Olanzapine Modulate Lipid Metabolism and Adipose Tissue Accumulation via Hepatic Muscarinic M3 Receptor-Mediated Alk-Related Signaling
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Experimental Design
2.2. Tissue Collection
2.3. Plasma Lipid Parameters Measurements
2.4. RNA Extraction and Gene Expression Analysis by Real Time PCR
2.5. Statistical Analysis
3. Results
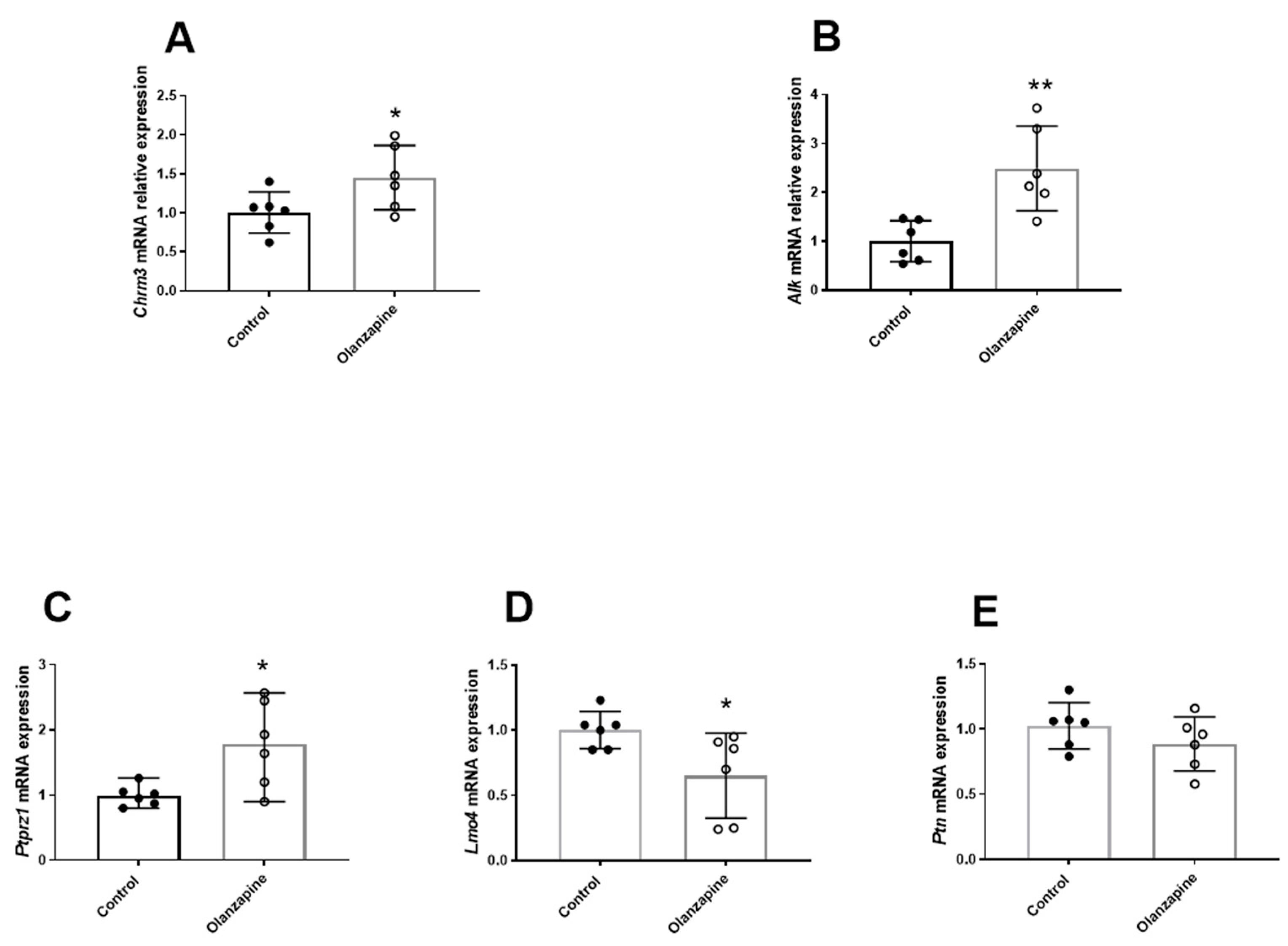
3.1. The Effect of Olanzapine on Lipid Metabolism and the Hepatic Alk Signaling
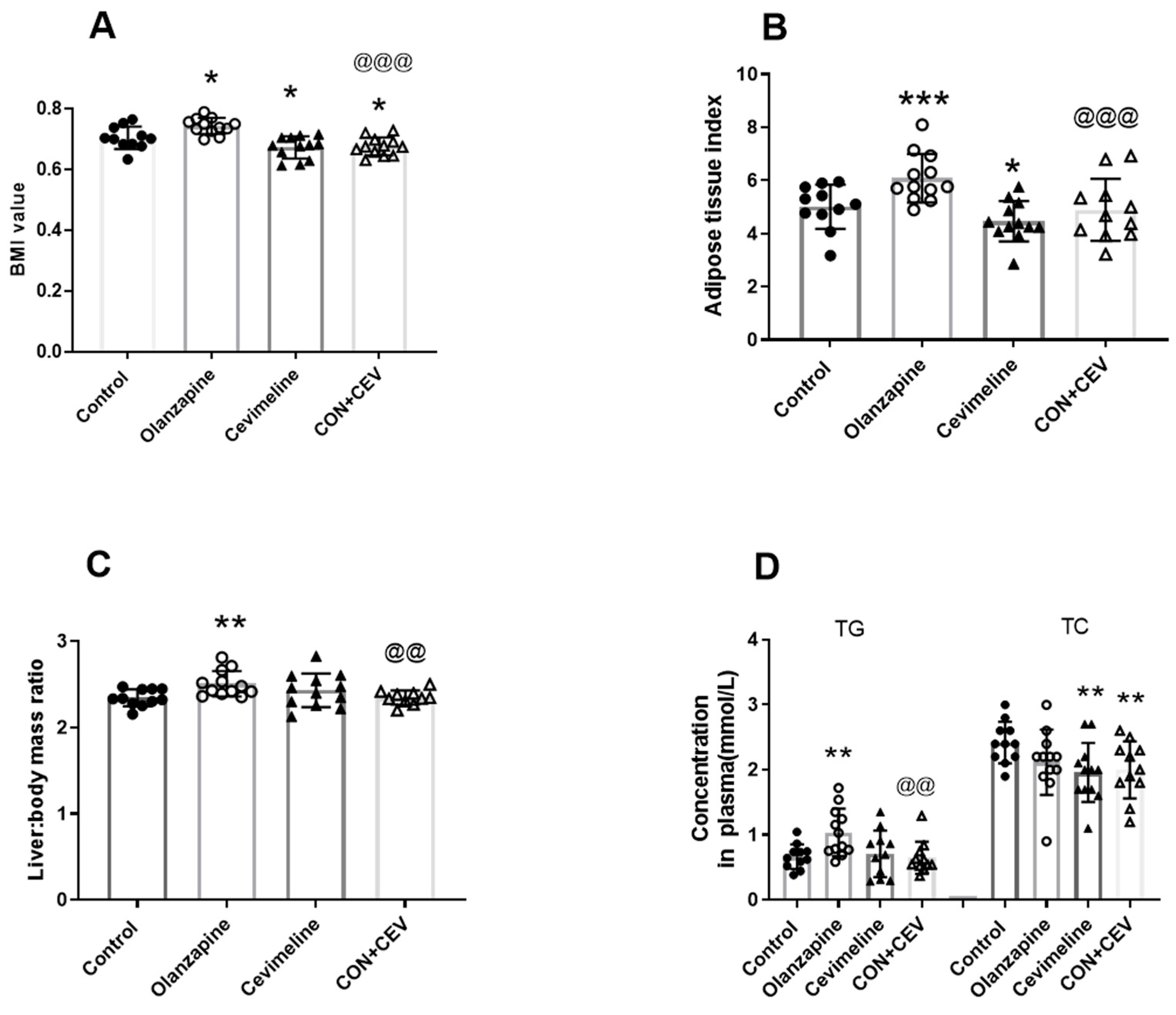
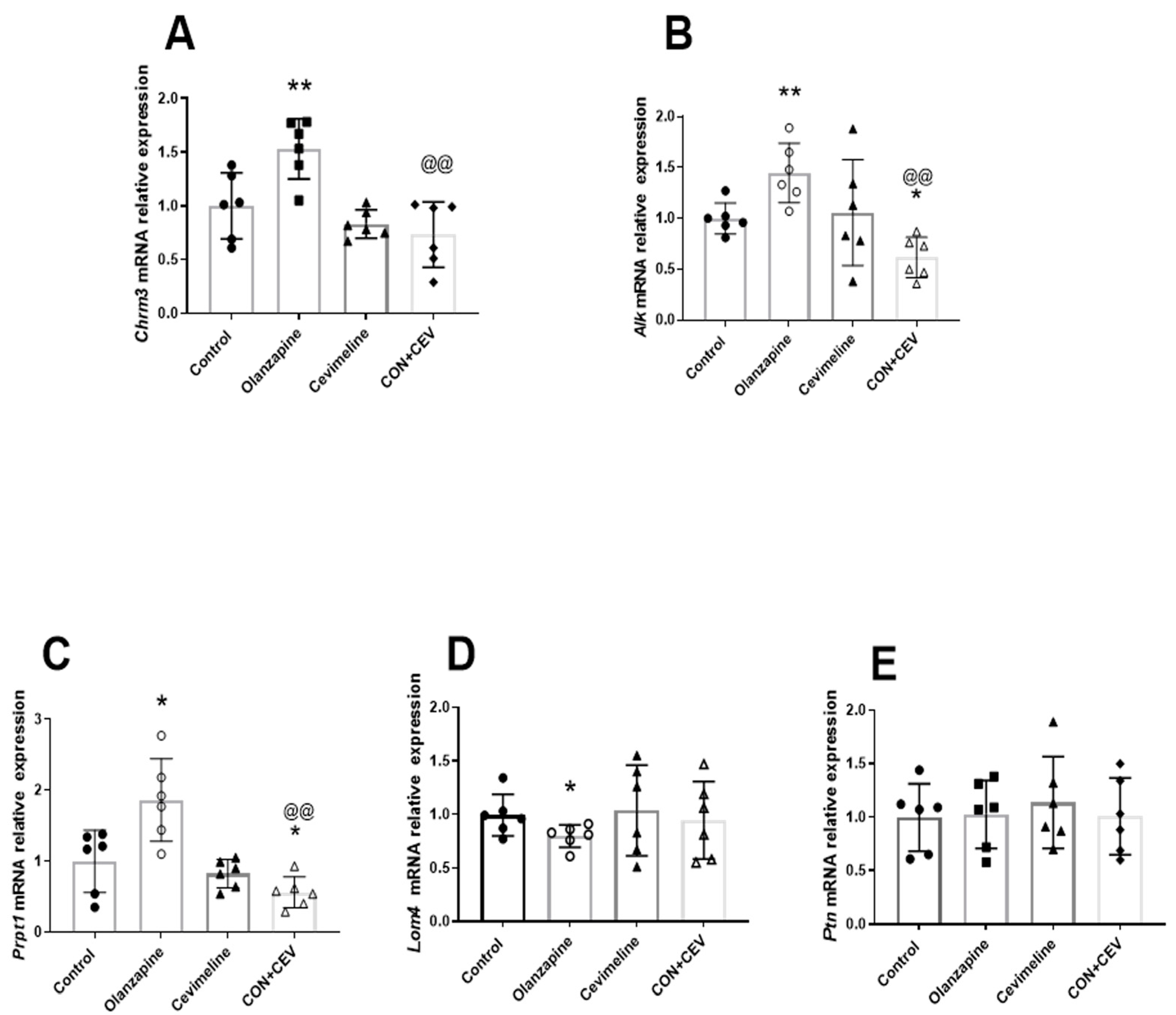
3.2. Cevimeline Cotreatment Ameliorates Olanzapine Effects on Lipid Metabolism and Related Hepatic Alk Pathway
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chiarle, R.; Voena, C.; Ambrogio, C.; Piva, R.; Inghirami, G. The anaplastic lymphoma kinase in the pathogenesis of cancer, Nature reviews. Cancer 2008, 8, 11–23. [Google Scholar]
- Huang, H. Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor with Many Faces. Int. J. Mol. Sci. 2018, 19, 3448. [Google Scholar] [CrossRef]
- Mangieri, R.A.; Maier, E.Y.; Buske, T.R.; Lasek, A.W.; Morrisett, R.A. Anaplastic Lymphoma Kinase Is a Regulator of Alcohol Consumption and Excitatory Synaptic Plasticity in the Nucleus Accumbens Shell. Front. Pharmacol. 2017, 8, 533. [Google Scholar] [CrossRef]
- Witek, B.; El Wakil, A.; Nord, C.; Ahlgren, U.; Eriksson, M.; Vernersson-Lindahl, E.; Helland, Å.; Alexeyev, O.A.; Hallberg, B.; Palmer, R.H. Targeted Disruption of ALK Reveals a Potential Role in Hypogonadotropic Hypogonadism. PLoS ONE 2015, 10, e0123542. [Google Scholar] [CrossRef]
- Li, W.D.; Jiao, H.; Wang, K.; Yang, F.; Grant, S.F.; Hakonarson, H.; Ahima, R.; Price, R.A. Pathway-Based Genome-wide Association Studies Reveal That the Rac1 Pathway Is Associated with Plasma Adiponectin Levels. Sci. Rep. 2015, 5, 13422. [Google Scholar] [CrossRef]
- Palmer, N.D.; Goodarzi, M.O.; Langefeld, C.D.; Wang, N.; Guo, X.; Taylor, K.D.; Fingerlin, T.E.; Norris, J.M.; Buchanan, T.A.; Xiang, A.H.; et al. Genetic Variants Associated with Quantitative Glucose Homeostasis Traits Translate to Type 2 Diabetes in Mexican Americans: The GUARDIAN (Genetics Underlying Diabetes in Hispanics) Consortium. Diabetes 2015, 64, 1853–1866. [Google Scholar] [CrossRef]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef]
