Induction of FoxP3 Pre-mRNA Alternative Splicing to Enhance the Suppressive Activity of Regulatory T Cells from Amyotrophic Lateral Sclerosis Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Demographic Characteristics of Study Participants
2.2. Blood Sampling and Treg Purification
2.3. Flow Cytometry of Treg-Associated Cell Markers
2.4. Treg Transfection and Cultivation
2.5. Quantitative PCR (qPCR) and Western Blotting
2.6. Suppression Assays and the Detection of Suppressive Cytokines
2.7. Statistics
3. Results
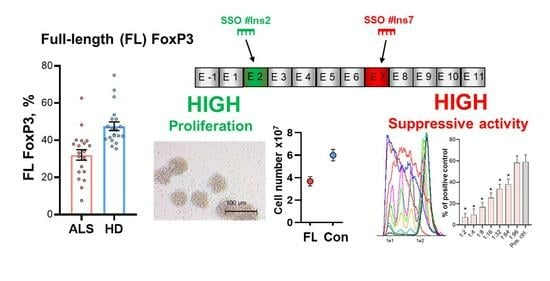
3.1. FoxP3 Full-Length Splice Variant Expression Is Reduced in Tregs from ALS Patients
3.2. The Induction of FL FoxP3 Splice Variant with Specific SSOs
3.3. Immunophenotype of Tregs with FL FoxP3
3.4. Proliferative Activity of Tregs with Selective Expression of FL FoxP3 Splice Variant
3.5. Suppressive Activity of Tregs with Selective Expression of FL FoxP3 Splice Variant
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.-F.; Zhang, S.-X.; Ma, X.-W.; Xue, Y.-L.; Gao, C.; Li, X.-Y.; Xu, A.-D. The Proportion of Peripheral Regulatory T Cells in Patients with Multiple Sclerosis: A Meta-Analysis. Mult. Scler. Relat. Disord. 2019, 28, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Venken, K.; Hellings, N.; Thewissen, M.; Somers, V.; Hensen, K.; Rummens, J.-L.; Medaer, R.; Hupperts, R.; Stinissen, P. Compromised CD4+ CD25(High) Regulatory T-Cell Function in Patients with Relapsing-Remitting Multiple Sclerosis Is Correlated with a Reduced Frequency of FOXP3-Positive Cells and Reduced FOXP3 Expression at the Single-Cell Level. Immunology 2008, 123, 79–89. [Google Scholar] [CrossRef]
- Bonelli, M.; Savitskaya, A.; von Dalwigk, K.; Steiner, C.W.; Aletaha, D.; Smolen, J.S.; Scheinecker, C. Quantitative and Qualitative Deficiencies of Regulatory T Cells in Patients with Systemic Lupus Erythematosus (SLE). Int. Immunol. 2008, 20, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Miyara, M.; Gorochov, G.; Ehrenstein, M.; Musset, L.; Sakaguchi, S.; Amoura, Z. Human FoxP3+ Regulatory T Cells in Systemic Autoimmune Diseases. Autoimmun. Rev. 2011, 10, 744–755. [Google Scholar] [CrossRef] [PubMed]
- Valencia, X.; Yarboro, C.; Illei, G.; Lipsky, P.E. Deficient CD4+CD25high T Regulatory Cell Function in Patients with Active Systemic Lupus Erythematosus. J. Immunol. 2007, 178, 2579–2588. [Google Scholar] [CrossRef]
- Haseda, F.; Imagawa, A.; Murase-Mishiba, Y.; Terasaki, J.; Hanafusa, T. CD4+ CD45RA−FoxP3high Activated Regulatory T Cells Are Functionally Impaired and Related to Residual Insulin-Secreting Capacity in Patients with Type 1 Diabetes. Clin. Exp. Immunol. 2013, 173, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Thiruppathi, M.; Rowin, J.; Li Jiang, Q.; Sheng, J.R.; Prabhakar, B.S.; Meriggioli, M.N. Functional Defect in Regulatory T Cells in Myasthenia Gravis. Ann. N. Y. Acad. Sci. 2012, 1274, 68–76. [Google Scholar] [CrossRef] [PubMed]
- van Roon, J.A.G.; Hartgring, S.A.Y.; van der Wurff-Jacobs, K.M.G.; Bijlsma, J.W.J.; Lafeber, F.P.J.G. Numbers of CD25+Foxp3+ T Cells That Lack the IL-7 Receptor Are Increased Intra-Articularly and Have Impaired Suppressive Function in RA Patients. Rheumatology 2010, 49, 2084–2089. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Villar, M.; Hafler, D.A. Regulatory T Cells in Autoimmune Disease. Nat. Immunol. 2018, 19, 665–673. [Google Scholar] [CrossRef]
- Chiò, A.; Logroscino, G.; Hardiman, O.; Swingler, R.; Mitchell, D.; Beghi, E.; Traynor, B.G. Prognostic Factors in ALS: A Critical Review. Amyotroph. Lateral Scler. 2009, 10, 310–323. [Google Scholar] [CrossRef]
