.O.G. and J.M.M.G.; software, S.V.G.; validation, J.E.S.F.; formal analysis, L.D.O.G.; investigation, F.P.M.; S.V.G.; data curation, S.V.G.; writing—original draft preparation, S.V.G. and J.M.M.G.; writing—review and editing, F.P.M.; supervision, R.G.P.S.; project administration, R.G.P.S.; funding acquisition, L.D.O.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by CES University and the Americas-AUNA clinic.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee for human research of the CES University (Section 122 of 8 May 2018).
Informed Consent Statement
The authors obtained informed consent from patients presented in this manuscript. The documentation is in the possession of the corresponding author.
Data Availability Statement
Data are available in the NCBI database under the following accession numbers: TP53-Y236C (2874534), TP53-V274G (2874538), TP53-V157F (2874541), TP53-C238W (2874549), TP53-A161S (2874551), TP53-T125M (2874552), TP53-R248Q (2874554), TP53-R248W (2874556), TP53-Y220H (2874558), TP53-R175H (2874572), TP53-C124T (2874573), TP53-G279E (2874575), TP53-H179Q (2874578), IDH1-R132H (2874579), and IDH2-R172K (2874580).
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
References
- Komori, T. The 2021 WHO classification of tumors, 5th edition, central nervous system tumors: The 10 basic principles. Brain Tumor Pathol. 2022, 39, 47–50. [Google Scholar] [CrossRef]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Primers 2015, 1, 15017. [Google Scholar] [CrossRef] [PubMed]
- Dekker, L.J.M.; Verheul, C.; Wensveen, N.; Leenders, W.; Lamfers, M.L.M.; Leenstra, S.; Luider, T.M. Effects of the IDH1 R132H Mutation on the Energy Metabolism: A Comparison between Tissue and Corresponding Primary Glioma Cell Cultures. ACS Omega 2022, 7, 3568–3578. [Google Scholar] [CrossRef] [PubMed]
- Palacios Paredes, L.F.; Silva, C.; García Matamoros, E.K. Gliomas de Alto Grado del Adulto: Biología Molecular (Parte I): Revisión Narrativa. Oncol. Ecuad. 2020, 30, 249–279. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Junaid, M.; Hamid, N.; Duan, J.-J.; Yang, X.; Pei, D.-S. Current understanding of gliomagenesis: From model to mechanism. Int. J. Med. Sci. 2022, 19, 2071–2079. [Google Scholar] [CrossRef]
- Zhang, Y.; Dube, C.; Gibert, M.; Cruickshanks, N.; Wang, B.; Coughlan, M.; Yang, Y.; Setiady, I.; Deveau, C.; Saoud, K.; et al. The p53 Pathway in Glioblastoma. Cancers 2018, 10, 297. [Google Scholar] [CrossRef]
- Krex, D.; Mohr, B.; Appelt, H.; Schackert, H.K.; Schackert, G. Genetic Analysis of a Multifocal Glioblastoma Multiforme: A Suitable Tool to Gain New Aspects in Glioma Development. Neurosurgery 2003, 53, 1377–1384. [Google Scholar] [CrossRef]
- Djuzenova, C.S.; Fiedler, V.; Memmel, S.; Katzer, A.; Hartmann, S.; Krohne, G.; Zimmermann, H.; Scholz, C.-J.; Polat, B.; Flentje, M.; et al. Actin cytoskeleton organization, cell surface modification and invasion rate of 5 glioblastoma cell lines differing in PTEN and p53 status. Exp. Cell Res. 2015, 330, 346–357. [Google Scholar] [CrossRef]
- Park, C.-M.; Park, M.-J.; Kwak, H.-J.; Moon, S.-I.; Yoo, D.-H.; Lee, H.-C.; Park, I.-C.; Rhee, C.H.; Hong, S.-I. Induction of p53-mediated apoptosis and recovery of chemosensitivity through p53 transduction in human glioblastoma cells by cisplatin. Int. J. Oncol. 2006, 28, 119–125. Available online: http://www.spandidos-publications.com/10.3892/ijo.28.1.119 (accessed on 20 September 2024). [CrossRef]
