Exploring Cardiac Exosomal RNAs of Acute Myocardial Infarction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Triphenyltetrazolium Chloride (TTC) Staining
2.3. Exosome Isolation
2.4. SmRNA Library Construction and miRNA Sequencing
2.5. Data Analysis of miRNA Sequencing
2.6. Transmission Electron Microscopy (TEM) Analysis
2.7. Immunoblot Analysis
2.8. Isolation of Primary Cardiomyocytes and Fibroblasts from Neonatal Mouse Heart
2.9. Transfection with miRNA Mimics
2.10. Cytotoxicity Assay
2.11. Immunofluorescence Analysis
2.12. Statistical Analysis
3. Results
3.1. Establishment of MI Animal Models and Isolation of Exosomes from Heart Tissues
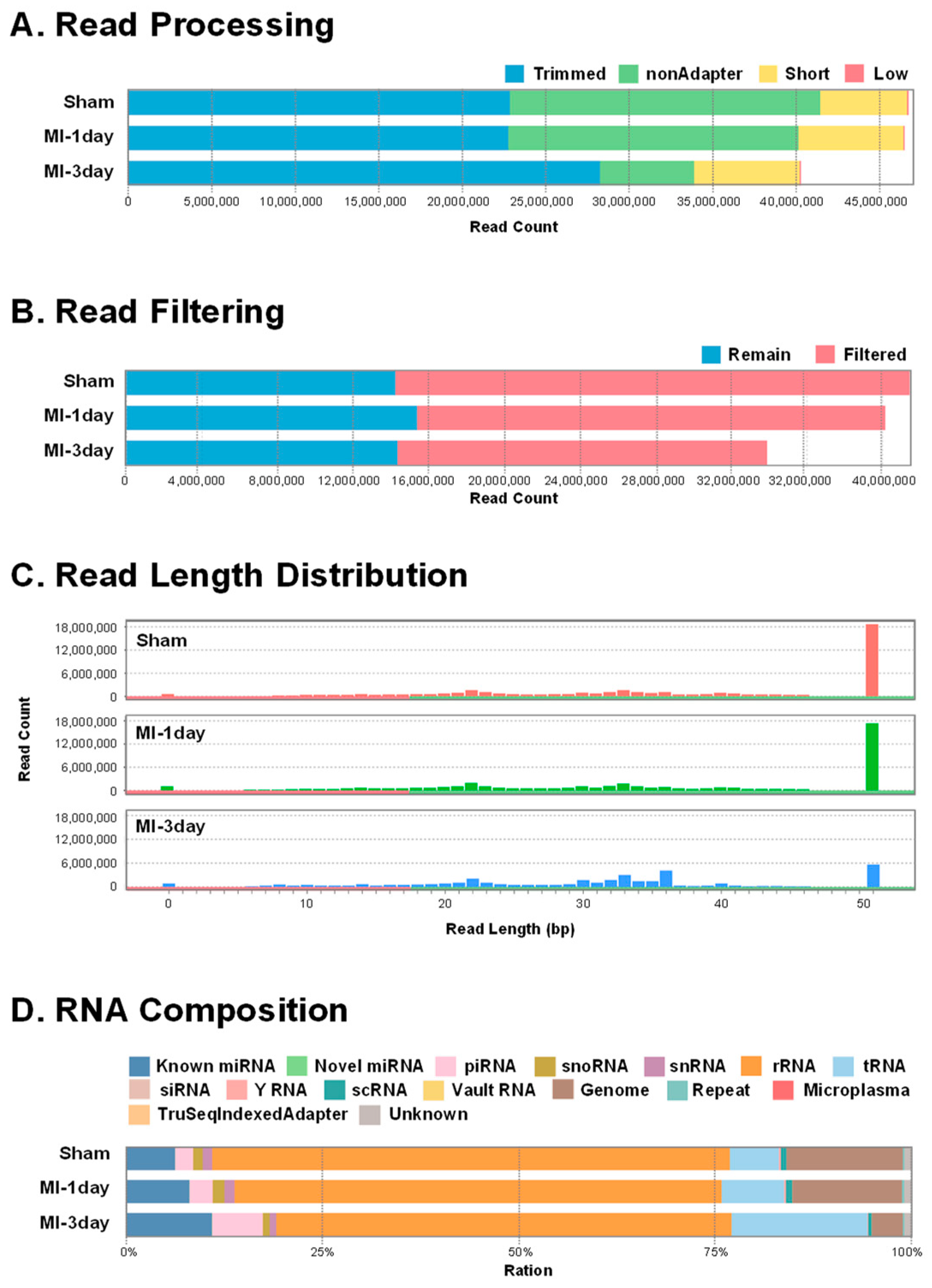
3.2. ExoRNA Sequencing and Data Processing
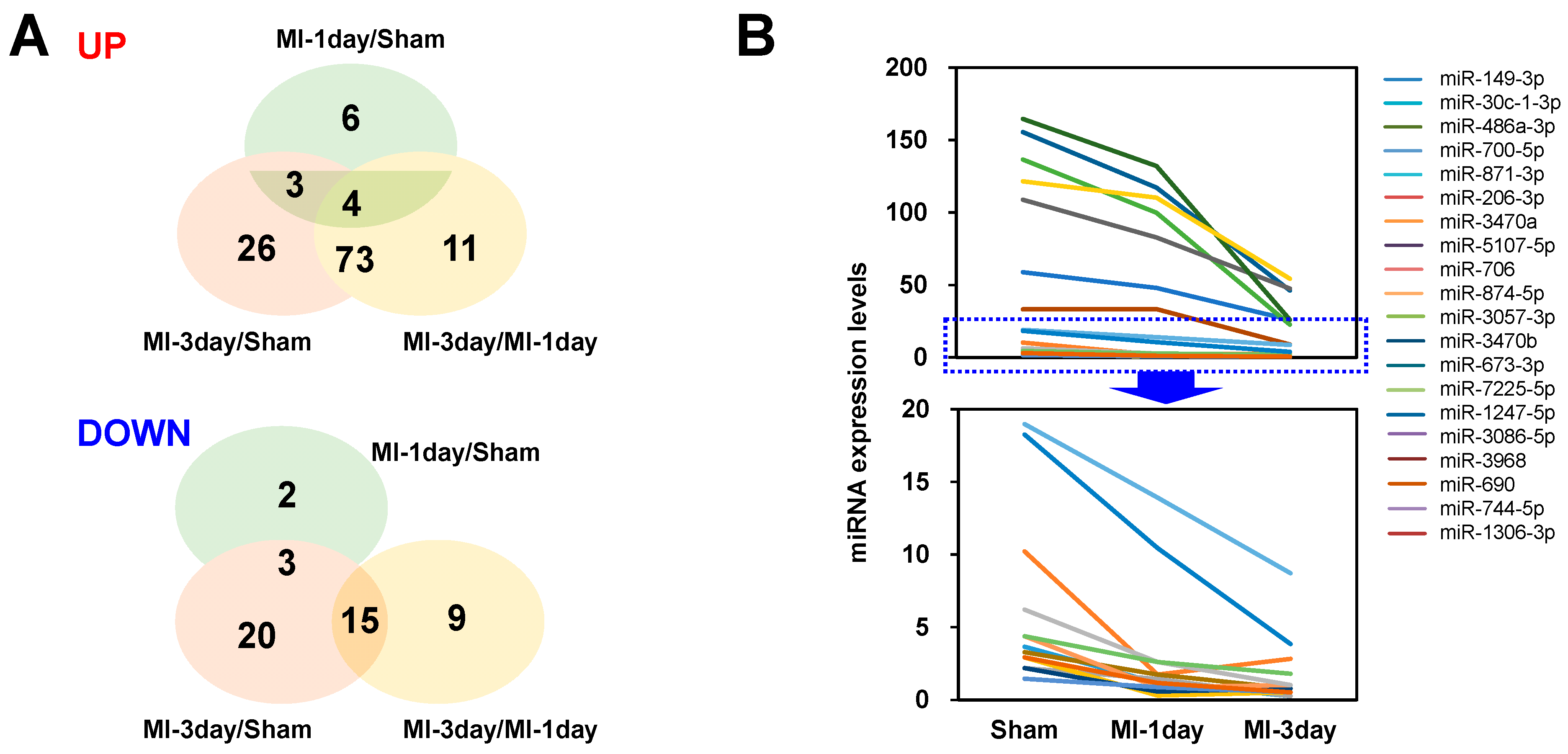
3.3. Differential Expressed (DE) miRNA Analysis
3.4. Selection of Differentially Expressed miRNAs
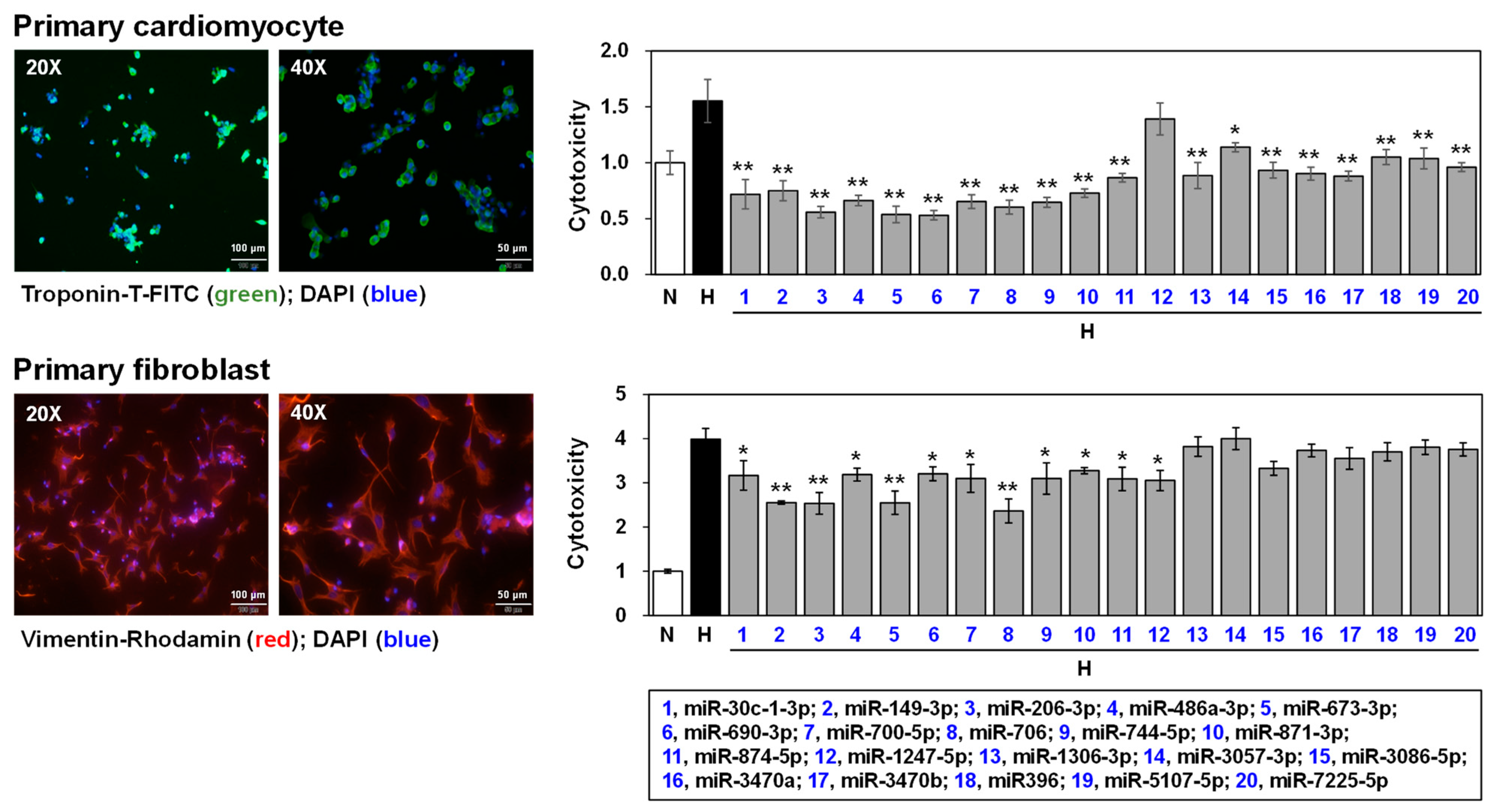
3.5. Effects of Selected miRNAs on Hypoxic Stress-Induced Cell Death
4. Discussion
4.1. Establishment of MI Model
4.2. Exosome Analysis
4.3. RNA Sequencing and Analysis
4.4. Functional Assay
4.5. Limitations, Strength, and Perspectives of This Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reed, G.W.; Rossi, J.E.; Cannon, C.P. Acute Myocardial Infarction. Lancet 2017, 389, 197–210. [Google Scholar] [CrossRef]
- Sachdeva, P.; Kaur, K.; Fatima, S.; Mahak, F.; Noman, M.; Siddenthi, S.M.; Surksha, M.A.; Munir, M.; Fatima, F.; Sultana, S.S.; et al. Advancements in Myocardial Infarction Management: Exploring Novel Approaches and Strategies. Cureus 2023, 15, e45578. [Google Scholar] [CrossRef]
- Sun, T.; Dong, Y.H.; Du, W.; Shi, C.Y.; Wang, K.; Tariq, M.A.; Wang, J.X.; Li, P.F. The Role of MicroRNAs in Myocardial Infarction: From Molecular Mechanism to Clinical Application. Int. J. Mol. Sci. 2017, 18, 745. [Google Scholar] [CrossRef]
