Expanding and Enriching the LncRNA Gene–Disease Landscape Using the GeneCaRNA Database
Abstract
:1. Introduction
2. Methods
2.1. Data Extraction and Text Mining
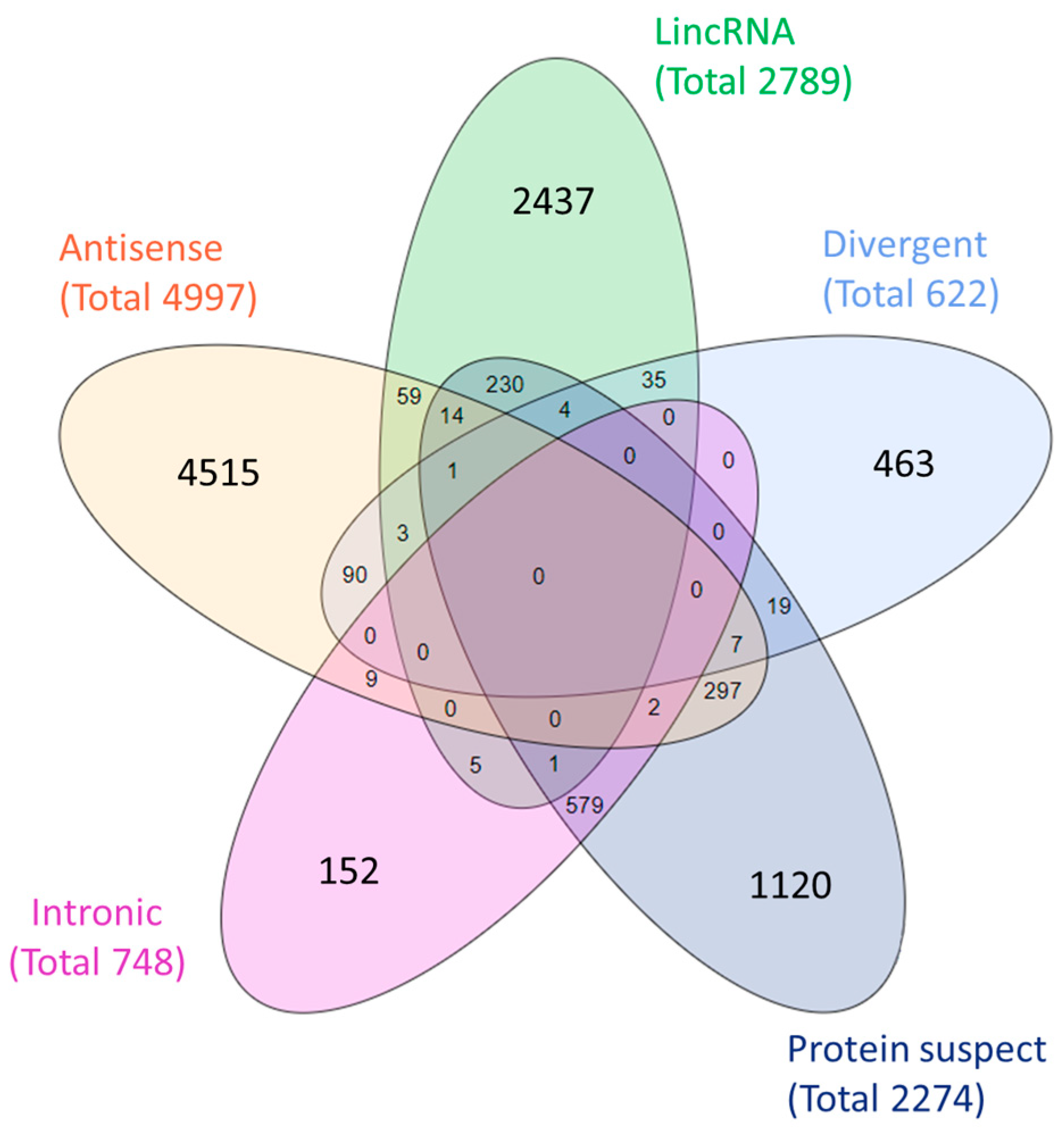
2.2. LncRNA Subclass Definition
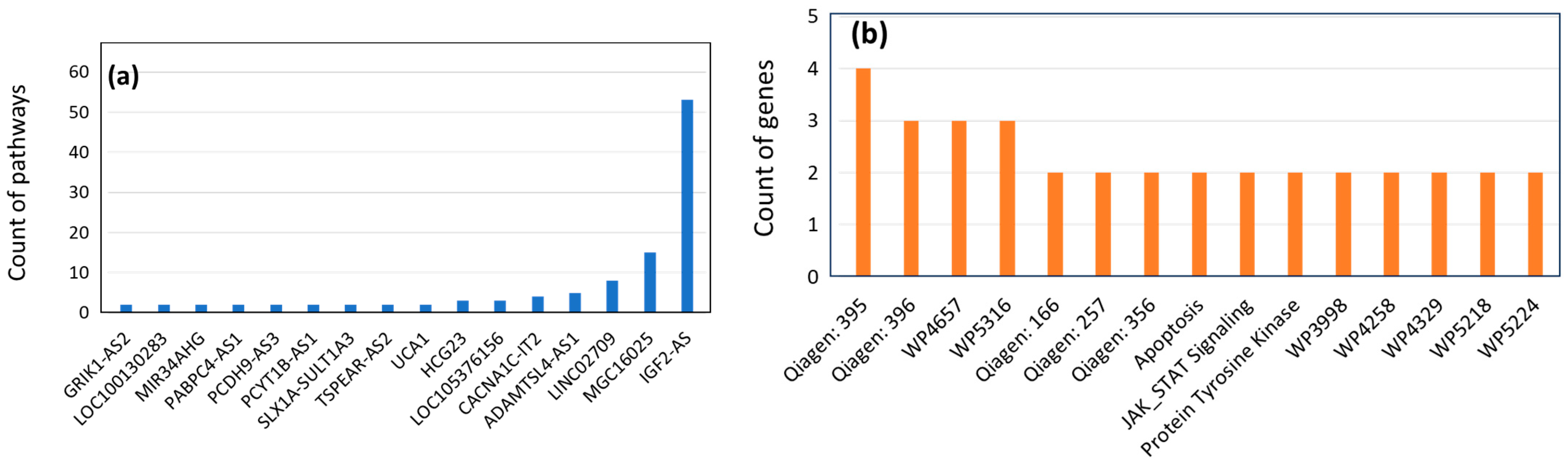
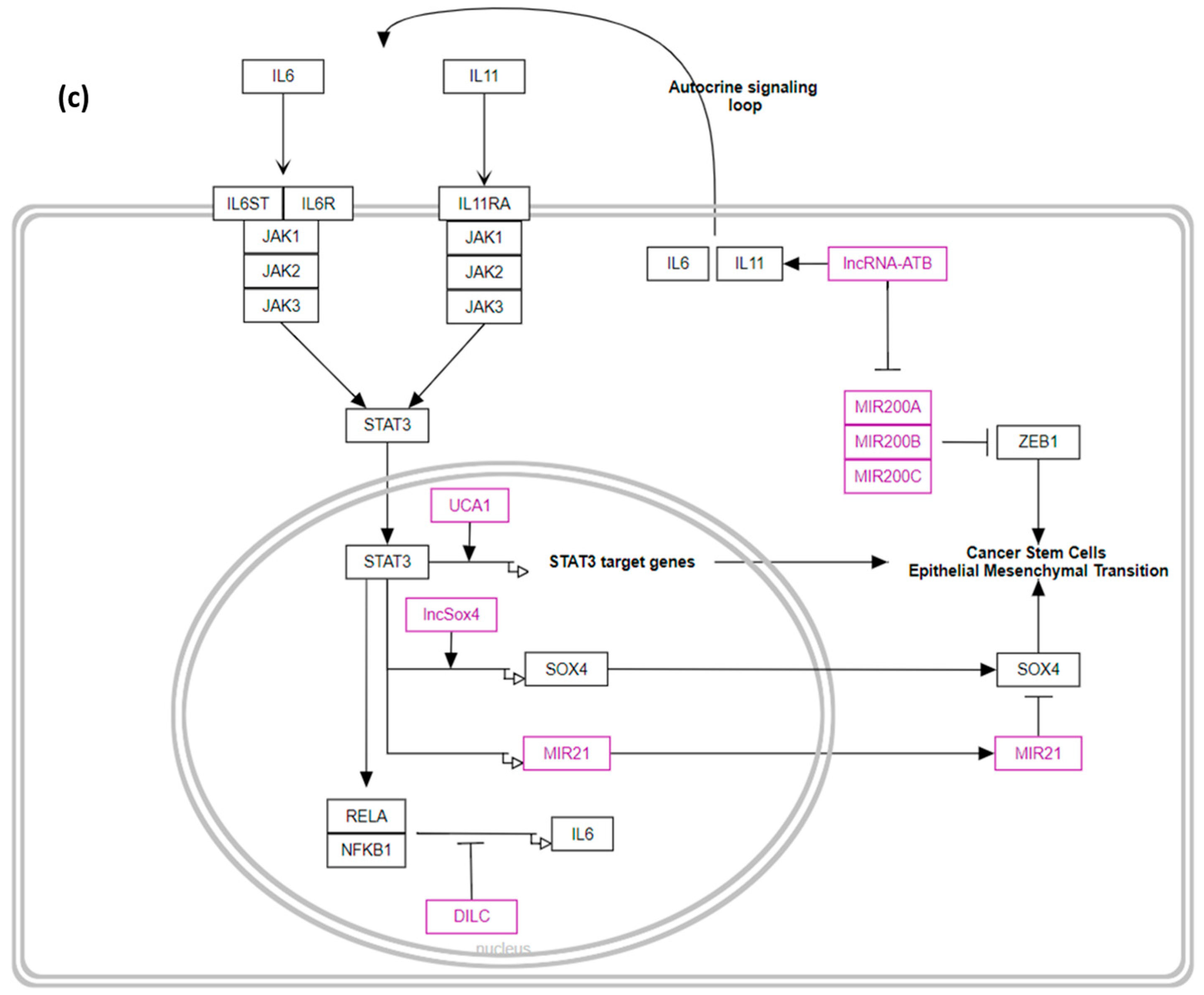
2.3. Gene–Disease and Gene-Pathway Associations
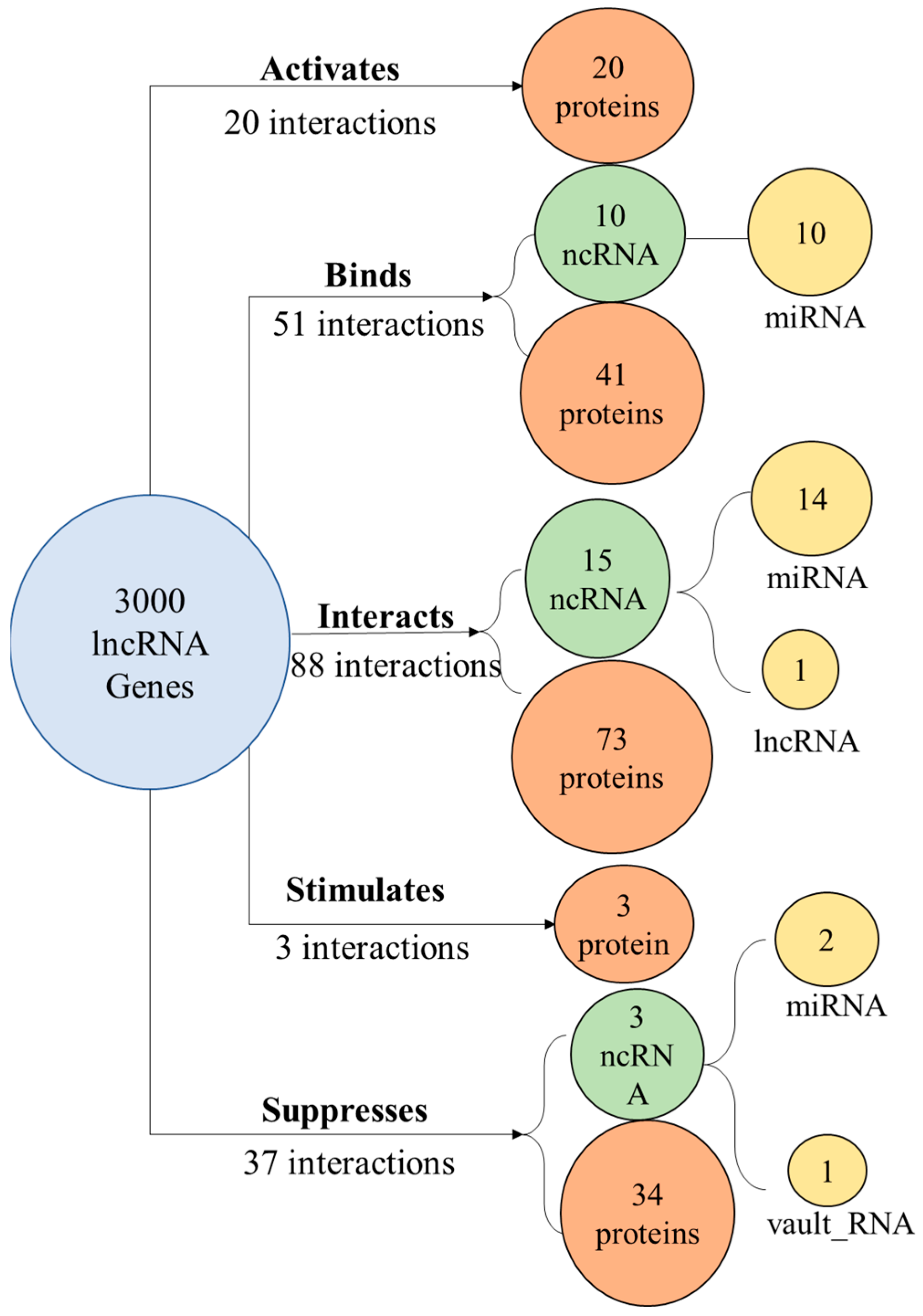
2.4. Gene Product Interactions
3. Results
3.1. Data Extraction and Text Mining
3.2. LncRNA Subclass Definition
3.3. Gene–Disease and Gene-Pathway Associations
3.4. Gene Product Interactions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LncRNA | Long non-coding RNA |
| ncRNA | Non-coding RNA |
| HGNC | HUGO Gene Nomenclature Committee |
| NCBI | National Center for Biotechnology Information |
| TRIGGs | Transcripts Inferred GeneCaRNA genes. |
| SQL | Structured Query Language |
| GIFtS | GeneCards Inferred Functionality Scores |
| LincRNA | Long intergenic non-coding RNA |
| MuSe | Multigene Search |
| ORF | Open Reading Frame |
| PE | Protein Evidence |
References
- Seal, R.L.; Chen, L.-L.; Griffiths-Jones, S.; Lowe, T.M.; Mathews, M.B.; O’Reilly, D.; Pierce, A.J.; Stadler, P.F.; Ulitsky, I.; Wolin, S.L.; et al. A Guide to Naming Human Non-Coding RNA Genes. EMBO J. 2020, 39, e103777. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.R.; Hem, V.; Katz, K.S.; Ovetsky, M.; Wallin, C.; Ermolaeva, O.; Tolstoy, I.; Tatusova, T.; Pruitt, K.D.; Maglott, D.R.; et al. Gene: A Gene-Centered Information Resource at NCBI. Nucleic Acids Res. 2015, 43, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.W.; Amode, M.R.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2024. Nucleic Acids Res. 2024, 52, D891–D899. [Google Scholar] [CrossRef] [PubMed]