- De Marchi, F.; Munitic, I.; Amedei, A.; Berry, J.D.; Feldman, E.L.; Aronica, E.; Nardo, G.; Van Weehaeghe, D.; Niccolai, E.; Prtenjaca, N.; et al. Interplay between Immunity and Amyotrophic Lateral Sclerosis: Clinical Impact. Neurosci. Biobehav. Rev. 2021, 127, 958–978. [Google Scholar] [CrossRef] [PubMed]
- Beers, D.R.; Appel, S.H. Immune Dysregulation in Amyotrophic Lateral Sclerosis: Mechanisms and Emerging Therapies. Lancet. Neurol. 2019, 18, 211–220. [Google Scholar] [CrossRef]
- Lyon, M.S.; Wosiski-Kuhn, M.; Gillespie, R.; Caress, J.; Milligan, C. Inflammation, Immunity, and Amyotrophic Lateral Sclerosis: I. Etiology and Pathology. Muscle Nerve 2019, 59, 10–22. [Google Scholar] [CrossRef] [PubMed]
- McCauley, M.E.; Baloh, R.H. Inflammation in ALS/FTD Pathogenesis. Acta Neuropathol. 2019, 137, 715–730. [Google Scholar] [CrossRef] [PubMed]
- Beers, D.R.; Zhao, W.; Wang, J.; Zhang, X.; Wen, S.; Neal, D.; Thonhoff, J.R.; Alsuliman, A.S.; Shpall, E.J.; Rezvani, K.; et al. ALS Patients’ Regulatory T Lymphocytes Are Dysfunctional, and Correlate with Disease Progression Rate and Severity. JCI Insight 2017, 2, e89530. [Google Scholar] [CrossRef]
- Henkel, J.S.; Beers, D.R.; Wen, S.; Rivera, A.L.; Toennis, K.M.; Appel, J.E.; Zhao, W.; Moore, D.H.; Powell, S.Z.; Appel, S.H. Regulatory T-Lymphocytes Mediate Amyotrophic Lateral Sclerosis Progression and Survival. EMBO Mol. Med. 2013, 5, 64–79. [Google Scholar] [CrossRef]
- Yazdani, S.; Seitz, C.; Cui, C.; Lovik, A.; Pan, L.; Piehl, F.; Pawitan, Y.; Kläppe, U.; Press, R.; Samuelsson, K.; et al. T Cell Responses at Diagnosis of Amyotrophic Lateral Sclerosis Predict Disease Progression. Nat. Commun. 2022, 13, 6733. [Google Scholar] [CrossRef]
- Lu, L.; Barbi, J.; Pan, F. The Regulation of Immune Tolerance by FOXP3. Nature reviews. Immunology 2017, 17, 703–717. [Google Scholar] [CrossRef]
- Pesenacker, A.M.; Cook, L.; Levings, M.K. The Role of FOXP3 in Autoimmunity. Curr. Opin. Immunol. 2016, 43, 16–23. [Google Scholar] [CrossRef]
- Tao, J.-H.; Cheng, M.; Tang, J.-P.; Liu, Q.; Pan, F.; Li, X.-P. Foxp3, Regulatory T Cell, and Autoimmune Diseases. Inflammation 2017, 40, 328–339. [Google Scholar] [CrossRef]
- Mercer, F.; Unutmaz, D. The Biology of FoxP3: A Key Player in Immune Suppression during Infections, Autoimmune Diseases and Cancer. Adv. Exp. Med. Biol. 2009, 665, 47–59. [Google Scholar] [CrossRef]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Rudensky, A.Y. Regulatory T Cells and Foxp3. Immunol. Rev. 2011, 241, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Liu, J.; Lee, E.; Perriman, R.; Roncarolo, M.G.; Bacchetta, R. Co-Expression of FOXP3FL and FOXP3Δ2 Isoforms Is Required for Optimal Treg-Like Cell Phenotypes and Suppressive Function. Front. Immunol. 2021, 12, 752394. [Google Scholar] [CrossRef] [PubMed]
- Blinova, V.G.; Novachly, N.S.; Gippius, S.N.; Hilal, A.; Gladilina, Y.A.; Eliseeva, D.D.; Zhdanov, D.D. Phenotypical and Functional Characteristics of Human Regulatory T Cells during Ex Vivo Maturation from CD4+ T Lymphocytes. Appl. Sci. 2021, 11, 154–165. [Google Scholar] [CrossRef]
- Du, J.; Wang, Q.; Yang, S.; Chen, S.; Fu, Y.; Spath, S.; Domeier, P.; Hagin, D.; Anover-Sombke, S.; Haouili, M.; et al. FOXP3 Exon 2 Controls T(Reg) Stability and Autoimmunity. Sci. Immunol. 2022, 7, eabo5407. [Google Scholar] [CrossRef]