- Kadesjo, E.; Roos, A.; Siddiqui, A.; Desta, L.; Lundback, M.; Holzmann, M.J. Acute versus Chronic Myocardial Injury and Long-term Outcomes. Heart 2019, 105, 1905–1912. [Google Scholar] [CrossRef]
- Chapman, A.R.; Adamson, P.D.; Mills, N.L. Assessment and Classification of Patients with Myocardial Injury and Infarction in Clinical Practice. Heart 2017, 103, 10–18. [Google Scholar] [CrossRef]
- Yasuda, S.; Shimokawa, H. Acute Myocardial Infarction: The Enduring Challenge for Cardiac Protection and Survival. Circ. J. 2009, 73, 2000–2008. [Google Scholar] [CrossRef]
- Ounpuu, S.; Negassa, A.; Yusuf, S. INTER-HEART: A Global Study of Risk Factors for Acute Myocardial Infarction. Am. Heart J. 2001, 141, 711–721. [Google Scholar] [CrossRef]
- Soares, R.O.S.; Losada, D.M.; Jordani, M.C.; Evora, P.; Castro, E.S.O. Ischemia/Reperfusion Injury Revisited: An Overview of the Latest Pharmacological Strategies. Int. J. Mol. Sci. 2019, 20, 5034. [Google Scholar] [CrossRef]
- Harishkumar, M.; Radha, M.; Yuichi, N.; Muthukalianan, G.K.; Kaoru, O.; Shiomori, K.; Sakai, K.; Nozomi, W. Designer Exosomes: Smart Nano-communication Tools for Translational Medicine. Bioengineering 2021, 8, 158. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yue, B.L.; Huang, Y.Z.; Lan, X.Y.; Liu, W.J.; Chen, H. Exosomal RNAs: Novel Potential Biomarkers for Diseases—A Review. Int. J. Mol. Sci. 2022, 23, 2461. [Google Scholar] [CrossRef]
- Narang, P.; Shah, M.; Beljanski, V. Exosomal RNAs in Diagnosis and Therapies. Non-Coding RNA Res. 2022, 7, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Dykes, I.M. Exosomes in Cardiovascular Medicine. Cardiol. Ther. 2017, 6, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Emanueli, C.; Shearn, A.I.; Angelini, G.D.; Sahoo, S. Exosomes and Exosomal MiRNAs in Cardiovascular Protection and Repair. Vascul. Pharmacol. 