- The RNAcentral Consortium; Sweeney, B.A.; Petrov, A.I.; Burkov, B.; Finn, R.D.; Bateman, A.; Szymanski, M.; Karlowski, W.M.; Gorodkin, J.; Seemann, S.E.; et al. RNAcentral: A Hub of Information for Non-Coding RNA Sequences. Nucleic Acids Res. 2019, 47, D221–D229. [Google Scholar] [CrossRef] [PubMed]
- Barshir, R.; Fishilevich, S.; Iny-Stein, T.; Zelig, O.; Mazor, Y.; Guan-Golan, Y.; Safran, M.; Lancet, D. GeneCaRNA: A Comprehensive Gene-Centric Database of Human Non-Coding RNAs in the GeneCards Suite. J. Mol. Biol. 2021, 433, 166913. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, S.; Meng, N.; He, Y.; Lu, R.; Yan, G.-R. ncRNA-Encoded Peptides or Proteins and Cancer. Mol. Ther. 2019, 27, 1718. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Aliperti, V.; Skonieczna, J.; Cerase, A. Long Non-Coding RNA (lncRNA) Roles in Cell Biology, Neurodevelopment and Neurological Disorders. Non-Coding RNA 2021, 7, 36. [Google Scholar] [CrossRef]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the Classification of Long Non-Coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. GENCODE 2021. Nucleic Acids Res. 2021, 49, D916–D923. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Hao, Q.; Prasanth, K.V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018, 34, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Lekka, E.; Hall, J. Noncoding RNAs in Disease. FEBS Lett. 2018, 592, 2884–2900. [Google Scholar] [CrossRef] [PubMed]
