Genetic and Epigenetic Aspects of Type 1 Diabetes Mellitus: Modern View on the Problem
Abstract
:1. Introduction
2. Major Histocompatibility Complex (HLA) Locus: Population Aspects
3. Candidate Gene Research
4. Multicenter and Genome-Wide Association Studies (GWAS)
5. The Polygenic Risk Score in Individuals with Type 1 Diabetes Mellitus
6. Functional Role of DNA Risk Loci Related to Developing T1DM in Signaling Pathway Changes
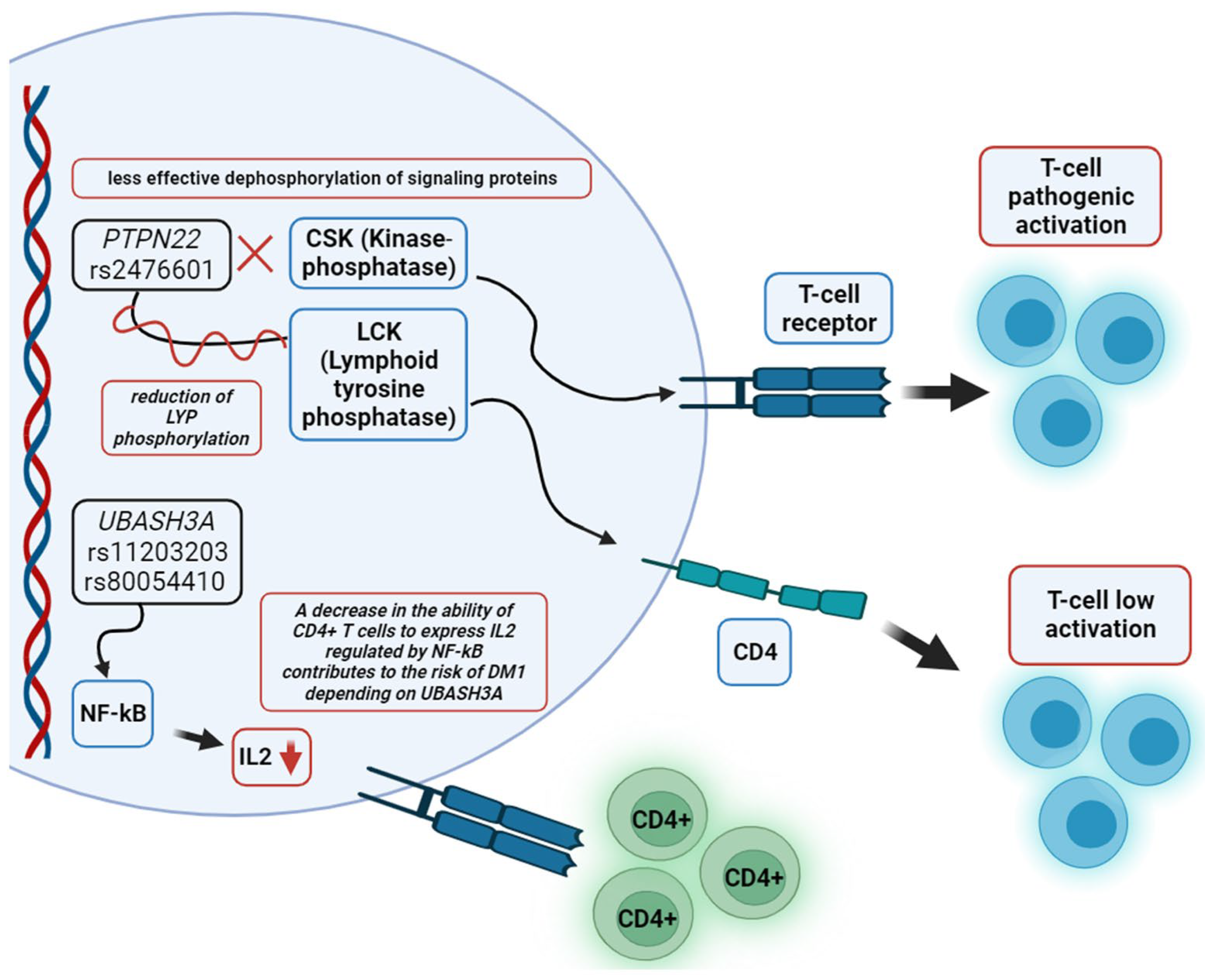
7. Molecular Pathogenesis of Autoimmune Diabetes in Adults
8. Epigenetic Factors in Type 1 Diabetes Mellitus
9. Prospects for Investigation of the Genetics of Type 1 Diabetes Mellitus
- Wide use of genome-wide association study analysis, genome-wide differential methylation analysis based on high-throughput parallel DNA sequencing to examine the disease in different populations and regions of the world;
- Development of new and selection of modern bioinformatics tools to search for biomarkers and perform geno-phenotypic correlations of the qualitative and quantitative features of T1DM;
- Replication of the most significant results that determine an increased susceptibility to the development of T1DM in independent samples, considering their ethnic origin;
- Functional annotation of the identified biomarkers of T1DM;
- Performing eQTL (expression quantitative trait loci) analysis for accurate detection of regulatory intercommunications and determining the significance of loci in the regulation of driver gene activity and gene expression.