- Yengo, L.; Sidorenko, J.; Kemper, K.E.; Zheng, Z.; Wood, A.R.; Weedon, M.N.; Frayling, T.M.; Hirschhorn, J.; Yang, J.; Visscher, P.M. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 2018, 27, 3641–3649. [Google Scholar] [CrossRef]
- Orthofer, M.; Valsesia, A.; Mägi, R.; Wang, Q.P.; Kaczanowska, J.; Kozieradzki, I.; Leopoldi, A.; Cikes, D.; Zopf, L.M.; Tretiakov, E.O.; et al. Identification of ALK in Thinness. Cell 2020, 181, 1246–1262.e22. [Google Scholar] [CrossRef]
- Tung, Y.C.L.; O’Rahilly, S. Constitutional Thinness: tALKing the tALK or wALKing the wALK? Cell Metab. 2020, 32, 8–10. [Google Scholar] [CrossRef]
- Ahmed, M.; Kaur, N.; Cheng, Q.; Shanabrough, M.; Tretiakov, E.O.; Harkany, T.; Horvath, T.L.; Schlessinger, J. A hypothalamic pathway for Augmentor α-controlled body weight regulation. Proc. Natl. Acad. Sci. USA 2022, 119, e2200476119. [Google Scholar] [CrossRef]
- Gibbons, C.H. Basics of autonomic nervous system function. Handb. Clin. Neurol. 2019, 160, 407–418. [Google Scholar]
- Nakagawara, A.; Li, Y.; Izumi, H.; Muramori, K.; Inada, H.; Nishi, M. Neuroblastoma. Jpn. J. Clin. Oncol. 2018, 48, 214–241. [Google Scholar] [CrossRef]
- Costa, J.; Moreira, A.; Moreira, P.; Delgado, L.; Silva, D. Effects of weight changes in the autonomic nervous system: A systematic review and meta-analysis. Clin. Nutr. 2019, 38, 110–126. [Google Scholar] [CrossRef]
- NamKoong, C.; Song, W.J.; Kim, C.Y.; Chun, D.H.; Shin, S.; Sohn, J.W.; Choi, H.J. Chemogenetic manipulation of parasympathetic neurons (DMV) regulates feeding behavior and energy metabolism. Neurosci. Lett. 2019, 712, 134356. [Google Scholar] [CrossRef]
- Rickels, M.R.; Perez, E.M.; Peleckis, A.J.; Alshehabi, E.; Nguyen, H.L.; Stefanovski, D.; Rickels, K.; Teff, K.L. Contribution of parasympathetic muscarinic augmentation of insulin secretion to olanzapine-induced hyperinsulinemia. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E250–E257. [Google Scholar] [CrossRef]