- Zheng, H.; Ying, H.; Yan, H.; Kimmelman, A.C.; Hiller, D.J.; Chen, A.-J.; Perry, S.R.; Tonon, G.; Chu, G.C.; Ding, Z.; et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 2008, 455, 1129–1133. [Google Scholar] [CrossRef]
- Chen, X.; Liu, J.; Li, Y.; Zeng, Y.; Wang, F.; Cheng, Z.; Duan, H.; Pan, G.; Yang, S.; Chen, Y.; et al. IDH1 mutation impairs antiviral response and potentiates oncolytic virotherapy in glioma. Nat. Commun. 2023, 14, 6781. Available online: https://www.nature.com/articles/s41467-023-42545-3 (accessed on 20 September 2024). [CrossRef] [PubMed]
- Grimi, A.; Bono, B.C.; Lazzarin, S.M.; Marcheselli, S.; Pessina, F.; Riva, M. Gliomagenesis, Epileptogenesis, and Remodeling of Neural Circuits: Relevance for Novel Treatment Strategies in Low- and High-Grade Gliomas. Int. J. Mol. Sci. 2024, 25, 8953. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH mutation in glioma: Molecular mechanisms and potential therapeutic targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef]
- Sim, N.-L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Capriotti, E.; Calabrese, R.; Casadio, R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 2006, 22, 2729–2734. [Google Scholar] [CrossRef]
- Calabrese, R.; Capriotti, E.; Fariselli, P.; Martelli, P.L.; Casadio, R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mutat. 2009, 30, 1237–1244. [Google Scholar] [CrossRef]
- Bava, K.A. ProTherm, version 4.0: Thermodynamic database for proteins and mutants. Nucleic Acids Res. 2004, 32, D120–D121. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Randall, A.; Baldi, P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins Struct. Funct. Bioinform. 2006, 62, 1125–1132. [Google Scholar] [CrossRef]
- Pejaver, V.; Urresti, J.; Lugo-Martinez, J.; Pagel, K.A.; Lin, G.N.; Nam, H.-J.; Mort, M.; Cooper, D.N.; Sebat, J.; Iakoucheva, L.M.; et al. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat. Commun. 2020, 11, 5918. [Google Scholar] [CrossRef]
- Venselaar, H.; Beek, T.A.T.; Kuipers, R.K.; Hekkelman, M.L.; Vriend, G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinform. 2010, 11, 548. [Google Scholar] [CrossRef]
- Csaba, G.; Birzele, F.; Zimmer, R. Systematic comparison of SCOP and CATH: A new gold standard for protein structure analysis. BMC Struct. Biol. 2009, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Skolnick, J. TM-Align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Meyer, M.J.; Lapcevic, R.; Romero, A.E.; Yoon, M.; Das, J.; Beltrán, J.F.; Mort, M.; Stenson, P.D.; Cooper, D.N.; Paccanaro, A.; et al. mutation3D: Cancer Gene Prediction Through Atomic Clustering of Coding Variants in the Structural Proteome. Hum. Mutat. 2016, 37, 447–456. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Chiang, Y.-T.; Chien, Y.-C.; Lin, Y.-H.; Wu, H.-H.; Lee, D.-F.; Yu, Y.-L. The Function of the Mutant p53-R175H in Cancer. Cancers 2021, 13, 4088. [Google Scholar] [CrossRef]
- Baugh, E.H.; Ke, H.; Levine, A.J.; Bonneau, R.A.; Chan, C.S. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018, 25, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Cheng, H.; Yu, L.; Zhang, J.; Wang, Y.; Liang, Y.; Lou, F.; Wang, H.; Cao, S. Mutation patterns and evolutionary action score of TP53 enable identification of a patient population with poor prognosis in advanced non-small cell lung cancer. Cancer Med. 2023, 12, 6649–6658. [Google Scholar] [CrossRef]
- Yokouchi, H.; Nishihara, H.; Harada, T.; Yamazaki, S.; Kikuchi, H.; Oizumi, S.; Uramoto, H.; Tanaka, F.; Harada, M.; Akie, K.; et al. Detection of somatic TP53 mutation in surgically resected small-cell lung cancer by targeted exome sequencing: Association with longer relapse-free survival. Heliyon 2020, 6, e04439. [Google Scholar] [CrossRef]