10. Conclusions
Funding
Conflicts of Interest
References
- American Diabetes Association Professional Practice Committee 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2022. Diabetes Care 2022, 45, S17–S38. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, J.M.; Divers, J.; Isom, S.; Saydah, S.; Imperatore, G.; Pihoker, C.; Marcovina, S.M.; Mayer-Davis, E.J.; Hamman, R.F.; Dolan, L.; et al. Trends in Prevalence of Type 1 and Type 2 Diabetes in Children and Adolescents in the US, 2001–2017. JAMA J. Am. Med. Assoc. 2021, 326, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Dedov, I.I.; Shestakova, M.V.; Vikulova, O.K.; Zheleznyakova, A.V.; Isakov, M.A.; Sazonova, D.V.; Mokrysheva, N.G. Diabetes Mellitus in the Russian Federation: Dynamics of Epidemiological Indicators According To the Federal Register of Diabetes Mellitus for the Period 2010–2022. Diabetes Mellit. 2023, 26, 104–123. [Google Scholar] [CrossRef]
- Landin-Olsson, M. Latent autoimmune diabetes in adults. Ann. N. Y. Acad. Sci. 2002, 958, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Redondo, M.J.; Steck, A.K.; Pugliese, A. Genetics of Type 1 Diabetes. Pediatr. Diabetes 2018, 19, 346–353. [Google Scholar] [CrossRef]
- Kawasaki, E. Type 1 Diabetes and Autoimmunity. Clin. Pediatr. Endocrinol. 2014, 23, 99–105. [Google Scholar] [CrossRef]
- Srinivasan, S.; Wu, P.; Mercader, J.M.; Udler, M.S.; Porneala, B.C.; Bartz, T.M.; Floyd, J.S.; Sitlani, C.; Guo, X.; Haessler, J.; et al. A Type 1 Diabetes Polygenic Score Is Not Associated with Prevalent Type 2 Diabetes in Large Population Studies. J. Endocr. Soc. 2023, 7, bvad123. [Google Scholar] [CrossRef]
- Noble, J.A.; Erlich, H.A. Genetics of Type 1 Diabetes. Cold Spring Harb. Perspect. Med. 2012, 2, a007732. [Google Scholar] [CrossRef]
- Elsherbini, A.M.; Alsamman, A.M.; Elsherbiny, N.M.; El-Sherbiny, M.; Ahmed, R.; Ebrahim, H.A.; Bakkach, J. Decoding Diabetes Biomarkers and Related Molecular Mechanisms by Using Machine Learning, Text Mining, and Gene Expression Analysis. Int. J. Environ. Res. Public Health 2022, 19, 13890. [Google Scholar] [CrossRef]
- Pugliese, A.; Awdeh, Z.L.; Alper, C.A.; Jackson, R.A.; Eisenbarth, G.S. The Paternally Inherited Insulin Gene B Allele (1,428 FokI Site) Confers Protection from Insulin-Dependent Diabetes in Families. J. Autoimmun. 1994, 7, 687–694. [Google Scholar] [CrossRef]
- Diez, J.; Park, Y.; Zeller, M.; Brown, D.; Garza, D.; Ricordi, C.; Hutton, J.; Eisenbarth, G.S.; Pugliese, A. Differential Splicing of the IA-2 MRNA in Pancreas and Lymphoid Organs as a Permissive Genetic Mechanism for Autoimmunity against the IA-2 Type 1 Diabetes Autoantigen. Diabetes 2001, 50, 895–900. [Google Scholar] [CrossRef]
- Stankov, K.; Benc, D.; Draskovic, D. Genetic and Epigenetic Factors in Etiology of Diabetes Mellitus Type 1. Pediatrics 2013, 132, 1112–1122. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Pinney, S.E. DNA Methylation and Its Role in the Pathogenesis of Diabetes. Pediatr. Diabetes 2017, 18, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Benoist, C.; Mathis, D. Retrovirus as Trigger, Precipitator or Marker? Nature 1997, 388, 833–834. [Google Scholar] [CrossRef] [PubMed]
- Conrad, B.; Weidmann, E.; Trucco, G.; Rudert, W.A.; Behboo, R.; Ricordi, C.; Rodriquez-Rilo, H.; Finegold, D.; Trucco, M. Evidence for Superantigen Involvement in Insulin-Dependent Diabetes Mellitus Aetiology. Nature 1994, 371, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Jerram, S.T.; Dang, M.N.; Leslie, R.D. The Role of Epigenetics in Type 1 Diabetes. Curr. Diab. Rep. 2017, 17, 89. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Ahmed, S.; Anderson, M.S.; Atkinson, M.A.; Becker, D.; Bingley, P.J.; Bosi, E.; Brusko, T.M.; DiMeglio, L.A.; Evans-Molina, C.; et al. Introducing the Endotype Concept to Address the Challenge of Disease Heterogeneity in Type 1 Diabetes. Diabetes Care 2020, 43, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Kulski, J.K.; Suzuki, S.; Shiina, T. Human Leukocyte Antigen Super-Locus: Nexus of Genomic Supergenes, SNPs, Indels, Transcripts, and Haplotypes. Hum. Genome Var. 2022, 9, 49. [Google Scholar] [CrossRef]