- Seitz, C.; Joly, A.-L.; Fang, F.; Frith, K.; Gray, P.; Andersson, J. The FOXP3 Full-Length Isoform Controls the Lineage-Stability of CD4(+)FOXP3(+) Regulatory T Cells. Clin. Immunol. 2022, 237, 108957. [Google Scholar] [CrossRef]
- Sambucci, M.; Gargano, F.; De Rosa, V.; De Bardi, M.; Picozza, M.; Placido, R.; Ruggieri, S.; Capone, A.; Gasperini, C.; Matarese, G.; et al. FoxP3 Isoforms and PD-1 Expression by T Regulatory Cells in Multiple Sclerosis. Sci. Rep. 2018, 8, 3674. [Google Scholar] [CrossRef]
- Blinova, V.G.; Gladilina, Y.A.; Abramova, A.A.; Eliseeva, D.D.; Vtorushina, V.V.; Shishparenok, A.N.; Zhdanov, D.D. Modulation of Suppressive Activity and Proliferation of Human Regulatory T Cells by Splice-Switching Oligonucleotides Targeting FoxP3 Pre-MRNA. Cells 2023, 13, 77. [Google Scholar] [CrossRef]
- Roche, J.C.; Rojas-Garcia, R.; Scott, K.M.; Scotton, W.; Ellis, C.E.; Burman, R.; Wijesekera, L.; Turner, M.R.; Leigh, P.N.; Shaw, C.E.; et al. A Proposed Staging System for Amyotrophic Lateral Sclerosis. Brain J. Neurol. 2012, 135, 847–852. [Google Scholar] [CrossRef]
- Cedarbaum, J.M.; Stambler, N.; Malta, E.; Fuller, C.; Hilt, D.; Thurmond, B.; Nakanishi, A. The ALSFRS-R: A Revised ALS Functional Rating Scale That Incorporates Assessments of Respiratory Function. BDNF ALS Study Group (Phase III). J. Neurol. Sci. 1999, 169, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Miller, R.G.; Swash, M.; Munsat, T.L. El Escorial Revisited: Revised Criteria for the Diagnosis of Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Other Mot. Neuron 2000, 1, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.F.; Lot, S.S.; Condon, T.P.; Cheng-Flournoy, S.; Lesnik, E.A.; Sasmor, H.M.; Bennett, C.F. 2′-O-(2-Methoxy)Ethyl-Modified Anti-Intercellular Adhesion Molecule 1 (ICAM-1) Oligonucleotides Selectively Increase the ICAM-1 MRNA Level and Inhibit Formation of the ICAM-1 Translation Initiation Complex in Human Umbilical Vein Endothelial Cells. J. Biol. Chem. 1997, 272, 11994–12000. [Google Scholar] [CrossRef] [PubMed]
- Zhdanov, D.D.; Plyasova, A.A.; Gladilina, Y.A.; Pokrovsky, V.S.; Grishin, D.V.; Grachev, V.A.; Orlova, V.S.; Pokrovskaya, M.V.; Alexandrova, S.S.; Lobaeva, T.A.; et al. Inhibition of Telomerase Activity by Splice-Switching Oligonucleotides Targeting the MRNA of the Telomerase Catalytic Subunit Affects Proliferation of Human CD4+ T Lymphocytes. Biochem. Biophys. Res. Commun. 2019, 509, 790–796. [Google Scholar] [CrossRef] [PubMed]
- Lifshitz, G.V.; Zhdanov, D.D.; Lokhonina, A.V.; Eliseeva, D.D.; Lyssuck, E.Y.; Zavalishin, I.A.; Bykovskaia, S.N. Ex Vivo Expanded Regulatory T Cells CD4 + CD25 + FoxP3 + CD127 Low Develop Strong Immunosuppressive Activity in Patients with Remitting-Relapsing Multiple Sclerosis. Autoimmunity 2016, 49, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Sherley, J.L.; Stadler, P.B.; Stadler, J.S. A Quantitative Method for the Analysis of Mammalian Cell Proliferation in Culture in Terms of Dividing and Non-Dividing Cells. Cell Prolif. 1995, 28, 137–144. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Pokrovsky, V.S.; Orlova, E.V.; Orlova, V.S.; Pokrovskaya, M.V.; Aleksandrova, S.S.; Sokolov, N.N. Intracellular Localization of Apoptotic Endonuclease EndoG and Splice-Variants of Telomerase Catalytic Subunit HTERT. Biochemistry 2017, 82, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Zhdanov, D.D.; Gladilina, Y.A.; Pokrovsky, V.S.; Grishin, D.V.; Grachev, V.A.; Orlova, V.S.; Pokrovskaya, M.V.; Alexandrova, S.S.; Plyasova, A.A.; Sokolov, N.N. Endonuclease G Modulates the Alternative Splicing of Deoxyribonuclease 1 MRNA in Human CD4+ T Lymphocytes and Prevents the Progression of Apoptosis. Biochimie 2019, 157, 158–176. [Google Scholar] [CrossRef] [PubMed]