- Plaschke, K.; Do, T.Q.M.; Uhle, F.; Brenner, T.; Weigand, M.A.; Kopitz, J. Ablation of the Right Cardiac Vagus Nerve Reduces Acetylcholine Content without Changing the Inflammatory Response during Endotoxemia. Int. J. Mol. Sci. 2018, 19, 442. [Google Scholar] [CrossRef]
- McCuskey, R.S. Anatomy of efferent hepatic nerves. Anat. Rec. Part A Discov. Mol. Cell. Evol. Biol. 2004, 280, 821–826. [Google Scholar] [CrossRef]
- Wessler, I.K.; Kirkpatrick, C.J. Activation of muscarinic receptors by non-neuronal acetylcholine. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2012; Volume 208, pp. 469–491. [Google Scholar]
- Ishii, M.; Kurachi, Y. Muscarinic acetylcholine receptors. Curr. Pharm. Des. 2006, 12, 3573–3581. [Google Scholar] [CrossRef]
- Tolaymat, M.; Sundel, M.H.; Alizadeh, M.; Xie, G.; Raufman, J.P. Potential Role for Combined Subtype-Selective Targeting of M1 and M3 Muscarinic Receptors in Gastrointestinal and Liver Diseases. Front. Pharmacol. 2021, 12, 786105. [Google Scholar] [CrossRef]
- Zhu, L.; Rossi, M.; Cohen, A.; Pham, J.; Zheng, H.; Dattaroy, D.; Mukaibo, T.; Melvin, J.E.; Langel, J.L.; Hattar, S.; et al. Allosteric modulation of β-cell M3 muscarinic acetylcholine receptors greatly improves glucose homeostasis in lean and obese mice. Proc. Natl. Acad. Sci. USA 2019, 116, 18684–18690. [Google Scholar] [CrossRef]
- Kovess-Masfety, V.; Balusson, F.; Leray, E.; Husky, M.; Scailteux, L. Prescription patterns of first- and second-generation antipsychotic drugs in the French population. Fundam. Clin. Pharmacol. 2020, 34, 603–611. [Google Scholar] [CrossRef]
- Correll, C. Current Treatment Options and Emerging Agents for Schizophrenia. J. Clin. Psychiatry 2020, 81, 26548. [Google Scholar] [CrossRef]
- Deng, C. Effects of antipsychotic medications on appetite, weight, and insulin resistance. Endocrinol. Metab. Clin. 2013, 42, 545–563. [Google Scholar] [CrossRef]
- Vantaggiato, C.; Panzeri, E.; Citterio, A.; Orso, G.; Pozzi, M. Antipsychotics Promote Metabolic Disorders Disrupting Cellular Lipid Metabolism and Trafficking. Trends Endocrinol. Metab. TEM 2019, 30, 189–210. [Google Scholar] [CrossRef]
- Deng, C.; Yao, J.K. Editorial: Metabolic Disturbances in Mental Illness: Neuropathogenetic Mechanisms and Therapeutic Implications. Front. Neurosci. 2020, 14, 522695. [Google Scholar] [CrossRef]