- Sparber, P.; Filatova, A.; Khantemirova, M.; Skoblov, M. The Role of Long Non-Coding RNAs in the Pathogenesis of Hereditary Diseases. BMC Med. Genom. 2019, 12, 42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [PubMed]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Abugessaisa, I., Kasukawa, T., Eds.; Springer Nature: Singapore, 2021; pp. 27–56. ISBN 9789811658129. [Google Scholar]
- Harel, A.; Inger, A.; Stelzer, G.; Strichman-Almashanu, L.; Dalah, I.; Safran, M.; Lancet, D. GIFtS: Annotation Landscape Analysis with GeneCards. BMC Bioinform. 2009, 10, 348. [Google Scholar] [CrossRef] [PubMed]
- Lane, L.; Bairoch, A.; Beavis, R.C.; Deutsch, E.W.; Gaudet, P.; Lundberg, E.; Omenn, G.S. Metrics for the Human Proteome Project—2013–2014 and Strategies for Finding Missing Proteins. J. Proteome Res. 2014, 13, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Seal, R.L.; Tweedie, S.; Bruford, E.A. A Standardised Nomenclature for Long Non-Coding RNAs. IUBMB Life 2023, 75, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Belinky, F.; Nativ, N.; Stelzer, G.; Zimmerman, S.; Iny Stein, T.; Safran, M.; Lancet, D. PathCards: Multi-Source Consolidation of Human Biological Pathways. Database J. Biol. Databases Curation 2015, 2015, bav006. [Google Scholar] [CrossRef]
- Rappaport, N.; Twik, M.; Plaschkes, I.; Nudel, R.; Iny Stein, T.; Levitt, J.; Gershoni, M.; Morrey, C.P.; Safran, M.; Lancet, D. MalaCards: An Amalgamated Human Disease Compendium with Diverse Clinical and Genetic Annotation and Structured Search. Nucleic Acids Res. 2017, 45, D877–D887. [Google Scholar] [CrossRef]