2015, 71, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Chopp, M.; Zhang, Z.G. Emerging Potential of Exosomes and Noncoding MicroRNAs for the Treatment of Neurological Injury/diseases. Expert Opin. Emerg. Drugs 2015, 20, 523–526. [Google Scholar] [CrossRef]
- O’Brien, K.; Lowry, M.C.; Corcoran, C.; Martinez, V.G.; Daly, M.; Rani, S.; Gallagher, W.M.; Radomski, M.W.; MacLeod, R.A.; O’Driscoll, L. MiR-134 in Extracellular Vesicles Reduces Triple-negative Breast Cancer Aggression and Increases Drug Sensitivity. Oncotarget 2015, 6, 32774–32789. [Google Scholar] [CrossRef]
- Tian, J.; Popal, M.S.; Zhao, Y.; Liu, Y.; Chen, K.; Liu, Y. Interplay between Exosomes and Autophagy in Cardiovascular Diseases: Novel Promising Target for Diagnostic and Therapeutic Application. Aging Dis. 2019, 10, 1302–1310. [Google Scholar] [CrossRef]
- Xue, R.; Tan, W.; Wu, Y.; Dong, B.; Xie, Z.; Huang, P.; He, J.; Dong, Y.; Liu, C. Role of Exosomal miRNAs in Heart Failure. Front. Cardiovasc. Med. 2020, 7, 592412. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Boon, R.A.; Dimmeler, S. MicroRNAs in Myocardial Infarction. Nat. Rev. Cardiol. 2015, 12, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Chen, J.; Zhao, Y.; Gao, D.; Wang, H.; Pu, J. A Modified Simple Method for Induction of Myocardial Infarction in Mice. J. Vis. Exp. 2021, 3, 178. [Google Scholar]
- Forte, E.; Skelly, D.A.; Chen, M.; Daigle, S.; Morelli, K.A.; Hon, O.; Philip, V.M.; Costa, M.W.; Rosenthal, N.A.; Furtado, M.B. Dynamic Interstitial Cell Response during Myocardial Infarction Predicts Resilience to Rupture in Genetically Diverse Mice. Cell Rep. 2020, 3, 3149–3163.e6. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Choi, J.W.; Moon, H.; Lee, C.Y.; Park, J.H.; Lee, J.; Seo, H.H.; Han, G.; Lim, S.; Kim, S.W.; et al. Stimultaneous Suppression of Multiple Programmed Cell Death Pathways by miRNA-105 in Cardiac Ischemic Injury. Mol. Ther. Nucleic Acids 2019, 1, 438–449. [Google Scholar] [CrossRef]
