Clinical Genetic Testing for Hearing Loss: Implications for Genetic Counseling and Gene-Based Therapies
Abstract
1. Introduction
2. Materials and Methods
2.1. Audiogram Data Review
2.2. Genetic Testing and Variant Analysis
3. Results
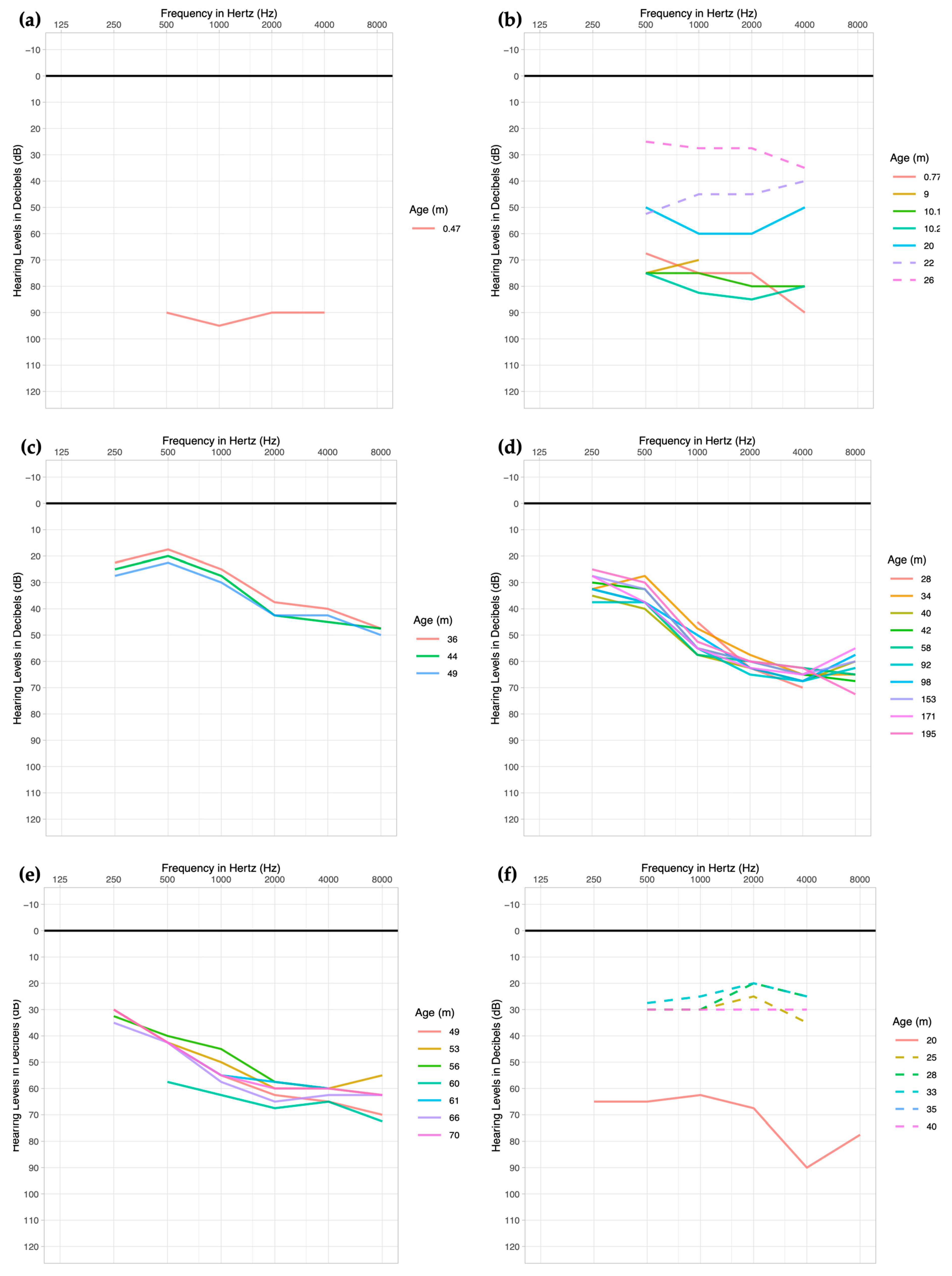
3.1. Clinical Profiles
Audiologic
3.2. Genetic Panel Testing
4. Discussion
5. Conclusions
- While the initial reports from genetic testing cores are based on the discussion and consensus of in-house experts on genetic analyses and hearing loss clinicians, their decision for each patient is based on the best evidence that is available at the time of the genetic diagnosis. As variant databases covering more sequence data from additional world populations and the bioinformatic prediction of pathogenicity from new software become available, some of these diagnoses are likely to change, whether it is due to the reclassification of pathogenicity of variants, or the putatively combined effects of multiple variants on the hearing function of a single patient.