- Lian, J.; Huang, X.-F.; Pai, N.; Deng, C. Ameliorating antipsychotic-induced weight gain by betahistine: Mechanisms and clinical implications. Pharmacol. Res. 2016, 106, 51–63. [Google Scholar] [CrossRef]
- Carli, M.; Kolachalam, S.; Longoni, B.; Pintaudi, A.; Baldini, M.; Aringhieri, S.; Fasciani, I.; Annibale, P.; Maggio, R.; Scarselli, M. Atypical Antipsychotics and Metabolic Syndrome: From Molecular Mechanisms to Clinical Differences. Pharmaceuticals 2021, 14, 238. [Google Scholar] [CrossRef]
- Silvestre, J.S.; Prous, J. Research on adverse drug events. I. Muscarinic M3 receptor binding affinity could predict the risk of antipsychotics to induce type 2 diabetes. Methods Find. Exp. Clin. Pharmacol. 2005, 27, 289–304. [Google Scholar] [CrossRef]
- Falk, S.; Lund, C.; Clemmensen, C. Muscarinic receptors in energy homeostasis: Physiology and pharmacology. Basic. Clin. Pharmacol. Toxicol. 2020, 126, 66–76. [Google Scholar] [CrossRef]
- Weston-Green, K.; Huang, X.F.; Deng, C. Second generation antipsychotic-induced type 2 diabetes: A role for the muscarinic M3 receptor. CNS Drugs 2013, 27, 1069–1080. [Google Scholar] [CrossRef]
- Han, M.; Lian, J.; Su, Y.; Deng, C. Cevimeline co-treatment attenuates olanzapine-induced metabolic disorders via modulating hepatic M3 muscarinic receptor: AMPKα signalling pathway in female rats. J. Psychopharmacol. 2022, 36, 202–213. [Google Scholar] [CrossRef]
- Lian, J.; Deng, C. The dosage-dependent effects of cevimeline in preventing olanzapine-induced metabolic side-effects in female rats. Pharmacol. Biochem. Behav. 2020, 191, 172878. [Google Scholar] [CrossRef]
- Charan, J.; Kantharia, N.D. How to calculate sample size in animal studies? J. Pharmacol. Pharmacother. 2013, 4, 303–306. [Google Scholar] [CrossRef]
- Lian, J.; Huang, X.-F.; Pai, N.; Deng, C. Betahistine ameliorates olanzapine-induced weight gain through modulation of histaminergic, NPY and AMPK pathways. Psychoneuroendocrinology 2014, 48, 77–86. [Google Scholar] [CrossRef]
- Liu, X.; Wu, Z.; Lian, J.; Hu, C.H.; Huang, X.F.; Deng, C. Time-dependent changes and potential mechanisms of glucose-lipid metabolic disorders associated with chronic clozapine or olanzapine treatment in rats. Sci. Rep. 2017, 7, 2762. [Google Scholar] [CrossRef]