- Hu, X.; Deutsch, A.J.; Lenz, T.L.; Onengut-Gumuscu, S.; Han, B.; Chen, W.M.; Howson, J.M.M.; Todd, J.A.; De Bakker, P.I.W.; Rich, S.S.; et al. Additive and Interaction Effects at Three Amino Acid Positions in HLA-DQ and HLA-DR Molecules Drive Type 1 Diabetes Risk. Nat. Genet. 2015, 47, 898–905. [Google Scholar] [CrossRef]
- Cudworth, A.G.; Woodrow, J.C. Hl-a Antigens and Diabetes Mellitus. Lancet 1974, 304, 1153. [Google Scholar] [CrossRef]
- Plasil, M.; Futas, J.; Jelinek, A.; Burger, P.A.; Horin, P. Comparative Genomics of the Major Histocompatibility Complex (MHC) of Felids. Front. Genet. 2022, 13, 829891. [Google Scholar] [CrossRef] [PubMed]
- Nunes, J.M.; Buhler, S.; Roessli, D.; Sanchez-Mazas, A.; Andreani, M.; Benhamamouch, S.; Boldyreva, M.; Canossi, A.; Chiaroni, J.; Darke, C.; et al. The HLA-Net GENE[RATE] Pipeline for Effective HLA Data Analysis and Its Application to 145 Population Samples from Europe and Neighbouring Areas. Tissue Antigens 2014, 83, 307–323. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.W.; Tallapragada, D.S.P.; Baptist, A.; Sharp, S.A.; Bhaskar, S.; Jog, K.S.; Patel, K.A.; Weedon, M.N.; Chandak, G.R.; Yajnik, C.S.; et al. Type 1 Diabetes Genetic Risk Score Is Discriminative of Diabetes in Non-Europeans: Evidence from a Study in India. Sci. Rep. 2020, 10, 9450. [Google Scholar] [CrossRef] [PubMed]
- Kuraeva, T.L.; Zubov, L.A.; Titovich, E.V.; Sibileva, E.N.; Ivanova, O.N.; Yur, T.; Peterkova, V.A.; Dedov, I.I. HLA-haplotypes and the risk of developing diabetes of type 1 diabetes in the native population of the Nenets Autonomous district. Diabetes Mellit. 2017, 20, 51–58. [Google Scholar] [CrossRef]
- Al-Balushi, M.; Al-Badi, S.; Al-Yaarubi, S.; Al-Riyami, H.; Al-Shidhani, A.; Al-Hinai, S.; Alshirawi, A.; Hasson, S.; Said, E.; Al-Jabri, A.; et al. The Association of Human Leukocyte Antigens Complex with Type 1 Diabetes in the Omani Population. Sultan Qaboos Univ. Med. J. 2023, 23, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Erlich, H.A.; Valdes, A.M.; McDevitt, S.L.; Simen, B.B.; Blake, L.A.; McGowan, K.R.; Todd, J.A.; Rich, S.S.; Noble, J.A. Next Generation Sequencing Reveals the Association of DRB3*02:02 with Type 1 Diabetes. Diabetes 2013, 62, 2618–2622. [Google Scholar] [CrossRef]
- Zhao, L.P.; Alshiekh, S.; Zhao, M.; Carlsson, A.; Larsson, H.E.; Forsander, G.; Ivarsson, S.A.; Ludvigsson, J.; Kockum, I.; Marcus, C.; et al. Next-Generation Sequencing Reveals That HLA-DRB3, -DRB4, and -DRB5 May Be Associated with Islet Autoantibodies and Risk for Childhood Type 1 Diabetes. Diabetes 2016, 65, 710–718. [Google Scholar] [CrossRef]
- Smith, A.G.; Pyo, C.W.; Nelson, W.; Gow, E.; Wang, R.; Shen, S.; Sprague, M.; Pereira, S.E.; Geraghty, D.E.; Hansen, J.A. Next Generation Sequencing to Determine HLA Class II Genotypes in a Cohort of Hematopoietic Cell Transplant Patients and Donors. Hum. Immunol. 2014, 75, 1040–1046. [Google Scholar] [CrossRef]
- Dilthey, A.T.; Gourraud, P.A.; Mentzer, A.J.; Cereb, N.; Iqbal, Z.; McVean, G. High-Accuracy HLA Type Inference from Whole-Genome Sequencing Data Using Population Reference Graphs. PLoS Comput. Biol. 2016, 12, e1005151. [Google Scholar] [CrossRef]
- Haris, B.; Ahmed, I.; Syed, N.; Almabrazi, H.; Saraswathi, S.; Al-Khawaga, S.; Saeed, A.; Mundekkadan, S.; Mohammed, I.; Sharari, S.; et al. Clinical Features, Epidemiology, Autoantibody Status, HLA Haplotypes and Genetic Mechanisms of Type 1 Diabetes Mellitus among Children in Qatar. Sci. Rep. 2021, 11, 18887. [Google Scholar] [CrossRef]
- Noble, J.A. Immunogenetics of Type 1 Diabetes: A Comprehensive Review. J. Autoimmun. 2015, 64, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Halagan, M.; Oliveira, D.C.; Maiers, M.; Fabreti-Oliveira, R.A.; Moraes, M.E.H.; Visentainer, J.E.L.; Pereira, N.F.; Romero, M.; Cardoso, J.F.; Porto, L.C. The Distribution of HLA Haplotypes in the Ethnic Groups That Make up the Brazilian Bone Marrow Volunteer Donor Registry (REDOME). Immunogenetics 2018, 70, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.B.; Rodrigues, V.; Santos, D.C.; Boas, P.R.V.; Silva, D.A.; de Sousa Azulay, R.S.; Dib, S.A.; Pavin, E.J.; Fernandes, V.O.; Montenegro Junior, R.M.; et al. Association between HLA Class II Alleles/Haplotypes and Genomic Ancestry in Brazilian Patients with Type 1 Diabetes: A Nationwide Exploratory Study. Genes 2023, 14, 991. [Google Scholar] [CrossRef]