- Blinova, V.G.; Gladilina, Y.A.; Eliseeva, D.D.; Lobaeva, T.A.; Zhdanov, D.D. [Increased suppressor activity of transformed ex vivo regulatory T-cells in comparison with unstimulated cells of the same donor]. Biomeditsinskaia Khimiia 2022, 68, 55–67. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Vasina, D.A.; Orlova, E.V.; Orlova, V.S.; Pokrovskaya, M.V.; Aleksandrova, S.S.; Sokolov, N.N. Apoptotic Endonuclease EndoG Regulates Alternative Splicing of Human Telomerase Catalytic Subunit HTERT. Biochem. (Mosc.) Suppl. Ser. B Biomed. Chem. 2017, 11, 154–165. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific Association of Human Telomerase Activity with Immortal Cells and Cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Zhdanov, D.D.; Gladilina, Y.A.; Pokrovsky, V.S.; Grishin, D.V.; Grachev, V.A.; Orlova, V.S.; Pokrovskaya, M.V.; Alexandrova, S.S.; Sokolov, N.N. Murine Regulatory T Cells Induce Death of Effector T, B, and NK Lymphocytes through a Contact-Independent Mechanism Involving Telomerase Suppression and Telomere-Associated Senescence. Cell. Immunol. 2018, 331, 146–160. [Google Scholar] [CrossRef] [PubMed]
- Zhdanov, D.D.; Gladilina, Y.A.; Grishin, D.V.; Grachev, V.A.; Orlova, V.S.; Pokrovskaya, M.V.; Alexandrova, S.S.; Pokrovsky, V.S.; Sokolov, N.N. Contact-Independent Suppressive Activity of Regulatory T Cells Is Associated with Telomerase Inhibition, Telomere Shortening and Target Lymphocyte Apoptosis. Mol. Immunol. 2018, 101, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Bluestone, J.A.; Buckner, J.H.; Fitch, M.; Gitelman, S.E.; Gupta, S.; Hellerstein, M.K.; Herold, K.C.; Lares, A.; Lee, M.R.; Li, K.; et al. Type 1 Diabetes Immunotherapy Using Polyclonal Regulatory T Cells. Sci. Transl. Med. 2015, 7, 315ra189. [Google Scholar] [CrossRef] [PubMed]
- Voskens, C.J.; Fischer, A.; Roessner, S.; Lorenz, C.; Hirschmann, S.; Atreya, R.; Neufert, C.; Atreya, I.; Neurath, M.F.; Schuler, G. Characterization and Expansion of Autologous GMP-Ready Regulatory T Cells for TREG-Based Cell Therapy in Patients with Ulcerative Colitis. Inflamm. Bowel Dis. 2017, 23, 1348–1359. [Google Scholar] [CrossRef] [PubMed]
- Ghobadinezhad, F.; Ebrahimi, N.; Mozaffari, F.; Moradi, N.; Beiranvand, S.; Pournazari, M.; Rezaei-Tazangi, F.; Khorram, R.; Afshinpour, M.; Robino, R.A.; et al. The Emerging Role of Regulatory Cell-Based Therapy in Autoimmune Disease. Front. Immunol. 2022, 13, 1075813. [Google Scholar] [CrossRef] [PubMed]
- Buc, M. Role of Regulatory T Cells in Pathogenesis and Biological Therapy of Multiple Sclerosis. Mediat. Inflamm. 2013, 2013, 963748. [Google Scholar] [CrossRef] [PubMed]
- Nussbaum, L.; Chen, Y.L.; Ogg, G.S. Role of Regulatory T Cells in Psoriasis Pathogenesis and Treatment. Br. J. Dermatol. 2021, 184, 14–24. [Google Scholar] [CrossRef]
- Dall’Era, M.; Pauli, M.L.; Remedios, K.; Taravati, K.; Sandova, P.M.; Putnam, A.L.; Lares, A.; Haemel, A.; Tang, Q.; Hellerstein, M.; et al. Adoptive Treg Cell Therapy in a Patient with Systemic Lupus Erythematosus. Arthritis Rheumatol. 2019, 71, 431–440. [Google Scholar] [CrossRef]
- Brunstein, C.G.; Miller, J.S.; McKenna, D.H.; Hippen, K.L.; DeFor, T.E.; Sumstad, D.; Curtsinger, J.; Verneris, M.R.; MacMillan, M.L.; Levine, B.L.; et al. Umbilical Cord Blood-Derived T Regulatory Cells to Prevent GVHD: Kinetics, Toxicity Profile, and Clinical Effect. Blood 2016, 127, 1044–1051. [Google Scholar] [CrossRef]