- Sylvester, E.; Yi, W.; Han, M.; Deng, C. Exercise intervention for preventing risperidone-induced dyslipidemia and gluco-metabolic disorders in female juvenile rats. Pharmacol. Biochem. Behav. 2020, 199, 173064. [Google Scholar] [CrossRef]
- Su, Y.; Liu, X.; Lian, J.; Deng, C. Epigenetic histone modulations of PPARγ and related pathways contribute to olanzapine-induced metabolic disorders. Pharmacol. Res. 2020, 155, 104703. [Google Scholar] [CrossRef]
- Ono, M.; Takamura, E.; Shinozaki, K.; Tsumura, T.; Hamano, T.; Yagi, Y.; Tsubota, K. Therapeutic effect of cevimeline on dry eye in patients with Sjögren’s syndrome: A randomized, double-blind clinical study. Am. J. Ophthalmol. 2004, 138, 6–17. [Google Scholar] [CrossRef]
- Ueda, H.; Mitoh, Y.; Fujita, M.; Kobashi, M.; Yamashiro, T.; Sugimoto, T.; Ichikawa, H.; Matsuo, R. Muscarinic receptor immunoreactivity in the superior salivatory nucleus neurons innervating the salivary glands of the rat. Neurosci. Lett. 2011, 499, 42–46. [Google Scholar] [CrossRef]
- Novelli, E.L.; Diniz, Y.S.; Galhardi, C.M.; Ebaid, G.M.; Rodrigues, H.G.; Mani, F.; Fernandes, A.A.; Cicogna, A.C.; Filho, J.L.N. Anthropometrical parameters and markers of obesity in rats. Lab. Anim. 2007, 41, 111–119. [Google Scholar] [CrossRef]
- Lasek, A.W.; Gesch, J.; Giorgetti, F.; Kharazia, V.; Heberlein, U. Alk is a transcriptional target of LMO4 and ERα that promotes cocaine sensitization and reward. J. Neurosci. Off. J. Soc. Neurosci. 2011, 31, 14134–14141. [Google Scholar] [CrossRef]
- Wellstein, A. ALK receptor activation, ligands and therapeutic targeting in glioblastoma and in other cancers. Front. Oncol. 2012, 2, 192. [Google Scholar] [CrossRef]
- Perez-Pinera, P.; Zhang, W.; Chang, Y.; Vega, J.A.; Deuel, T.F. Anaplastic lymphoma kinase is activated through the pleiotrophin/receptor protein-tyrosine phosphatase beta/zeta signaling pathway: An alternative mechanism of receptor tyrosine kinase activation. J. Biol. Chem. 2007, 282, 28683–28690. [Google Scholar] [CrossRef]