- Fishilevich, S.; Nudel, R.; Rappaport, N.; Hadar, R.; Plaschkes, I.; Iny Stein, T.; Rosen, N.; Kohn, A.; Twik, M.; Safran, M.; et al. GeneHancer: Genome-Wide Integration of Enhancers and Target Genes in GeneCards. Database J. Biol. Databases Curation 2017, 2017, bax028. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Rinn, J.L. Discovery and Annotation of Long Noncoding RNAs. Nat. Struct. Mol. Biol. 2015, 22, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, T.; Siqueira, E.; Guil, S. Techniques for Investigating lncRNA Transcript Functions in Neurodevelopment. Mol. Psychiatry 2024, 29, 874–890. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qi, F.; Gao, F.; Cao, H.; Xu, D.; Salehi-Ashtiani, K.; Kapranov, P. Hovlinc Is a Recently Evolved Class of Ribozyme Found in Human lncRNA. Nat. Chem. Biol. 2021, 17, 601–607. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Wu, S.; Xue, M.; Chen, W. Long Non-Coding RNA UCA1 Promotes Glycolysis by Upregulating Hexokinase 2 through the mTOR–STAT3/microRNA143 Pathway. Cancer Sci. 2014, 105, 951–955. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.; Chang, H.Y. Long Noncoding RNAs and Human Disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Jin, L.; Tong, W.M.; Leung, Y.Y.; Gu, M.; Yang, Y. Identification and Integrated Analysis of Differentially Expressed Long Non-coding RNAs Associated with Periodontitis in Humans. J. Periodontal Res. 2021, 56, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Plaschkes, I.; Oz-Levi, D.; Alkelai, A.; Olender, T.; Zimmerman, S.; Twik, M.; Belinky, F.; Fishilevich, S.; Nudel, R.; et al. VarElect: The Phenotype-Based Variation Prioritizer of the GeneCards Suite. BMC Genom. 2016, 17, 444. [Google Scholar] [CrossRef]