- Giovannelli, I.; Heath, P.; Shaw, P.J.; Kirby, J. The Involvement of Regulatory T Cells in Amyotrophic Lateral Sclerosis and Their Therapeutic Potential. Amyotroph. Lateral Scler. Front. Degener. 2020, 21, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Rajabinejad, M.; Ranjbar, S.; Afshar Hezarkhani, L.; Salari, F.; Gorgin Karaji, A.; Rezaiemanesh, A. Regulatory T Cells for Amyotrophic Lateral Sclerosis/Motor Neuron Disease: A Clinical and Preclinical Systematic Review. J. Cell. Physiol. 2020, 235, 5030–5040. [Google Scholar] [CrossRef] [PubMed]
- Beers, D.R.; Zhao, W.; Appel, S.H. The Role of Regulatory T Lymphocytes in Amyotrophic Lateral Sclerosis. JAMA Neurol. 2018, 75, 656–658. [Google Scholar] [CrossRef] [PubMed]
- Delgoffe, G.M.; Kole, T.P.; Zheng, Y.; Zarek, P.E.; Matthews, K.L.; Xiao, B.; Worley, P.F.; Kozma, S.C.; Powell, J.D. The MTOR Kinase Differentially Regulates Effector and Regulatory T Cell Lineage Commitment. Immunity 2009, 30, 832–844. [Google Scholar] [CrossRef] [PubMed]
- Staats, K.A.; Hernandez, S.; Schönefeldt, S.; Bento-Abreu, A.; Dooley, J.; Van Damme, P.; Liston, A.; Robberecht, W.; Van Den Bosch, L. Rapamycin Increases Survival in ALS Mice Lacking Mature Lymphocytes. Mol. Neurodegener. 2013, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Vallarola, A.; Sironi, F.; Tortarolo, M.; Gatto, N.; De Gioia, R.; Pasetto, L.; De Paola, M.; Mariani, A.; Ghosh, S.; Watson, R.; et al. RNS60 Exerts Therapeutic Effects in the SOD1 ALS Mouse Model through Protective Glia and Peripheral Nerve Rescue. J. Neuroinflamm. 2018, 15, 65. [Google Scholar] [CrossRef]
- Paganoni, S.; Alshikho, M.J.; Luppino, S.; Chan, J.; Pothier, L.; Schoenfeld, D.; Andres, P.L.; Babu, S.; Zürcher, N.R.; Loggia, M.L.; et al. A Pilot Trial of RNS60 in Amyotrophic Lateral Sclerosis. Muscle Nerve 2019, 59, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Camu, W.; Tremblier, B.; Plassot, C.; Alphandery, S.; Salsac, C.; Pageot, N.; Juntas-Morales, R.; Scamps, F.; Daures, J.-P.; Raoul, C. Vitamin D Confers Protection to Motoneurons and Is a Prognostic Factor of Amyotrophic Lateral Sclerosis. Neurobiol. Aging 2014, 35, 1198–1205. [Google Scholar] [CrossRef]
- Karam, C.; Barrett, M.J.; Imperato, T.; MacGowan, D.J.L.; Scelsa, S. Vitamin D Deficiency and Its Supplementation in Patients with Amyotrophic Lateral Sclerosis. J. Clin. Neurosci. J. Neurosurg. Soc. Australas. 2013, 20, 1550–1553. [Google Scholar] [CrossRef]
- Schlöder, J.; Berges, C.; Luessi, F.; Jonuleit, H. Dimethyl Fumarate Therapy Significantly Improves the Responsiveness of T Cells in Multiple Sclerosis Patients for Immunoregulation by Regulatory T Cells. Int. J. Mol. Sci. 2017, 18, 271. [Google Scholar] [CrossRef]
- Kornberg, M.D.; Bhargava, P.; Kim, P.M.; Putluri, V.; Snowman, A.M.; Putluri, N.; Calabresi, P.A.; Snyder, S.H. Dimethyl Fumarate Targets GAPDH and Aerobic Glycolysis to Modulate Immunity. Science 2018, 360, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Tenspolde, M.; Zimmermann, K.; Weber, L.C.; Hapke, M.; Lieber, M.; Dywicki, J.; Frenzel, A.; Hust, M.; Galla, M.; Buitrago-Molina, L.E.; et al. Regulatory T Cells Engineered with a Novel Insulin-Specific Chimeric Antigen Receptor as a Candidate Immunotherapy for Type 1 Diabetes. J. Autoimmun. 2019, 103, 102289. [Google Scholar] [CrossRef] [PubMed]