- Song, P.; Zhang, F.; Li, Y.; Yang, G.; Li, W.; Ying, J.; Gao, S. Concomitant TP53 mutations with response to crizotinib treatment in patients with ALK-rearranged non-small-cell lung cancer. Cancer Med. 2019, 8, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Barta, J.A.; Pauley, K.; Kossenkov, A.V.; McMahon, S.B. The lung-enriched p53 mutants V157F and R158L/P regulate a gain of function transcriptome in lung cancer. Carcinogenesis 2020, 41, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A.; Hoshida, Y. Depicting the role of TP53 in hepatocellular carcinoma progression. J. Hepatol. 2011, 55, 724–725. [Google Scholar] [CrossRef]
- Mena, A.C.; Pulido, E.G.; Guillen-Ponce, C. Understanding the molecular-based mechanism of action of the tyrosine kinase inhibitor: Sunitinib. Anticancer. Drugs 2010, 21 (Suppl. S1), S3–S11. [Google Scholar] [CrossRef]
- Bougeard, G.; Renaux-Petel, M.; Flaman, J.-M.; Charbonnier, C.; Fermey, P.; Belotti, M.; Gauthier-Villars, M.; Stoppa-Lyonnet, D.; Consolino, E.; Brugières, L.; et al. Revisiting Li-Fraumeni Syndrome From TP53 Mutation Carriers. J. Clin. Oncol. 2015, 33, 2345–2352. [Google Scholar] [CrossRef]
- Lai, Z.-Y.; Tsai, K.-Y.; Chang, S.-J.; Chuang, Y.-J. Gain-of-Function Mutant TP53 R248Q Overexpressed in Epithelial Ovarian Carcinoma Alters AKT-Dependent Regulation of Intercellular Trafficking in Responses to EGFR/MDM2 Inhibitor. Int. J. Mol. Sci. 2021, 22, 8784. [Google Scholar] [CrossRef] [PubMed]
- Allende-Vega, N.; Villalba, M. Metabolic stress controls mutant p53 R248Q stability in acute myeloid leukemia cells. Sci. Rep. 2019, 9, 5637. [Google Scholar] [CrossRef]
- Klemke, L.; Fehlau, C.F.; Winkler, N.; Toboll, F.; Singh, S.K.; Moll, U.M.; Schulz-Heddergott, R. The Gain-of-Function p53 R248W Mutant Promotes Migration by STAT3 Deregulation in Human Pancreatic Cancer Cells. Front. Oncol. 2021, 11, 642603. [Google Scholar] [CrossRef]
- Yoshikawa, K.; Hamada, J.-I.; Tada, M.; Kameyama, T.; Nakagawa, K.; Suzuki, Y.; Ikawa, M.; Hassan, N.M.M.; Kitagawa, Y.; Moriuchi, T. Mutant p53 R248Q but not R248W enhances in vitro invasiveness of human lung cancer NCI-H1299 cells. Biomed. Res. 2010, 31, 401–411. [Google Scholar] [CrossRef]
- Bauer, M.R.; Krämer, A.; Settanni, G.; Jones, R.N.; Ni, X.; Tareque, R.K.; Fersht, A.R.; Spencer, J.; Joerger, A.C. Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: The Y220X Paradigm. ACS Chem. Biol. 2020, 15, 657–668. [Google Scholar] [CrossRef]
- Gomes, A.S.; Ramos, H.; Inga, A.; Sousa, E.; Saraiva, L. Structural and Drug Targeting Insights on Mutant p53. Cancers 2021, 13, 3344. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Gao, S.-J.; Soubise, B.; Douet-Guilbert, N.; Liu, Z.-L.; Troadec, M.-B. TP53 in Myelodysplastic Syndromes. Cancers 2021, 13, 5392. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, X.; Tang, Y.; Lao, Z.; Lei, J.; Wei, G. Common cancer mutations R175H and R273H drive the p53 DNA-binding domain towards aggregation-prone conformations. Phys. Chem. Chem. Phys. 2020, 22, 9225–9232. [Google Scholar] [CrossRef] [PubMed]
- Sarma, P.P.; Dutta, D.; Mirza, Z.; Saikia, K.K.; Baishya, B.K. Point mutations in the DNA binding domain of p53 contribute to glioma progression and poor prognosis. Mol. Biol. 2017, 51, 293–299. [Google Scholar] [CrossRef]
- Schaefer, K.N.; Geil, W.M.; Sweredoski, M.J.; Moradian, A.; Hess, S.; Barton, J.K. Oxidation of p53 through DNA Charge Transport Involves a Network of Disulfides within the DNA-Binding Domain. Biochemistry 2015, 54, 932–941. [Google Scholar] [CrossRef]