- Redondo, M.J.; Gignoux, C.R.; Dabelea, D.; Hagopian, W.A.; Onengut-Gumuscu, S.; Oram, R.A.; Rich, S.S. Type 1 Diabetes in Diverse Ancestries and the Use of Genetic Risk Scores. Lancet Diabetes Endocrinol. 2022, 10, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Immel, A.; Key, F.M.; Szolek, A.; Barquera, R.; Robinson, M.K.; Harrison, G.F.; Palmer, W.H.; Spyrou, M.A.; Susat, J.; Krause-Kyora, B.; et al. Analysis of Genomic DNA from Medieval Plague Victims Suggests Long-Term Effect of Yersinia Pestis on Human Immunity Genes. Mol. Biol. Evol. 2021, 38, 4059–4076. [Google Scholar] [CrossRef] [PubMed]
- Vehik, K.; Hamman, R.F.; Lezotte, D.; Norris, J.M.; Klingensmith, G.J.; Rewers, M.; Dabelea, D. Trends in High-Risk HLA Susceptibility Genes among Colorado Youth with Type 1 Diabetes. Diabetes Care 2008, 31, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Hermann, R.; Knip, M.; Veijola, R.; Simell, O.; Laine, A.P.; Åkerblom, H.K.; Groop, P.H.; Forsblom, C.; Pettersson-Fernholm, K.; Ilonen, J. Temporal Changes in the Frequencies of HLA Genotypes in Patients with Type 1 Diabetes—Indication of an Increased Environmental Pressure? Diabetologia 2003, 46, 420–425. [Google Scholar] [CrossRef]
- Ferrara, C.T.; Geyer, S.M.; Liu, Y.F.; Evans-Molina, C.; Libman, I.M.; Besser, R.; Becker, D.J.; Rodriguez, H.; Moran, A.; Gitelman, S.E.; et al. Excess BMI in Childhood: A Modifiable Risk Factor for Type 1 Diabetes Development? Diabetes Care 2017, 40, 698–701. [Google Scholar] [CrossRef]
- Bottini, N.; Musumeci, L.; Alonso, A.; Rahmouni, S.; Nika, K.; Rostamkhani, M.; MacMurray, J.; Meloni, G.F.; Lucarelli, P.; Pellecchia, M.; et al. A Functional Variant of Lymphoid Tyrosine Phosphatase Is Associated with Type I Diabetes. Nat. Genet. 2004, 36, 337–338. [Google Scholar] [CrossRef]
- Ivanova, O.N.; Prokof’ev, S.A.; Smirnova, N.B.; Tishina, J.V.; Bardymova, T.P.; Danilova, G.I.; Kovalenko, T.V.; Titovich, E.V.; Kuraeva, T.L.; Peterkova, V.A.; et al. Ptpn22 Polymorphisms Associated with Type 1 Diabetes Mellitus in Ethnic Populations of Russian Federation. Diabetes Mellit. 2013, 16, 4–10. [Google Scholar] [CrossRef]
- Nistico, L.; Buzzetti, R.; Pritchard, L.E.; Van Der Auwera, B.; Giovannini, C.; Bosi, E.; Martinez Larrad, M.T.; Rios, M.S.; Chow, C.C.; Cockram, C.S.; et al. The CTLA-4 Gene Region of Chromosome 2q33 Is Linked to, and Associated with, Type 1 Diabetes. Hum. Mol. Genet. 1996, 5, 1075–1080. [Google Scholar] [CrossRef]
- Kavvoura, F.K.; Ioannidis, J.P.A. CTLA-4 Gene Polymorphisms and Susceptibility to Type 1 Diabetes Mellitus: A HuGE Review and Meta-Analysis. Am. J. Epidemiol. 2005, 162, 3–16. [Google Scholar] [CrossRef]
- Lowe, C.E.; Cooper, J.D.; Brusko, T.; Walker, N.M.; Smyth, D.J.; Bailey, R.; Bourget, K.; Plagnol, V.; Field, S.; Atkinson, M.; et al. Large-Scale Genetic Fine Mapping and Genotype-Phenotype Associations Implicate Polymorphism in the IL2RA Region in Type 1 Diabetes. Nat. Genet. 2007, 39, 1074–1082. [Google Scholar] [CrossRef] [PubMed]
- Rich, S.S.; Akolkar, B.; Concannon, P.; Erlich, H.; Hilner, J.E.; Julier, C.; Morahan, G.; Nerup, J.; Nierras, C.; Pociot, F.; et al. Overview of the Type i Diabetes Genetics Consortium. Genes Immun. 2009, 10, S1–S4. [Google Scholar] [CrossRef] [PubMed]
- Hagopian, W.A.; Erlich, H.; Lernmark, A.; Rewers, M.; Ziegler, A.G.; Simell, O.; Akolkar, B.; Vogt, R.; Blair, A.; Ilonen, J.; et al. The Environmental Determinants of Diabetes in the Young (TEDDY): Genetic Criteria and International Diabetes Risk Screening of 421 000 Infants. Pediatr. Diabetes 2011, 12, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Rewers, M.; Bugawan, T.L.; Norris, J.M.; Blair, A.; Beaty, B.; Hoffman, M.; McDuffie, R.S.; Hamman, R.F.; Klingensmith, G.; Eisenbarth, G.S.; et al. Newborn Screening for HLA Markers Associated with IDDM: Diabetes Autoimmunity Study in the Young (DAISY). Diabetologia 1996, 39, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, A.G.; Hummel, M.; Schenker, M.; Bonifacio, E. Autoantibody Appearance and Risk for Development of Childhood Diabetes in Offspring of Parents with Type 1 Diabetes: The 2-Year Analysis of the German BABYDIAB Study. Diabetes 1999, 48, 460–468. [Google Scholar] [CrossRef] [PubMed]