- Elinav, E.; Adam, N.; Waks, T.; Eshhar, Z. Amelioration of Colitis by Genetically Engineered Murine Regulatory T Cells Redirected by Antigen-Specific Chimeric Receptor. Gastroenterology 2009, 136, 1721–1731. [Google Scholar] [CrossRef] [PubMed]
- Fransson, M.; Piras, E.; Burman, J.; Nilsson, B.; Essand, M.; Lu, B.; Harris, R.A.; Magnusson, P.U.; Brittebo, E.; Loskog, A.S. CAR/FoxP3-Engineered T Regulatory Cells Target the CNS and Suppress EAE upon Intranasal Delivery. J. Neuroinflamm. 2012, 9, 112. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Schmidt, A.; Zhang, A.-H.; Königs, C.; Kim, Y.C.; Scott, D.W. FVIII-Specific Human Chimeric Antigen Receptor T-Regulatory Cells Suppress T- and B-Cell Responses to FVIII. Blood 2017, 129, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Mukhatayev, Z.; Dellacecca, E.R.; Cosgrove, C.; Shivde, R.; Jaishankar, D.; Pontarolo-Maag, K.; Eby, J.M.; Henning, S.W.; Ostapchuk, Y.O.; Cedercreutz, K.; et al. Antigen Specificity Enhances Disease Control by Tregs in Vitiligo. Front. Immunol. 2020, 11, 581433. [Google Scholar] [CrossRef]
- Pierini, A.; Iliopoulou, B.P.; Peiris, H.; Pérez-Cruz, M.; Baker, J.; Hsu, K.; Gu, X.; Zheng, P.-P.; Erkers, T.; Tang, S.-W.; et al. T Cells Expressing Chimeric Antigen Receptor Promote Immune Tolerance. JCI Insight 2017, 2, e92865. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, K.G.; Hoeppli, R.E.; Huang, Q.; Gillies, J.; Luciani, D.S.; Orban, P.C.; Broady, R.; Levings, M.K. Alloantigen-Specific Regulatory T Cells Generated with a Chimeric Antigen Receptor. J. Clin. Investig. 2016, 126, 1413–1424. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.; Cheekatla, S.S.; Paidipally, P.; Radhakrishnan, R.K.; Welch, E.; Thandi, R.S.; Tvinnereim, A.R.; Vankayalapati, R. C-Jun N-Terminal Kinase 1 Defective CD4+CD25+FoxP3+ Cells Prolong Islet Allograft Survival in Diabetic Mice. Sci. Rep. 2018, 8, 3310. [Google Scholar] [CrossRef]
- Lynn, R.C.; Weber, E.W.; Sotillo, E.; Gennert, D.; Xu, P.; Good, Z.; Anbunathan, H.; Lattin, J.; Jones, R.; Tieu, V.; et al. C-Jun Overexpression in CAR T Cells Induces Exhaustion Resistance. Nature 2019, 576, 293–300. [Google Scholar] [CrossRef]
- Hirai, T.; Ramos, T.L.; Lin, P.-Y.; Simonetta, F.; Su, L.L.; Picton, L.K.; Baker, J.; Lin, J.-X.; Li, P.; Seo, K.; et al. Selective Expansion of Regulatory T Cells Using an Orthogonal IL-2/IL-2 Receptor System Facilitates Transplantation Tolerance. J. Clin. Investig. 2021, 131, e139991. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hresko, M.E.; Picton, L.K.; Su, L.; Hollander, M.J.; Nunez-Cruz, S.; Zhang, Z.; Assenmacher, C.-A.; Sockolosky, J.T.; Garcia, K.C.; et al. A Human Orthogonal IL-2 and IL-2Rβ System Enhances CAR T Cell Expansion and Antitumor Activity in a Murine Model of Leukemia. Sci. Transl. Med. 2021, 13, eabg6986. [Google Scholar] [CrossRef] [PubMed]
- Honaker, Y.; Hubbard, N.; Xiang, Y.; Fisher, L.; Hagin, D.; Sommer, K.; Song, Y.; Yang, S.J.; Lopez, C.; Tappen, T.; et al. Gene Editing to Induce FOXP3 Expression in Human CD4+ T Cells Leads to a Stable Regulatory Phenotype and Function. Sci. Transl. Med. 2020, 12, eaay6422. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, M.; Lee, E.; Lakshmanan, U.; Shipp, S.; Froessl, L.; Barzaghi, F.; Passerini, L.; Narula, M.; Sheikali, A.; Lee, C.M.; et al. CRISPR-Based Gene Editing Enables FOXP3 Gene Repair in IPEX Patient Cells. Sci. Adv. 2020, 6, eaaz0571. [Google Scholar] [CrossRef]