- Chang, X.; Zhang, K.; Zhou, R.; Luo, F.; Zhu, L.; Gao, J.; He, H.; Wei, T.; Yan, T.; Ma, C. Cardioprotective Effects of Salidroside on Myocardial Ischemia-reperfusion Injury in Coronary Artery Occlusion-induced Rats and Langendorff-perfused Rat Hearts. Int. J. Cardiol. 2016, 15, 532–544. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, S.R.; Lee, C.; Jun, Y.I.; Bae, S.; Yoon, Y.J.; Kim, O.Y.; Gho, Y.S. Extracellular Vesicles from In Vivo Liver Tissue Accelerate Recovery of Liver Necrosis Induced by Carbon Tetrachloride. J. Extracell. Vesicles 2021, 10, e12133. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Yuan, W.; Zhou, Y.; Ma, R.; Ge, Z. Small RNA Sequencing and Bioinformatics Analysis of RAW264.7-derived Exosomes after Mycobacterium Bovis Bacillus Calmette-Guerin Infection. BMC Genomics 2022, 7, 355. [Google Scholar] [CrossRef] [PubMed]
- Cizmar, P.; Yuana, Y. Detection and Characterization of Extracellular Vesicles by Transmission and Cryo-Transmission Electron Microscopy. Methods Mol. Biol. 2017, 1660, 221–232. [Google Scholar] [PubMed]
- Choi, J.W.; Lim, S.; Kang, J.H.; Hwang, S.H.; Hwang, K.C.; Kim, S.W.; Lee, S. Proteome Analysis of Human Natural Killer Cell Derived Extracellular Vesicles for Identification of Anticancer Effectors. Molecules 2020, 9, 5216. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.; Tiwari, S.; Gomes, A.V. Protein Purification and Analysis: Next Generation Western Blotting Techniques. Expert Rev. Proteomics 2017, 14, 1037–1053. [Google Scholar] [CrossRef]