- The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007, 447, 661–678. [Google Scholar] [CrossRef] [PubMed]
- Skyler, J.S.; Greenbaum, C.J.; Lachin, J.M.; Leschek, E.; Rafkin-Mervis, L.; Savage, P.; Spain, L. Type 1 Diabetes TrialNet—An International Collaborative Clinical Trials Network. Ann. N. Y. Acad. Sci. 2008, 1150, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Mahon, J.L.; Sosenko, J.M.; Rafkin-Mervis, L.; Krause-Steinrauf, H.; Lachin, J.M.; Thompson, C.; Bingley, P.J.; Bonifacio, E.; Palmer, J.P.; Eisenbarth, G.S.; et al. The TrialNet Natural History Study of the Development of Type 1 Diabetes: Objectives, Design, and Initial Results. Pediatr. Diabetes 2009, 10, 97–104. [Google Scholar] [CrossRef]
- Leslie, R.D.G.; Kolb, H.; Schloot, N.C.; Buzzetti, R.; Mauricio, D.; De Leiva, A.; Yderstraede, K.; Sarti, C.; Thivolet, C.; Hadden, D.; et al. Diabetes Classification: Grey Zones, Sound and Smoke: Action LADA 1. Diabetes. Metab. Res. Rev. 2008, 24, 511–519. [Google Scholar] [CrossRef]
- Soltesz, G. Familial Risk of Type I Diabetes in European Children. Diabetologia 1998, 41, 1151–1156. [Google Scholar] [CrossRef]
- Barrett, J.C.; Clayton, D.G.; Concannon, P.; Akolkar, B.; Cooper, J.D.; Erlich, H.A.; Julier, C.; Morahan, G.; Nerup, J.; Nierras, C.; et al. Genome-Wide Association Study and Meta-Analysis Find That over 40 Loci Affect Risk of Type 1 Diabetes. Nat. Genet. 2009, 41, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Senee, V.; Chelala, C.; Duchatelet, S.; Feng, D.; Blanc, H.; Cossec, J.C.; Charon, C.; Nicolino, M.; Boileau, P.; Cavener, D.R.; et al. Mutations in GLIS3 Are Responsible for a Rare Syndrome with Neonatal Diabetes Mellitus and Congenital Hypothyroidism. Nat. Genet. 2006, 38, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Shiow, L.R.; Rosen, D.B.; Brdickova, N.; Xu, Y.; An, J.; Lanier, L.L.; Cyster, J.G.; Matloubian, M. CD69 Acts Downstream of Interferon-α/β to Inhibit S1P 1 and Lymphocyte Egress from Lymphoid Organs. Nature 2006, 440, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Onengut-Gumuscu, S.; Chen, W.M.; Burren, O.; Cooper, N.J.; Quinlan, A.R.; Mychaleckyj, J.C.; Farber, E.; Bonnie, J.K.; Szpak, M.; Schofield, E.; et al. Fine Mapping of Type 1 Diabetes Susceptibility Loci and Evidence for Colocalization of Causal Variants with Lymphoid Gene Enhancers. Nat. Genet. 2015, 47, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Torn, C.; Hadley, D.; Lee, H.S.; Hagopian, W.; Lernmark, Å.; Simell, O.; Rewers, M.; Ziegler, A.; Schatz, D.; Akolkar, B.; et al. Role of Type 1 Diabetes- Associated Snps on Risk of Autoantibody Positivity in the TEDDY Study. Diabetes 2015, 64, 1818–1829. [Google Scholar] [CrossRef] [PubMed]
- Ilonen, J.; Kiviniemi, M.; Lempainen, J.; Simell, O.; Toppari, J.; Veijola, R.; Knip, M. Genetic Susceptibility to Type 1 Diabetes in Childhood—Estimation of HLA Class II Associated Disease Risk and Class II Effect in Various Phases of Islet Autoimmunity. Pediatr. Diabetes 2016, 17, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Steck, A.K.; Dong, F.; Wong, R.; Fouts, A.; Liu, E.; Romanos, J.; Wijmenga, C.; Norris, J.M.; Rewers, M.J. Improving Prediction of Type 1 Diabetes by Testing Non-HLA Genetic Variants in Addition to HLA Markers. Pediatr. Diabetes 2014, 15, 355–362. [Google Scholar] [CrossRef]
- Winkler, C.; Krumsiek, J.; Lempainen, J.; Achenbach, P.; Grallert, H.; Giannopoulou, E.; Bunk, M.; Theis, F.J.; Bonifacio, E.; Ziegler, A.G. A Strategy for Combining Minor Genetic Susceptibility Genes to Improve Prediction of Disease in Type 1 Diabetes. Genes Immun. 2012, 13, 549–555. [Google Scholar] [CrossRef]