- Du, Z.; Fei, T.; Verhaak, R.G.W.; Su, Z.; Zhang, Y.; Brown, M.; Chen, Y.; Liu, X.S. Integrative Genomic Analyses Reveal Clinically Relevant Long Noncoding RNAs in Human Cancer. Nat. Struct. Mol. Biol. 2013, 20, 908–913. [Google Scholar] [CrossRef]



| S.no | Input LncRNA Genes | Action (Keyword) | Target Genes | ||||
|---|---|---|---|---|---|---|---|
| Symbol | Description | Symbol | Description | Category | Class | ||
| 1 | BCAR4 | Breast Cancer Anti-Estrogen Resistance 4 | Activates | GLI2 | GLI Family Zinc Finger 2 | Protein-Coding | transcription regulator |
| 2 | HOTAIR | HOX Transcript Antisense RNA | Activates | DNMT1 | DNA Methyltransferase 1 | Protein-Coding | methyltransferase |
| 3 | HOXA11-AS | HOXA11 Antisense RNA | Activates | ITGB3 | Integrin Subunit Beta 3 | Protein-Coding | receptor |
| 4 | UCA1 | Urothelial Cancer Associated 1 | Activates | MTOR | Mechanistic Target Of Rapamycin Kinase | Protein-Coding | kinase |
| 5 | SNHG1 | Small Nucleolar RNA Host Gene 1 | Activates | HOXA1 | Homeobox A1 | Protein-Coding | transcription factor |
| 6 | LINC00261 | Long Intergenic Non-Protein Coding RNA 261 | Binds | MIR8485 | MicroRNA 8485 | RNA Gene | miRNA |
| 7 | ZFAS1 | ZNFX1 Antisense RNA 1 | Binds | MIR548E | MicroRNA 548e | RNA Gene | miRNA |
| 8 | H19 | H19 Imprinted Maternally Expressed Transcript | Binds | EZH2 | Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit | Protein-Coding | enzyme |
| 9 | HOTAIR | HOX Transcript Antisense RNA | Binds | AR | Androgen Receptor | Protein- Coding | receptor |
| 10 | LINC01413 | Long Intergenic Non-Protein Coding RNA 1413 | Binds | HNRNPK | Heterogeneous Nuclear Ribonucleoprotein K | Protein- Coding | nuclear ribonucleoprotein |
| 11 | NORAD | Non-Coding RNA Activated by DNA Damage | Binds | WEE1 | WEE1 G2 Checkpoint Kinase | Protein- Coding | kinase |