- Jung, S.E.; Kim, S.W.; Jeong, S.; Moon, H.; Choi, W.S.; Lim, S.; Lee, S.; Hwang, K.C.; Choi, J.W. MicroRNA-26a/b-5p Promotes Myocardial Infarction-induced Cell Death by Downregulation Cytochrome C Oxidase 5a. Exp. Mol. Med. 2021, 53, 1332–1343. [Google Scholar] [CrossRef]
- Pokrowiecki, R.; Zaręba, T.; Szaraniec, B.; Pałka, K.; Mielczarek, A.; Menaszek, E.; Tyski, S. In Vitro Studies of Nanosilver-doped Titanium Implants for Oral and Maxillofacial Surgery. Int. J. Nanomed. 2017, 6, 4285–4297. [Google Scholar] [CrossRef]
- Oliveria, S.R.; Dionisio, P.A.; Brito, H.; Franco, L.; Rodrigues, C.A.B.; Guedes, R.C.; Afonso, C.A.M.; Amaral, J.D.; Rodrigues, C.M.P. Phenotypic Screening Identifies a New Oxazolone Inhibitor of Necroptosis and Neuroinflammation. Cell Death. Dicov. 2018, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Katsumata, O. Heat-Induced Antigen Retrieval in Immunohistochemistry: Mechanisms and Applications. Methods Mol. Bio. 2017, 1630, 147–161. [Google Scholar]
- Wu, Y.; Yin, X.; Wijaya, C.; Huang, M.H.; McConnell, B.K. Acute Myocardial Infarction in Rats. J. Vis. Exp. 2011, 16, 2464. [Google Scholar]
- Hashmi, S.; Al-Salam, S. Acute Myocardial Infarction and Myocardial Ischemia-reperfusion Injury: A Comparison. Int. J. Clin. Exp. Pathol. 2015, 8, 8786–8796. [Google Scholar]
- Lindsey, M.L.; Brunt, K.R.; Kirk, J.A.; Kleinbongard, P.; Calvert, J.W.; de Castro Bras, L.E.; DeLeon-Pennell, K.Y.; Del Re, D.P.; Frangogiannis, N.G.; Frantz, S.; et al. Guidelines for In Vivo Mouse Models of Myocardial Infarction. Am. J. Physiol. Heart Circ. Physiol. 2021, 321, H1056–H1073. [Google Scholar] [CrossRef]
- Weil, B.R.; Neelamegham, S. Selectins and Immune Cells in Acute Myocardial Infarction and Post-infarction Ventricular Remodeling: Pathophysiology and Novel Treatments. Front. Immunol. 2019, 10, 300. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, S.N.; Olcese, J.M.; Meckes, D.G., Jr. Extraction of Extracellular Vesicles from Whole Tissue. J. Vis. Exp. 2019, 144, e59143. [Google Scholar]