- Frohnert, B.I.; Laimighofer, M.; Krumsiek, J.; Theis, F.J.; Winkler, C.; Norris, J.M.; Ziegler, A.G.; Rewers, M.J.; Steck, A.K. Prediction of Type 1 Diabetes Using a Genetic Risk Model in the Diabetes Autoimmunity Study in the Young. Pediatr. Diabetes 2018, 19, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Hudgins, A.D.; Yang, J.; Zhu, Y.; Tu, Z.; Rosenfeld, M.G.; DiLorenzo, T.P.; Suh, Y. A Comprehensive Integrated Post-GWAS Analysis of Type 1 Diabetes Reveals Enhancerbased Immune Dysregulation. PLoS ONE 2021, 16, e0257265. [Google Scholar] [CrossRef]
- Wand, H.; Lambert, S.A.; Tamburro, C.; Iacocca, M.A.; O’Sullivan, J.W.; Sillari, C.; Kullo, I.J.; Rowley, R.; Dron, J.S.; Brockman, D.; et al. Improving Reporting Standards for Polygenic Scores in Risk Prediction Studies. Nature 2021, 591, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Winkler, C.; Krumsiek, J.; Buettner, F.; Angermüller, C.; Giannopoulou, E.Z.; Theis, F.J.; Ziegler, A.G.; Bonifacio, E. Feature Ranking of Type 1 Diabetes Susceptibility Genes Improves Prediction of Type 1 Diabetes. Diabetologia 2014, 57, 2521–2529. [Google Scholar] [CrossRef] [PubMed]
- Oram, R.A.; Patel, K.; Hill, A.; Shields, B.; McDonald, T.J.; Jones, A.; Hattersley, A.T.; Weedon, M.N. A Type 1 Diabetes Genetic Risk Score Can Aid Discrimination between Type 1 and Type 2 Diabetes in Young Adults. Diabetes Care 2016, 39, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Bonifacio, E.; Beyerlein, A.; Hippich, M.; Winkler, C.; Vehik, K.; Weedon, M.N.; Laimighofer, M.; Hattersley, A.T.; Krumsiek, J.; Frohnert, B.I.; et al. Genetic Scores to Stratify Risk of Developing Multiple Islet Autoantibodies and Type 1 Diabetes: A Prospective Study in Children. PLoS Med. 2018, 15, e1002548. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, M.R.; Thirawatananond, P.; Peters, L.; Sharp, R.C.; Ogundare, S.; Posgai, A.L.; Perry, D.J.; Brusko, T.M. De-Coding Genetic Risk Variants in Type 1 Diabetes. Immunol. Cell Biol. 2021, 99, 496–508. [Google Scholar] [CrossRef] [PubMed]
- Koskinen, M.K.; Mikk, M.L.; Laine, A.P.; Lempainen, J.; Löyttyniemi, E.; Vähäsalo, P.; Hekkala, A.; Härkönen, T.; Kiviniemi, M.; Simell, O.; et al. Longitudinal Pattern of First-Phase Insulin Response Is Associated with Genetic Variants Outside the Class II HLA Region in Children with Multiple Autoantibodies. Diabetes 2020, 69, 12–19. [Google Scholar] [CrossRef]
- Smyth, D.J.; Cooper, J.D.; Howson, J.M.M.; Clarke, P.; Downes, K.; Mistry, T.; Stevens, H.; Walker, N.M.; Todd, J.A. FUT2 Nonsecretor Status Links Type 1 Diabetes Susceptibility and Resistance to Infection. Diabetes 2011, 60, 3081–3084. [Google Scholar] [CrossRef]
- Marroqui, L.; Dos Santos, R.S.; Floyel, T.; Grieco, F.A.; Santin, I.; Op De Beeck, A.; Marselli, L.; Marchetti, P.; Pociot, F.; Eizirik, D.L. TYK2, a Candidate Gene for Type 1 Diabetes, Modulates Apoptosis and the Innate Immune Response in Human Pancreatic β-Cells. Diabetes 2015, 64, 3808–3817. [Google Scholar] [CrossRef]
- Buhling, F.; Kouadio, M.; Chwieralski, C.E.; Kern, U.; Hohlfeld, J.M.; Klemm, N.; Friedrichs, N.; Roth, W.; Deussing, J.M.; Peters, C.; et al. Gene Targeting of the Cysteine Peptidase Cathepsin H Impairs Lung Surfactant in Mice. PLoS ONE 2011, 6, e26247. [Google Scholar] [CrossRef] [PubMed]
- Faraco, J.; Lin, L.; Kornum, B.R.; Kenny, E.E.; Trynka, G.; Einen, M.; Rico, T.J.; Lichtner, P.; Dauvilliers, Y.; Arnulf, I.; et al. ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy. PLoS Genet. 2013, 9, e1003270. [Google Scholar] [CrossRef] [PubMed]
- Inshaw, J.R.J.; Cutler, A.J.; Crouch, D.J.M.; Wicker, L.S.; Todd, J.A. Genetic Variants Predisposing Most Strongly to Type 1 Diabetes Diagnosed under Age 7 Years Lie near Candidate Genes That Function in the Immune System and in Pancreatic B-Cells. Diabetes Care 2020, 43, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Flyoel, T.; Brorsson, C.; Nielsen, L.B.; Miani, M.; Bang-Berthelsen, C.H.; Friedrichsen, M.; Overgaard, A.J.; Berchtold, L.A.; Wiberg, A.; Poulsen, P.; et al. CTSH Regulates β-Cell Function and Disease Progression in Newly Diagnosed Type 1 Diabetes Patients. Proc. Natl. Acad. Sci. USA 2014, 111, 10305–10310. [Google Scholar] [CrossRef] [PubMed]