| 12 | MIR222HG | MiR222/221 Cluster Host Gene | Interacts | DNM3OS | DNM3 Opposite Strand/Antisense RNA | RNA Gene | lncRNA |
| 13 | SNHG16 | Small Nucleolar RNA Host Gene 16 | Interacts | MIR195 | MicroRNA 195 | RNA Gene | miRNA |
| 14 | FOXD2-AS1 | FOXD2 Adjacent Opposite Strand RNA 1 | Interacts | EZH2 | Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit | Protein- Coding | enzyme |
| 15 | CASC11 | Cancer Susceptibility 11 | Interacts | HNRNPK | Heterogeneous Nuclear Ribonucleoprotein K | Protein- Coding | Nuclear Ribonucleoprotein |
| 16 | DSCAM-AS1 | DSCAM Antisense RNA 1 | Interacts | YBX1 | Y-Box Binding Protein 1 | Protein- Coding | RNA-binding protein |
| 17 | HOTAIR | HOX Transcript Antisense RNA | Interacts | AR | Androgen Receptor | Protein- Coding | receptor |
| 18 | CRNDE | Colorectal Neoplasia Differentially Expressed | Interacts | TLR3 | Toll Like Receptor 3 | Protein- Coding | receptor |
| 19 | PCAT1 | Prostate Cancer Associated Transcript 1 | Interacts | AR | Androgen Receptor | Protein- Coding | receptor |
| 20 | RMST | Rhabdomyosarcoma 2 Associated Transcript | Interacts | SOX2 | SRY-Box Transcription Factor 2 | Protein- Coding | transcription factor |
| 21 | H19 | H19 Imprinted Maternally Expressed Transcript | Stimulates | STAR | Steroidogenic Acute Regulatory Protein | Protein- Coding | regulator of steroid hormone synthesis |
| 22 | HULC | Hepatocellular Carcinoma Up-Regulated Long Non-Coding RNA | Stimulates | EMT | EMT ITK (IL2 Inducible T Cell Kinase) | Protein- Coding | kinase |
| 23 | SNHG8 | Small Nucleolar RNA Host Gene 8 | Stimulates | NPC | NPC (Intracellular Cholesterol Transporter) | Protein- Coding | transporter |