- Walkowski, B.; Kleibert, M.; Majka, M.; Wojciechowska, M. Insight into the Role of the PI3K/Akt Pathway in Ischemic Injury and Post-Infarct Left Ventricular Remodeling in Normal and Diabetic Heart. Cells 2022, 11, 1553. [Google Scholar] [CrossRef]
- Colombe, A.S.; Pidoux, G. Cardiac cAMP-PKA Signaling Compartmentalization in Myocardial Infarction. Cells 2021, 10, 922. [Google Scholar] [CrossRef]
- Guo, H.; Zhao, X.; Li, H.; Liu, K.; Jiang, H.; Zeng, X.; Chang, J.; Ma, C.; Fu, Z.; Lv, X.; et al. GDF15 Promotes Cardiac Fibrosis and Proliferation of Cardiac Fibroblasts via the MAPK/ERK1/2 Pathway after Irradiation in Rats. Radiat. Res. 2021, 196, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Lyu, L.; Wang, H.; Li, B.; Qin, Q.; Qi, L.; Nagarkatti, M.; Nagarkatti, P.; Janicki, J.S.; Wang, X.L.; Cui, T. A Critical Role of Cardiac Fibroblast-derived Exosomes in Activating Renin Angiotensin System in Cardiomyocytes. J. Mol. Cell. Cardiol. 2015, 89, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Yu, J. Lidocaine Protects H9c2 Cells from Hypoxia-induced Injury through Regulation of the MAPK/ERK/NFκB Signaling Pathway. Exp. Ther. Med. 2019, 18, 4125–4131. [Google Scholar] [PubMed]
- Cai, Y.; Li, Y. Upregulation of MiR-29b-3p Protects Cardiomyocytes from Hypoxia-induced Apoptosis by Targeting TRAF5. Cell. Mol. Biol. Lett. 2019, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Pyo, J.O.; Nah, J.; Kim, H.J.; Chang, J.W.; Song, Y.W.; Yang, D.K.; Jo, D.G.; Kim, H.R.; Chae, H.J.; Chae, S.W.; et al. Protection of Cardiomyocytes from Ischemic/hypoxia Cell Death via Drbp1 and pMe2GlyDH in Cardio-specific ARC transgenic mice. J. Biol. Chem. 2008, 7, 30707–30714. [Google Scholar] [CrossRef]