- Wilk, C.; Effenberg, L.; Abberger, H.; Steenpass, L.; Hansen, W.; Zeschnigk, M.; Kirschning, C.; Buer, J.; Kehrmann, J. CRISPR/Cas9-Mediated Demethylation of FOXP3-TSDR toward Treg-Characteristic Programming of Jurkat T Cells. Cell. Immunol. 2022, 371, 104471. [Google Scholar] [CrossRef] [PubMed]
- Ozay, E.I.; Shanthalingam, S.; Sherman, H.L.; Torres, J.A.; Osborne, B.A.; Tew, G.N.; Minter, L.M. Cell-Penetrating Anti-Protein Kinase C Theta Antibodies Act Intracellularly to Generate Stable, Highly Suppressive Regulatory T Cells. Mol. Ther. 2020, 28, 1987–2006. [Google Scholar] [CrossRef]
- Charbonnier, L.-M.; Wang, S.; Georgiev, P.; Sefik, E.; Chatila, T.A. Control of Peripheral Tolerance by Regulatory T Cell–Intrinsic Notch Signaling. Nat. Immunol. 2015, 16, 1162–1173. [Google Scholar] [CrossRef]
- Havens, M.A.; Hastings, M.L. Splice-Switching Antisense Oligonucleotides as Therapeutic Drugs. Nucleic Acids Res. 2016, 44, 6549–6563. [Google Scholar] [CrossRef]
- Sergeeva, O.V.; Shcherbinina, E.Y.; Shomron, N.; Zatsepin, T.S. Modulation of RNA Splicing by Oligonucleotides: Mechanisms of Action and Therapeutic Implications. Nucleic Acid Ther. 2022, 32, 123–138. [Google Scholar] [CrossRef]
- Gladilina, Y.A.; Bey, L.; Hilal, A.; Neborak, E.V.; Blinova, V.G.; Zhdanov, D.D. Cytoprotective Activity of Polyamines Is Associated with the Alternative Splicing of RAD51A Pre-MRNA in Normal Human CD4+ T Lymphocytes. Int. J. Mol. Sci. 2022, 23, 1863. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Plyasova, A.A.; Pokrovsky, V.S.; Pokrovskaya, M.V.; Alexandrova, S.S.; Gladilina, Y.A.; Sokolov, N.N. Inhibition of Nuclease Activity by a Splice-Switching Oligonucleotide Targeting Deoxyribonuclease 1 MRNA Prevents Apoptosis Progression and Prolong Viability of Normal Human CD4(+) T-Lymphocytes. Biochimie 2020, 174, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Hoff, H.; Kolar, P.; Ambach, A.; Radbruch, A.; Brunner-Weinzierl, M.C. CTLA-4 (CD152) Inhibits T Cell Function by Activating the Ubiquitin Ligase Itch. Mol. Immunol. 2010, 47, 1875–1881. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.-Q.; Ji, N.-F.; Gu, C.-J.; Wang, Y.-L.; Huang, M.; Zhang, M.-S. Coexpression of Helios in Foxp3(+) Regulatory T Cells and Its Role in Human Disease. Dis. Markers 2021, 2021, 5574472. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Oppenheim, J.J. Resolving the Identity Myth: Key Markers of Functional CD4+FoxP3+ Regulatory T Cells. Int. Immunopharmacol. 2011, 11, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Chougnet, C.; Hildeman, D. Helios-Controller of Treg Stability and Function. Transl. Cancer Res. 2016, 5, S338–S341. [Google Scholar] [CrossRef] [PubMed]
- Lam, A.J.; Uday, P.; Gillies, J.K.; Levings, M.K. Helios Is a Marker, Not a Driver, of Human Treg Stability. Eur. J. Immunol. 2022, 52, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Yang, J.; Hao, J.; Ren, Y.; Chen, L.; Li, G.; Xie, R.; Yang, Y.; Gao, F.; Liu, M. Comparative Study of Regulatory T Cells Expanded Ex Vivo from Cord Blood and Adult Peripheral Blood. Immunology 2012, 136, 218–230. [Google Scholar] [CrossRef]
- Klein, M.; Bopp, T. Cyclic AMP Represents a Crucial Component of Treg Cell-Mediated Immune Regulation. Front. Immunol. 2016, 7, 315. [Google Scholar] [CrossRef]
- Liang, B.; Workman, C.; Lee, J.; Chew, C.; Dale, B.M.; Colonna, L.; Flores, M.; Li, N.; Schweighoffer, E.; Greenberg, S.; et al. Regulatory T Cells Inhibit Dendritic Cells by Lymphocyte Activation Gene-3 Engagement of MHC Class II. J. Immunol. 2008, 180, 5916–5926. [Google Scholar] [CrossRef]