- Banni, S.; Carta, G.; Murru, E.; Cordeddu, L.; Giordano, E.; Marrosu, F.; Puligheddu, M.; Floris, G.; Asuni, G.P.; Cappai, A.L.; et al. Vagus nerve stimulation reduces body weight and fat mass in rats. PLoS ONE 2012, 7, e44813. [Google Scholar] [CrossRef]
- Correll, C.U. From receptor pharmacology to improved outcomes: Individualising the selection, dosing, and switching of antipsychotics. Eur. Psychiatry J. Assoc. Eur. Psychiatr. 2010, 25, S12–S21. [Google Scholar] [CrossRef]
- Singh, R.R.; Barnes, C.J.; Talukder, A.H.; Fuqua, S.A.; Kumar, R. Negative regulation of estrogen receptor alpha transactivation functions by LIM domain only 4 protein. Cancer Res. 2005, 65, 10594–10601. [Google Scholar] [CrossRef]
- Pandey, N.R.; Zhou, X.; Zaman, T.; Cruz, S.A.; Qin, Z.; Lu, M.; Keyhanian, K.; Brunel, J.M.; Stewart, A.F.; Chen, H.H. LMO4 is required to maintain hypothalamic insulin signaling. Biochem. Biophys. Res. Commun. 2014, 450, 666–672. [Google Scholar] [CrossRef]
- Zhou, X.; Gomez-Smith, M.; Qin, Z.; Duquette, P.M.; Cardenas-Blanco, A.; Rai, P.S.; Harper, M.E.; Tsai, E.C.; Anisman, H.; Chen, H.H. Ablation of LMO4 in glutamatergic neurons impairs leptin control of fat metabolism. Cell. Mol. Life Sci. CMLS 2012, 69, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Strathe, A.B.; Ostersen, T.; Jensen, J.; Mark, T.; Kadarmideen, H.N. Genome-wide association study reveals genetic architecture of eating behavior in pigs and its implications for humans obesity by comparative mapping. PLoS ONE 2013, 8, e71509. [Google Scholar] [CrossRef] [PubMed]
- Kapur, S.; VanderSpek, S.C.; Brownlee, B.A.; Nobrega, J. Antipsychotic Dosing in Preclinical Models Is Often Unrepresentative of the Clinical Condition: A Suggested Solution Based on In Vivo Occupancy. J. Pharmacol. Exp. Ther. 2003, 305, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Kozielska, M.; Reddy, V.P.; Vermeulen, A.; Li, C.; Grimwood, S.; de Greef, R.; Groothuis, G.M.; Danhof, M.; Proost, J.H. Mechanism-based pharmacokinetic-pharmacodynamic modeling of the dopamine D2 receptor occupancy of olanzapine in rats. Pharm. Res. 2011, 28, 2490–2504. [Google Scholar] [CrossRef] [PubMed]