| 24 | MALAT1 | Metastasis-Associated Lung Adenocarcinoma Transcript 1 | Suppresses | MIR211 | MicroRNA 211 | RNA Gene | miRNA |
| 25 | SNHG1 | Small Nucleolar RNA Host Gene 1 | Suppresses | SOX9 | SRY-Box Transcription Factor 9 | Protein- Coding | transcription factor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aggarwal, S.; Rosenblum, C.; Gould, M.; Ziman, S.; Barshir, R.; Zelig, O.; Guan-Golan, Y.; Iny-Stein, T.; Safran, M.; Pietrokovski, S.; et al. Expanding and Enriching the LncRNA Gene–Disease Landscape Using the GeneCaRNA Database. Biomedicines 2024, 12, 1305. https://doi.org/10.3390/biomedicines12061305
Aggarwal S, Rosenblum C, Gould M, Ziman S, Barshir R, Zelig O, Guan-Golan Y, Iny-Stein T, Safran M, Pietrokovski S, et al. Expanding and Enriching the LncRNA Gene–Disease Landscape Using the GeneCaRNA Database. Biomedicines. 2024; 12(6):1305. https://doi.org/10.3390/biomedicines12061305
Chicago/Turabian StyleAggarwal, Shalini, Chana Rosenblum, Marshall Gould, Shahar Ziman, Ruth Barshir, Ofer Zelig, Yaron Guan-Golan, Tsippi Iny-Stein, Marilyn Safran, Shmuel Pietrokovski, and et al. 2024. "Expanding and Enriching the LncRNA Gene–Disease Landscape Using the GeneCaRNA Database" Biomedicines 12, no. 6: 1305. https://doi.org/10.3390/biomedicines12061305
APA StyleAggarwal, S., Rosenblum, C., Gould, M., Ziman, S., Barshir, R., Zelig, O., Guan-Golan, Y., Iny-Stein, T., Safran, M., Pietrokovski, S., & Lancet, D. (2024). Expanding and Enriching the LncRNA Gene–Disease Landscape Using the GeneCaRNA Database. Biomedicines, 12(6), 1305. https://doi.org/10.3390/biomedicines12061305