- Datta Chaudhuri, R.; Banik, A.; Mandal, B.; Sarkar, S. Cardia-specific overexpression of HIF-1α during acute myocardial infarction ameliorates cardiomyocyte apoptosis via differential regulation of hypoxia-inducible pro-apoptotic and anti-oxidative genes. Biochem. Biophys. Res. Commun. 2021, 22, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Pu, W.T. Recounting Cardiac Cellular Composition. Circ. Res. 2016, 118, 368–370. [Google Scholar] [CrossRef]
- Banerjee, I.; Fuseler, J.W.; Price, R.L.; Borg, T.K.; Baudino, T.A. Determination of Cell Types and Numbers during Cardiac Development in the Neonatal and Adult Rat and Mouse. Am. J. Physiol. Heart Circ. Physiol. 2007, 293, H1883–H1891. [Google Scholar] [CrossRef]
- Zhang, L.S.; Zhou, Y.D.; Peng, Y.Q.; Zeng, H.L.; Yoshida, S.; Zhao, T.T. Identification of Altered MicroRNAs in Retinas of Mice with Oxygen-induced Retinopathy. Int. J. Ophthalmol. 2019, 12, 739–745. [Google Scholar]
- Lu, M.; Xu, L.; Wang, M.; Guo, T.; Luo, F.; Su, N.; Yi, S.; Chen, T. MiR-149 Promotes the Myocardial Differentiation of Mouse Bone Marrow Stem Cells by Targeting Dab2. Mol. Med. Rep. 2018, 17, 8502–8509. [Google Scholar] [CrossRef]
- Yin, R.; Guo, D.; Zhang, S.; Zhang, X. MiR-706 Inhibits the Oxidative Stress-induced Activation of PKCalpha/TAOK1 in Liver Fibrogenesis. Sci. Rep. 2016, 6, 37509. [Google Scholar] [CrossRef]
- Guo, M.; Li, R.; Yang, L.; Zhu, Q.; Han, M.; Chen, Z.; Ruan, F.; Yuan, Y.; Liu, Z.; Huang, B.; et al. Evaluation of Exosomal MiRNAs as Potential Diagnostic Biomarkers for Acute Myocardial Infarction Using Next-generation Sequencing. Ann. Transl. Med. 2021, 9, 219. [Google Scholar] [CrossRef]



| No. | Target Name | Accession# | Sequence (5′–3′) |
|---|---|---|---|
| 1 | mmu-miR-30c-1-3p | MIMAT0004616 | CUGGGAGAGGGUUGUUUACUCC |