- Tauscher, J.; Jones, C.; Remington, G.; Zipursky, R.B.; Kapur, S. Significant dissociation of brain and plasma kinetics with antipsychotics. Mol. Psychiatry 2002, 7, 317–321. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aravagiri, M.; Teper, Y.; Marder, S.R. Pharmacokinetics and tissue distribution of olanzapine in rats. Biopharm. Drug Dispos. 1999, 20, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Reagan-Shaw, S.; Nihal, M.; Ahmad, N. Dose translation from animal to human studies revisited. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2008, 22, 659–661. [Google Scholar] [CrossRef] [PubMed]
- FDA. Estimating the Safe Starting Dose in Clinical Trials for Therapeutics in Adult Healthy Volunteers; U.S. FDA Center for Drug Evaluation and Research, Health and Human Services Department: Washington, DC, USA, 2005. [Google Scholar]
- Weber, J.; Keating, G.M. Cevimeline. Drugs 2008, 68, 1691–1698. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.J.; Rivers, C.I.; Serra, L.M.; Singh, A.K. Long-term outcomes of interventions for radiation-induced xerostomia: A review. World J. Clin. Oncol. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.E.; Kim, Y.J.; Hwang, M.W.; Park, Y.-J.; Yang, J. Cevimeline-induced anti-inflammatory effect through upregulations of mucins in the ocular surface of a dry eye mouse model. Biomed. Pharmacother. 2021, 139, 111571. [Google Scholar] [CrossRef] [PubMed]
- Arisawa, H.; Imai, E.; Fujise, N.; Fukui, K.; Masunaga, H. General pharmacological profile of the novel muscarinic receptor agonist SNI-2011, a drug for xerostomia in Sjogren’s syndrome. 2nd communication: Effects on general behaviours and central nervous system. Arzneimittelforschung 2002, 52, 14–20. [Google Scholar]
- Weston-Green, K.; Huang, X.F.; Deng, C. Sensitivity of the female rat to olanzapine-induced weight gain-Far from the clinic? Schizophr. Res. 2010, 116, 299–300. [Google Scholar] [CrossRef]
- Seeman, M.V. Men and women respond differently to antipsychotic drugs. Neuropharmacology 2020, 163, 107631. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; DiSilvio, B.; Unangst, J.; Fernstrom, J.D. Effect of chronic infusion of olanzapine and clozapine on food intake and body weight gain in male and female rats. Life Sci. 2007, 81, 1024–1030. [Google Scholar] [CrossRef] [PubMed]