- Schmidt, A.; Oberle, N.; Krammer, P.H. Molecular Mechanisms of Treg-Mediated T Cell Suppression. Front. Immunol. 2012, 3, 51. [Google Scholar] [CrossRef]
- Wang, J.; Ioan-Facsinay, A.; van der Voort, E.I.H.; Huizinga, T.W.J.; Toes, R.E.M. Transient Expression of FOXP3 in Human Activated Nonregulatory CD4+ T Cells. Eur. J. Immunol. 2007, 37, 129–138. [Google Scholar] [CrossRef]
- Gavin, M.A.; Torgerson, T.R.; Houston, E.; DeRoos, P.; Ho, W.Y.; Stray-Pedersen, A.; Ocheltree, E.L.; Greenberg, P.D.; Ochs, H.D.; Rudensky, A.Y. Single-Cell Analysis of Normal and FOXP3-Mutant Human T Cells: FOXP3 Expression without Regulatory T Cell Development. Proc. Natl. Acad. Sci. USA 2006, 103, 6659–6664. [Google Scholar] [CrossRef] [PubMed]
- Allan, S.E.; Crome, S.Q.; Crellin, N.K.; Passerini, L.; Steiner, T.S.; Bacchetta, R.; Roncarolo, M.G.; Levings, M.K. Activation-Induced FOXP3 in Human T Effector Cells Does Not Suppress Proliferation or Cytokine Production. Int. Immunol. 2007, 19, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Lui, P.P.; Cho, I.; Ali, N. Tissue Regulatory T Cells. Immunology 2020, 161, 4–17. [Google Scholar] [CrossRef] [PubMed]







| Parameter | ALS (n = 20) | Healthy Donors (n = 20) |
|---|---|---|
| Number of female patients | 2 (10%) | 2 (10%) |
| Age at study enrollment, mean ± SD * | 53.2 ± 12.2 | 50.6 ± 14.4 |
| Age range | 27–70 | 26–70 |
| Parameter | ALS Group (n = 20) |
|---|---|
| Disease duration (months) | |
| ALS onset, patients (%) | |
| spinal onset | 17 (85%) |
| bulbar onset | 3 (15%) |
| ALS stage (King’s staging system) | |
| Stage 1 | - |
| Stage 2 | 7 (35%) |
| Stage 3 | 10 (50%) |
| Stage 4 | 3 (15%) |
| ALSFRS-R | 40.4 ± 3.0 (35–46) |
| Cell Marker | Con | FL |
|---|---|---|
| CD4 | 115.1 ± 4.2 | 167.4 ± 15.7 |
| CD25 | 167.9 ± 6.1 | 180.3 ± 2.8 |
| CD127 | 40.7 ± 8.9 | 48.8 ± 3.3 |
| CD152 | 57.7 ± 9.4 | 141.7 ± 13.4 |
| Helios | 34.5 ± 9.3 | 111.4 ± 19.9 |
| Suppressive Molecule | Con | FL |
|---|---|---|
| CD39 | 141.2 ± 13.3 | 153.6 ± 14.2 |
| CD223 | 130.4 ± 28 | 143.1 ± 21.8 |
| Granzyme A | 54.1 ± 10.0 | 100.5 ± 6.3 |
| Granzyme B | 43.0 ± 10.6 | 93.1 ± 6.9 |
| Perforin | 61.9 ± 8.1 | 110.6 ± 8.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhdanov, D.D.; Gladilina, Y.A.; Blinova, V.G.; Abramova, A.A.; Shishparenok, A.N.; Eliseeva, D.D. Induction of FoxP3 Pre-mRNA Alternative Splicing to Enhance the Suppressive Activity of Regulatory T Cells from Amyotrophic Lateral Sclerosis Patients. Biomedicines 2024, 12, 1022. https://doi.org/10.3390/biomedicines12051022
Zhdanov DD, Gladilina YA, Blinova VG, Abramova AA, Shishparenok AN, Eliseeva DD. Induction of FoxP3 Pre-mRNA Alternative Splicing to Enhance the Suppressive Activity of Regulatory T Cells from Amyotrophic Lateral Sclerosis Patients. Biomedicines. 2024; 12(5):1022. https://doi.org/10.3390/biomedicines12051022
Chicago/Turabian StyleZhdanov, Dmitry D., Yulia A. Gladilina, Varvara G. Blinova, Anna A. Abramova, Anastasia N. Shishparenok, and Daria D. Eliseeva. 2024. "Induction of FoxP3 Pre-mRNA Alternative Splicing to Enhance the Suppressive Activity of Regulatory T Cells from Amyotrophic Lateral Sclerosis Patients" Biomedicines 12, no. 5: 1022. https://doi.org/10.3390/biomedicines12051022
APA StyleZhdanov, D. D., Gladilina, Y. A., Blinova, V. G., Abramova, A. A., Shishparenok, A. N., & Eliseeva, D. D. (2024). Induction of FoxP3 Pre-mRNA Alternative Splicing to Enhance the Suppressive Activity of Regulatory T Cells from Amyotrophic Lateral Sclerosis Patients. Biomedicines, 12(5), 1022. https://doi.org/10.3390/biomedicines12051022