| 2 | mmu-miR-149-3p | MIMAT0016990 | GAGGGAGGGACGGGGGCGGUGC |
| 3 | mmu-miR-206-3p | MIMAT0000239 | UGGAAUGUAAGGAAGUGUGUGG |
| 4 | mmu-miR-486a-3p | MIMAT0017206 | CGGGGCAGCUCAGUACAGGAU |
| 5 | mmu-miR-673-3p | MIMAT0004824 | UCCGGGGCUGAGUUCUGUGCACC |
| 6 | mmu-miR-690 | MIMAT0003469 | AAAGGCUAGGCUCACAACCAAA |
| 7 | mmu-miR-700-5p | MIMAT0017256 | UAAGGCUCCUUCCUGUGCUUGC |
| 8 | mmu-miR-706 | MIMAT0003496 | AGAGAAACCCUGUCUCAAAAAA |
| 9 | mmu-miR-744-5p | MIMAT0004187 | UGCGGGGCUAGGGCUAACAGCA |
| 10 | mmu-miR-871-3p | MIMAT0017265 | UGACUGGCACCAUUCUGGAUAAU |
| 11 | mmu-miR-874-5p | MIMAT0017268 | CGGCCCCACGCACCAGGGUAAG |
| 12 | mmu-miR-1247-5p | MIMAT0014800 | ACCCGUCCCGUUCGUCCCCGGA |
| 13 | mmu-miR-1306-3p | MIMAT0009411 | ACGUUGGCUCUGGUGGUGAUG |
| 14 | mmu-miR-3057-3p | MIMAT0014823 | UCCCACAGGCCCAGCUCAUAGC |
| 15 | mmu-miR-3086-5p | MIMAT0014880 | UAGAUUGUAGGCCCAUUGGA |
| 16 | mmu-miR-3470a | MIMAT0015640 | UCACUUUGUAGACCAGGCUGG |
| 17 | mmu-miR-3470b | MIMAT0015641 | UCACUCUGUAGACCAGGCUGG |
| 18 | mmu-miR-3968 | MIMAT0019352 | CGAAUCCCACUCCAGACACCA |
| 19 | mmu-miR-5107-5p | MIMAT0020615 | UGGGCAGAGGAGGCAGGGACA |
| 20 | mmu-miR-7225-5p | MIMAT0028418 | ACGUAGACUGUGUAGAAGCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, S.E.; Kim, S.W.; Choi, J.-W. Exploring Cardiac Exosomal RNAs of Acute Myocardial Infarction. Biomedicines 2024, 12, 430. https://doi.org/10.3390/biomedicines12020430
Jung SE, Kim SW, Choi J-W. Exploring Cardiac Exosomal RNAs of Acute Myocardial Infarction. Biomedicines. 2024; 12(2):430. https://doi.org/10.3390/biomedicines12020430
Chicago/Turabian StyleJung, Seung Eun, Sang Woo Kim, and Jung-Won Choi. 2024. "Exploring Cardiac Exosomal RNAs of Acute Myocardial Infarction" Biomedicines 12, no. 2: 430. https://doi.org/10.3390/biomedicines12020430