- Van Der Zwaal, E.M.; Janhunen, S.K.; La Fleur, S.E.; Adan, R.A.H. Modelling olanzapine-induced weight gain in rats. Int. J. Neuropsychopharmacol. 2014, 17, 169–186. [Google Scholar] [CrossRef] [PubMed]
- Koseoglu, M.; Hur, A.; Atay, A.; Cuhadar, S. Effects of hemolysis interferences on routine biochemistry parameters. Biochem. Med. 2011, 21, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Reed, M.; Rosales, A.-L.S.; Chioda, M.D.; Parker, L.; Devgan, G.; Kettle, J. Consensus Recommendations for Management and Counseling of Adverse Events Associated with Lorlatinib: A Guide for Healthcare Practitioners. Adv. Ther. 2020, 37, 3019–3030. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Lin, J.; Liao, G.; Tian, Y.; Liang, Y.; Li, R.; Liu, M.; Yuan, Y. ALK Inhibitors in the Treatment of ALK Positive NSCLC. Front. Oncol. 2019, 8, 557. [Google Scholar] [CrossRef]
- Wu, J.; Savooji, J.; Liu, D. Second- and third-generation ALK inhibitors for non-small cell lung cancer. J. Hematol. Oncol. 2016, 9, 19. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, Y.; Cao, C.; Chen, S.; Lian, J.; Han, M.; Liu, X.; Deng, C. Olanzapine Modulate Lipid Metabolism and Adipose Tissue Accumulation via Hepatic Muscarinic M3 Receptor-Mediated Alk-Related Signaling. Biomedicines 2024, 12, 1403. https://doi.org/10.3390/biomedicines12071403
Su Y, Cao C, Chen S, Lian J, Han M, Liu X, Deng C. Olanzapine Modulate Lipid Metabolism and Adipose Tissue Accumulation via Hepatic Muscarinic M3 Receptor-Mediated Alk-Related Signaling. Biomedicines. 2024; 12(7):1403. https://doi.org/10.3390/biomedicines12071403
Chicago/Turabian StyleSu, Yueqing, Chenyun Cao, Shiyan Chen, Jiamei Lian, Mei Han, Xuemei Liu, and Chao Deng. 2024. "Olanzapine Modulate Lipid Metabolism and Adipose Tissue Accumulation via Hepatic Muscarinic M3 Receptor-Mediated Alk-Related Signaling" Biomedicines 12, no. 7: 1403. https://doi.org/10.3390/biomedicines12071403
APA StyleSu, Y., Cao, C., Chen, S., Lian, J., Han, M., Liu, X., & Deng, C. (2024). Olanzapine Modulate Lipid Metabolism and Adipose Tissue Accumulation via Hepatic Muscarinic M3 Receptor-Mediated Alk-Related Signaling. Biomedicines, 12(7), 1403. https://doi.org/10.3390/biomedicines12071403