- Krischer, J.P.; Liu, X.; Vehik, K.; Akolkar, B.; Hagopian, W.A.; Rewers, M.J.; She, J.X.; Toppari, J.; Ziegler, A.G.; Lernmark, A. Predicting Islet Cell Autoimmunity and Type 1 Diabetes: An 8-Year Teddy Study Progress Report. Diabetes Care 2019, 42, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Galvani, G.; Fousteri, G. PTPN22 and Islet-Specific Autoimmunity: What Have the Mouse Models Taught Us? World J. Diabetes 2017, 8, 330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zahir, N.; Jiang, Q.; Miliotis, H.; Heyraud, S.; Meng, X.; Dong, B.; Xie, G.; Qiu, F.; Hao, Z.; et al. The Autoimmune Diseaseg-Associated PTPN22 Variant Promotes Calpain-Mediated Lyp/Pep Degradation Associated with Lymphocyte and Dendritic Cell Hyperresponsiveness. Nat. Genet. 2011, 43, 902–907. [Google Scholar] [CrossRef] [PubMed]
- Bottini, N.; Vang, T.; Cucca, F.; Mustelin, T. Role of PTPN22 in Type 1 Diabetes and Other Autoimmune Diseases. Semin. Immunol. 2006, 18, 207–213. [Google Scholar] [CrossRef]
- Sharp, R.C.; Beg, S.A.; Naser, S.A. Polymorphisms in Protein Tyrosine Phosphatase Non-Receptor Type 2 and 22 (PTPN2/22) Are Linked to Hyper-Proliferative T-Cells and Susceptibility to Mycobacteria in Rheumatoid Arthritis. Front. Cell. Infect. Microbiol. 2018, 8, 11. [Google Scholar] [CrossRef]
- Ge, Y.; Paisie, T.K.; Newman, J.R.B.; McIntyre, L.M.; Concannon, P. UBASH3A Mediates Risk for Type 1 Diabetes through Inhibition of T-Cell Receptor-Induced NF-ΚB Signaling. Diabetes 2017, 66, 2033–2043. [Google Scholar] [CrossRef]
- Suomi, T.; Starskaia, I.; Kalim, U.U.; Rasool, O.; Jaakkola, M.K.; Grönroos, T.; Välikangas, T.; Brorsson, C.; Mazzoni, G.; Bruggraber, S.; et al. Gene Expression Signature Predicts Rate of Type 1 Diabetes Progression. eBioMedicine 2023, 92, 104625. [Google Scholar] [CrossRef] [PubMed]
- Golodnikov, I.I.; Rusyaeva, N.V.; Nikonova, T.V.; Kononenko, I.V.; Shestakova, M.V. Modern Understanding of Latent Autoimmune Diabetes of Adults. Diabetes Mellit. 2023, 26, 262–274. [Google Scholar] [CrossRef]
- Cousminer, D.L.; Ahlqvist, E.; Mishra, R.; Andersen, M.K.; Chesi, A.; Hawa, M.I.; Davis, A.; Hodge, K.M.; Bradfield, J.P.; Zhou, K.; et al. First Genome-Wide Association Study of Latent Autoimmune Diabetes in Adults Reveals Novel Insights Linking Immune and Metabolic Diabetes. Diabetes Care 2018, 41, 2396–2403. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.K. New Insights into the Genetics of Latent Autoimmune Diabetes in Adults. Curr. Diab. Rep. 2020, 20, 43. [Google Scholar] [CrossRef] [PubMed]
- Weinhold, B. Epigenetics: The Science of Change. Environ. Health Perspect. 2006, 114, A160–A167. [Google Scholar] [CrossRef] [PubMed]
- Yalaev, B.I.; Tyurin, A.V.; Mirgalieva, R.Y.; Khusainova, R.I. The Role of DNA Methylation in the Disorders of Bone Metabolism. Vavilovskii Zhurnal Genet. Sel. 2019, 23, 67–74. [Google Scholar] [CrossRef]
- Rakyan, V.K.; Beyan, H.; Down, T.A.; Hawa, M.I.; Maslau, S.; Aden, D.; Daunay, A.; Busato, F.; Mein, C.A.; Manfras, B.; et al. Identification of Type 1 Diabetes-Associated DNA Methylation Variable Positions That Precede Disease Diagnosis. PLoS Genet. 2011, 7, e1002300. [Google Scholar] [CrossRef] [PubMed]
- Starskaia, I.; Laajala, E.; Grönroos, T.; Härkönen, T.; Junttila, S.; Kattelus, R.; Kallionpää, H.; Laiho, A.; Suni, V.; Tillmann, V.; et al. Early DNA Methylation Changes in Children Developing Beta Cell Autoimmunity at a Young Age. Diabetologia 2022, 65, 844–860. [Google Scholar] [CrossRef]
- Olsson, A.H.; Volkov, P.; Bacos, K.; Dayeh, T.; Hall, E.; Nilsson, E.A.; Ladenvall, C.; Rönn, T.; Ling, C. Genome-Wide Associations between Genetic and Epigenetic Variation Influence MRNA Expression and Insulin Secretion in Human Pancreatic Islets. PLoS Genet. 2014, 10, e1004735. [Google Scholar] [CrossRef]
- Paul, D.S.; Teschendorff, A.E.; Dang, M.A.N.; Lowe, R.; Hawa, M.I.; Ecker, S.; Beyan, H.; Cunningham, S.; Fouts, A.R.; Ramelius, A.; et al. Increased DNA Methylation Variability in Type 1 Diabetes across Three Immune Effector Cell Types. Nat. Commun. 2016, 7, 13555. [Google Scholar] [CrossRef]
- Johnson, R.K.; Vanderlinden, L.A.; Dong, F.; Carry, P.M.; Seifert, J.; Waugh, K.; Shorrosh, H.; Fingerlin, T.; Frohnert, B.I.; Yang, I.V.; et al. Longitudinal DNA Methylation Differences Precede Type 1 Diabetes. Sci. Rep. 2020, 10, 3721. [Google Scholar] [CrossRef] [PubMed]