- Synoradzki, K.J.; Bartnik, E.; Czarnecka, A.M.; Fiedorowicz, M.; Firlej, W.; Brodziak, A.; Stasinska, A.; Rutkowski, P.; Grieb, P. TP53 in Biology and Treatment of Osteosarcoma. Cancers 2021, 13, 4284. [Google Scholar] [CrossRef]
- Carson, C.; Omolo, B.; Chu, H.; Zhou, Y.; Sambade, M.J.; Peters, E.C.; Tompkins, P.; Simpson, D.A.; Thomas, N.E.; Fan, C.; et al. A prognostic signature of defective p53-dependent G1 checkpoint function in melanoma cell lines. Pigment. Cell Melanoma Res. 2012, 25, 514–526. [Google Scholar] [CrossRef]
- Bendahou, M.A.; Arrouchi, H.; Lakhlili, W.; Allam, L.; Aanniz, T.; Cherradi, N.; Ibrahimi, A.; Boutarbouch, M. Computational Analysis of IDH1, IDH2, and TP53 Mutations in Low-Grade Gliomas Including Oligodendrogliomas and Astrocytomas. Cancer Inform. 2020, 19, 117693512091583. [Google Scholar] [CrossRef]
- Zhan, D.; Ma, D.; Wei, S.; Lal, B.; Fu, Y.; Eberhart, C.; Laterra, J.; Ying, M.; Li, Y.; Meeker, A.; et al. Monoallelic IDH1 R132H Mutation Mediates Glioma Cell Response to Anticancer Therapies via Induction of Senescence. Mol. Cancer Res. 2021, 19, 1878–1888. [Google Scholar] [CrossRef]
- Kayabolen, A.; Yilmaz, E.; Bagci-Onder, T. IDH Mutations in Glioma: Double-Edged Sword in Clinical Applications? Biomedicines 2021, 9, 799. [Google Scholar] [CrossRef]
- Ahmed, H.U.; Paul, A.; Mahmud, Z.; Rahman, T.; Hosen, I. Comprehensive characterization of the single nucleotide polymorphisms located in the isocitrate dehydrogenase isoform 1 and 2 genes using in silico approach. Gene Rep. 2021, 24, 101259. [Google Scholar] [CrossRef]
- Najafi, S.; Esmaeili, S.; Zhaleh, H.; Rahmati, Y. The role of IDH1 mutation on gene expression in glioblastoma. Inform. Med. Unlocked 2022, 28, 100812. [Google Scholar] [CrossRef]
- Lemonnier, F.; Cairns, R.A.; Inoue, S.; Li, W.Y.; Dupuy, A.; Broutin, S.; Martin, N.; Fataccioli, V.; Pelletier, R.; Wakeham, A.; et al. The IDH2 R172K mutation associated with angioimmunoblastic T-cell lymphoma produces 2HG in T cells and impacts lymphoid development. Proc. Natl. Acad. Sci. USA 2016, 113, 15084–15089. [Google Scholar] [CrossRef] [PubMed]
- Kotredes, K.P.; Razmpour, R.; Lutton, E.; Alfonso-Prieto, M.; Ramirez, S.H.; Gamero, A.M. Characterization of cancer-associated IDH2 mutations that differ in tumorigenicity, chemosensitivity and 2-hydroxyglutarate production. Oncotarget 2019, 10, 2675–2692. [Google Scholar] [CrossRef]
- Sporikova, Z.; Slavkovsky, R.; Tuckova, L.; Kalita, O.; Houdova, M.M.; Ehrmann, J.; Hajduch, M.; Hrabalek, L.; Vaverka, M. IDH1/2 Mutations in Patients With Diffuse Gliomas: A Single Centre Retrospective Massively Parallel Sequencing Analysis. Appl. Immunohistochem. Mol. Morphol. 2022, 30, 178–183. [Google Scholar] [CrossRef]
- Alfadul, S.M.; Matnurov, E.M.; Varakutin, A.E.; Babak, M.V. Metal-Based Anticancer Complexes and p53: How Much Do We Know? Cancers 2023, 15, 2834. [Google Scholar] [CrossRef]
- Cho, Y.; Gorina, S.; Jeffrey, P.D.; Pavletich, N.P. Crystal Structure of a p53 Tumor Suppressor-DNA Complex: Understanding Tumorigenic Mutations. Science 1994, 265, 346–355. [Google Scholar] [CrossRef]
- Joerger, A.C.; Fersht, A.R. Structural Biology of the Tumor Suppressor p53 and Cancer-Associated Mutants. In Advances in Cancer Research [Internet]; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–23. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0065230X06970018 (accessed on 17 September 2024).