- Lakhter, A.J.; Pratt, R.E.; Moore, R.E.; Doucette, K.K.; Maier, B.F.; DiMeglio, L.A.; Sims, E.K. Beta Cell Extracellular Vesicle MiR-21-5p Cargo Is Increased in Response to Inflammatory Cytokines and Serves as a Biomarker of Type 1 Diabetes. Diabetologia 2018, 61, 1124–1134. [Google Scholar] [CrossRef]
- Samandari, N.; Mirza, A.H.; Nielsen, L.B.; Kaur, S.; Hougaard, P.; Fredheim, S.; Mortensen, H.B.; Pociot, F. Circulating MicroRNA Levels Predict Residual Beta Cell Function and Glycaemic Control in Children with Type 1 Diabetes Mellitus. Diabetologia 2017, 60, 354–363. [Google Scholar] [CrossRef]
- Scherm, M.G.; Daniel, C. MiRNA-Mediated Immune Regulation in Islet Autoimmunity and Type 1 Diabetes. Front. Endocrinol. 2020, 11, 606322. [Google Scholar] [CrossRef]
- Margaritis, K.; Margioula-siarkou, G.; Giza, S.; Kotanidou, E.P.; Tsinopoulou, V.R.; Christoforidis, A.; Galli-tsinopoulou, A. Micro-RNA Implications in Type-1 Diabetes Mellitus: A Review of Literature. Int. J. Mol. Sci. 2021, 22, 12165. [Google Scholar] [CrossRef] [PubMed]
- Mcclelland, A.D.; Kantharidis, P. MicroRNA in the Development of Diabetic Complications. Clin. Sci. 2014, 126, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.; Chen, Z.; Zhang, L.; Liu, Z.; Wu, X.; Yuan, Y.-C.; Natarajan, R. Profiles of Epigenetic Histone Post-translational Modifications at Type 1 Diabetes Susceptible Genes. J. Biol. Chem. 2012, 287, 16335–16345. [Google Scholar] [CrossRef]
- Cerna, M. Epigenetic Regulation in Etiology of Type 1 Diabetes Mellitus. Int. J. Mol. Sci. 2020, 21, 36. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Y.; Jiang, Q.; Song, W.; Zhang, L. Therapeutic potential of selective histone deacetylase 3 inhibition. Eur. J. Med. Chem. 2019, 162, 534–542. [Google Scholar] [CrossRef]
- Hu, Q.; Che, G.; Yang, Y.; Xie, H.; Tian, J. Histone Deacetylase 3 Aggravates Type 1 Diabetes Mellitus by Inhibiting Lymphocyte Apoptosis Through the microRNA-296-5p/Bcl-xl Axis. Front. Genet. 2020, 11, 536854. [Google Scholar] [CrossRef]
- Chen, S.S.; Jenkins, A.J.; Majewski, H. Elevated plasma prostaglandins and acetylated histone in monocytes in type 1 diabetes patients. Diabet. Med. 2009, 26, 182–186. [Google Scholar] [CrossRef]
- Christensen, D.P.; Dahllof, M.; Lundh, M.; Rasmussen, D.N.; Nielsen, M.D.; Billestrup, N.; Grunnet, L.G.; Mandrup-Poulsen, T. Histone Deacetylase (HDAC) Inhibition as a Novel Treatment for Diabetes Mellitus. Mol. Med. 2011, 17, 378–390. [Google Scholar] [CrossRef]



| Loci | Gene | Odds Ratio and Condifedence Interval | p |
|---|---|---|---|
| rs9273368 | HLA-DQB1 | 3.12 (2.86–3.40) | 7.9 × 10−143 |
| rs2476601 | PTPN22 | 1.62 (1.48–1.78) | <0.0001 |
| rs689 | INS | 1.39 (1.29–1.48) | <0.0001 |
| rs7310615 | SH2B3 | 1.28 (1.19–1.38) | 4.9 × 10−11 |
| rs7903146 | TCF7L2 | 1.19 (1.00–1.40) | 0.04 |
| rs1983890 | PFKFB3 | 1.16 (1.14–1.32) | 3.0 × 10−8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minniakhmetov, I.; Yalaev, B.; Khusainova, R.; Bondarenko, E.; Melnichenko, G.; Dedov, I.; Mokrysheva, N. Genetic and Epigenetic Aspects of Type 1 Diabetes Mellitus: Modern View on the Problem. Biomedicines 2024, 12, 399. https://doi.org/10.3390/biomedicines12020399
Minniakhmetov I, Yalaev B, Khusainova R, Bondarenko E, Melnichenko G, Dedov I, Mokrysheva N. Genetic and Epigenetic Aspects of Type 1 Diabetes Mellitus: Modern View on the Problem. Biomedicines. 2024; 12(2):399. https://doi.org/10.3390/biomedicines12020399
Chicago/Turabian StyleMinniakhmetov, Ildar, Bulat Yalaev, Rita Khusainova, Ekaterina Bondarenko, Galina Melnichenko, Ivan Dedov, and Natalia Mokrysheva. 2024. "Genetic and Epigenetic Aspects of Type 1 Diabetes Mellitus: Modern View on the Problem" Biomedicines 12, no. 2: 399. https://doi.org/10.3390/biomedicines12020399
APA StyleMinniakhmetov, I., Yalaev, B., Khusainova, R., Bondarenko, E., Melnichenko, G., Dedov, I., & Mokrysheva, N. (2024). Genetic and Epigenetic Aspects of Type 1 Diabetes Mellitus: Modern View on the Problem. Biomedicines, 12(2), 399. https://doi.org/10.3390/biomedicines12020399