- Butler, J.S.; Loh, S.N. Structure, Function, and Aggregation of the Zinc-Free Form of the p53 DNA Binding Domain. Biochemistry 2003, 42, 2396–2403. [Google Scholar] [CrossRef]
- Bullock, A.N.; Henckel, J.; Fersht, A.R. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: Definition of mutant states for rescue in cancer therapy. Oncogene 2000, 19, 1245–1256. [Google Scholar] [CrossRef]
- Loh, S.N. The missing Zinc: p53 misfolding and cancer. Metallomics 2010, 2, 442–449. [Google Scholar] [CrossRef]
- Pavletich, N.P.; Chambers, K.A.; Pabo, C.O. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes. Dev. 1993, 7, 2556–2564. [Google Scholar] [CrossRef] [PubMed]
- Joerger, A.; Fersht, A.R. Structure–function–rescue: The diverse nature of common p53 cancer mutants. Oncogene 2007, 26, 2226–2242. [Google Scholar] [CrossRef] [PubMed]
- Freed-Pastor, W.A.; Prives, C. Mutant p53: One name, many proteins. Genes. Dev. 2012, 26, 1268–1286. [Google Scholar] [CrossRef]
- Kogan, S.; Carpizo, D.R. Zinc Metallochaperones as Mutant p53 Reactivators: A New Paradigm in Cancer Therapeutics. Cancers 2018, 10, 166. [Google Scholar] [CrossRef]
- Blanden, A.R.; Yu, X.; Wolfe, A.J.; Gilleran, J.A.; Augeri, D.J.; O’dell, R.S.; Olson, E.C.; Kimball, S.D.; Emge, T.J.; Movileanu, L.; et al. Synthetic Metallochaperone ZMC1 Rescues Mutant p53 Conformation by Transporting Zinc into Cells as an Ionophore. Mol. Pharmacol. 2015, 87, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gao, T.; Wang, Y.; Yang, G. Investigating the Influence of Magnesium Ions on p53–DNA Binding Using Atomic Force Microscopy. Int. J. Mol. Sci. 2017, 18, 1585. [Google Scholar] [CrossRef]
- Méplan, C.; Richard, M.-J.; Hainaut, P. Metalloregulation of the tumor suppressor protein p53: Zinc mediates the renaturation of p53 after exposure to metal chelators in vitro and in intact cells. Oncogene 2000, 19, 5227–5236. [Google Scholar] [CrossRef]
- Butler, J.S.; Loh, S.N. Zn2+-Dependent Misfolding of the p53 DNA Binding Domain. Biochemistry 2007, 46, 2630–2639. [Google Scholar] [CrossRef]
- Hainaut, P.; Milner, J. A structural role for metal ions in the “wild-type” conformation of the tumor suppressor protein p53. Cancer Res. 1993, 53, 1739–1742. [Google Scholar]
- Liu, S.; Abboud, M.I.; John, T.; Mikhailov, V.; Hvinden, I.; Walsby-Tickle, J.; Liu, X.; Pettinati, I.; Cadoux-Hudson, T.; McCullagh, J.S.O.; et al. Roles of metal ions in the selective inhibition of oncogenic variants of isocitrate dehydrogenase 1. Commun. Biol. 2021, 4, 1243. Available online: https://www.nature.com/articles/s42003-021-02743-5 (accessed on 20 September 2024). [CrossRef]
- Murnyak, B.; Huang, L.E. Association of TP53 Alteration with Tissue Specificity and Patient Outcome of IDH1-Mutant Glioma. Cells 2021, 10, 2116. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).