- Additionally, as the price of genome sequencing drops, access to the available genome data may allow for reanalyses, i.e., using updated databases and software, of other variants that lie within novel genes for SNHL, the information on which might not be available at the time of initial diagnoses. The limiting step in such a case would be reconsenting the family or patient for the reanalysis of the available genome data. There are many factors why a family might be more receptive or not to a reanalysis, though it is more likely that a previous negative report from genetic testing might favor agreement to reanalyze the genome data. Nonetheless, informed consent is essential before any reanalysis should be performed, followed by genetic counseling using the updated genetic information.
- Because most genetic diagnoses for SNHL are sought when a child is young, the progression of hearing loss or development of additional features (e.g., retinitis pigmentosa as part of Usher Syndrome) should trigger a request for the re-evaluation of previous genetic diagnoses [51]. While most of the literature on genetic hearing loss is focused on novel genes and variants, it will be more helpful if longitudinal analyses of hearing loss profiles and information on additional clinical features that arise over time in patients with genetic variants are published. These profiles may inform other patients with similar variants or variants within the same genes on the long-term prediction of audiologic and clinical profiles as well as the response to habilitation [68,69]. Over the past three decades, the identification of rare genetic variants for hearing loss has contributed to the cumulative information on the prevalence by population and potential mechanism of hearing and hearing loss by a gene or gene domain, which in turn facilitated the prioritization of gene therapies that are currently being developed. Updated genetic diagnoses based on the best available evidence to date will also facilitate referral once inner ear therapies for specific genes are available [70]. In the future, it will be even more important to determine pathogenicity of variants not just with bioinformatics tools but also by functional experiments to ensure that gene therapy is targeted to the pathogenic variant(s) or gene that is truly involved in the patient’s hearing loss.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, X.C.; Friedman, R.A. Nonsyndromic Hereditary Hearing Loss. Otolaryngol. Clin. North Am. 2002, 35, 275–285. [Google Scholar] [CrossRef]
- Walls, W.; Azaiez, H.; Smith, R. Hereditary Hearing Loss Homepage. Available online: https://hereditaryhearingloss.org/ (accessed on 24 April 2024).
- Chen, W.; Kahrizi, K.; Meyer, N.C.; Riazalhosseini, Y.; Van Camp, G.; Najmabadi, H.; Smith, R.J.H. Mutation of COL11A2 Causes Autosomal Recessive Non-Syndromic Hearing Loss at the DFNB53 Locus. J. Med. Genet. 2005, 42, e61. [Google Scholar] [CrossRef] [PubMed]
- McGuirt, W.T.; Prasad, S.D.; Griffith, A.J.; Kunst, H.P.; Green, G.E.; Shpargel, K.B.; Runge, C.; Huybrechts, C.; Mueller, R.F.; Lynch, E.; et al. Mutations in COL11A2 Cause Non-Syndromic Hearing Loss (DFNA13). Nat. Genet. 1999, 23, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Vikkula, M.; Mariman, E.C.; Lui, V.C.; Zhidkova, N.I.; Tiller, G.E.; Goldring, M.B.; van Beersum, S.E.; de Waal Malefijt, M.C.; van den Hoogen, F.H.; Ropers, H.H. Autosomal Dominant and Recessive Osteochondrodysplasias Associated with the COL11A2 Locus. Cell 1995, 80, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Prezant, T.R.; Agapian, J.V.; Bohlman, M.C.; Bu, X.; Öztas, S.; Qiu, W.-Q.; Arnos, K.S.; Cortopassi, G.A.; Jaber, L.; Rotter, J.I.; et al. Mitochondrial Ribosomal RNA Mutation Associated with Both Antibiotic–Induced and Non–Syndromic Deafness. Nat. Genet. 1993, 4, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Yousaf, R.; Ahmed, Z.M.; Giese, A.P.; Morell, R.J.; Lagziel, A.; Dabdoub, A.; Wilcox, E.R.; Riazuddin, S.; Friedman, T.B.; Riazuddin, S. Modifier Variant of METTL13 Suppresses Human GAB1-Associated Profound Deafness. J. Clin. Investig. 2018, 128, 1509–1522. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.; Karroum, S.B.; Barake, R.; Dunya, G.; Abou-Rizk, S.; Kamar, A.; Nemer, G.; Bassim, M. Post-Lingual Non-Syndromic Hearing Loss Phenotype: A Polygenic Case with 2 Biallelic Mutations in MYO15A and MITF. BMC Med. Genet. 2020, 21, 1. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, T.; Kamiya, K.; Gotoh, S.; Sugitani, Y.; Suzuki, M.; Noda, T.; Minowa, O.; Ikeda, K. Perinatal Gjb2 Gene Transfer Rescues Hearing in a Mouse Model of Hereditary Deafness. Human. Mol. Genet. 2015, 24, 3651–3661. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Nist-Lund, C.; Solanes, P.; Goldberg, H.; Wu, J.; Pan, B.; Schneider, B.L.; Holt, J.R. Efficient Viral Transduction in Mouse Inner Ear Hair Cells with Utricle Injection and AAV9-PHP.B. Hear. Res. 2020, 394, 107882. [Google Scholar] [CrossRef]
- Isgrig, K.; Shteamer, J.W.; Belyantseva, I.A.; Drummond, M.C.; Fitzgerald, T.S.; Vijayakumar, S.; Jones, S.M.; Griffith, A.J.; Friedman, T.B.; Cunningham, L.L.; et al. Gene Therapy Restores Balance and Auditory Functions in a Mouse Model of Usher Syndrome. Mol. Ther. 2017, 25, 780–791. [Google Scholar] [CrossRef]
- György, B.; Meijer, E.J.; Ivanchenko, M.V.; Tenneson, K.; Emond, F.; Hanlon, K.S.; Indzhykulian, A.A.; Volak, A.; Karavitaki, K.D.; Tamvakologos, P.I.; et al. Gene Transfer with AAV9-PHP.B Rescues Hearing in a Mouse Model of Usher Syndrome 3A and Transduces Hair Cells in a Non-Human Primate. Mol. Ther. Methods Clin. Dev. 2018, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Akouos, Inc. A Phase 1/2 Trial of AAVAnc80-hOTOF Gene Therapy in Individuals With Sensorineural Hearing Loss Due to Biallelic Otoferlin Gene Mutations; clinicaltrials.gov; Akouos, Inc.: Boston, MA, USA, 2024.
- Regeneron Pharmaceuticals A Phase 1/2, Open-Label, Multicenter Trial with a Single Ascending Dose Cohort with Unilateral Intracochlear Injection Followed by a Bilateral Injection Expansion Cohort to Evaluate the Safety, Tolerability, and Efficacy of Db-Oto in Children and Infants with Biallelic Hotof Mutations; clinicaltrials.gov; Akouos, Inc.: Boston, MA, USA, 2024.
- Sloan-Heggen, C.M.; Bierer, A.O.; Shearer, A.E.; Kolbe, D.L.; Nishimura, C.J.; Frees, K.L.; Ephraim, S.S.; Shibata, S.B.; Booth, K.T.; Campbell, C.A.; et al. Comprehensive Genetic Testing in the Clinical Evaluation of 1119 Patients with Hearing Loss. Hum. Genet. 2016, 135, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Zazo Seco, C.; Wesdorp, M.; Feenstra, I.; Pfundt, R.; Hehir-Kwa, J.Y.; Lelieveld, S.H.; Castelein, S.; Gilissen, C.; de Wijs, I.J.; Admiraal, R.J.; et al. The Diagnostic Yield of Whole-Exome Sequencing Targeting a Gene Panel for Hearing Impairment in The Netherlands. Eur. J. Hum. Genet. 2017, 25, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Wei, X.; Chai, Y.; Li, L.; Wu, H. Genetic Etiology Study of the Non-Syndromic Deafness in Chinese Hans by Targeted next-Generation Sequencing. Orphanet J. Rare Dis. 2013, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Cabanillas, R.; Diñeiro, M.; Cifuentes, G.A.; Castillo, D.; Pruneda, P.C.; Álvarez, R.; Sánchez-Durán, N.; Capín, R.; Plasencia, A.; Viejo-Díaz, M.; et al. Comprehensive Genomic Diagnosis of Non-Syndromic and Syndromic Hereditary Hearing Loss in Spanish Patients. BMC Med. Genom. 2018, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of Protein-Coding Genetic Variation in 60,706 Humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.M.; Halees, A.; Itan, Y.; Spencer, E.G.; He, Y.; Azab, M.A.; Gabriel, S.B.; Belkadi, A.; Boisson, B.; Abel, L.; et al. Characterization of Greater Middle Eastern Genetic Variation for Enhanced Disease Gene Discovery. Nat. Genet. 2016, 48, 1071–1076. [Google Scholar] [CrossRef]
- All of Us Research Program Investigators. The “All of Us” Research Program. N. Engl. J. Med. 2019, 381, 668–676. [Google Scholar] [CrossRef]
- Taliun, D.; Harris, D.N.; Kessler, M.D.; Carlson, J.; Szpiech, Z.A.; Torres, R.; Taliun, S.A.G.; Corvelo, A.; Gogarten, S.M.; Kang, H.M.; et al. Sequencing of 53,831 Diverse Genomes from the NHLBI TOPMed Program. Nature 2021, 590, 290–299. [Google Scholar] [CrossRef]
- Azaiez, H.; Booth, K.T.; Ephraim, S.S.; Crone, B.; Black-Ziegelbein, E.A.; Marini, R.J.; Shearer, A.E.; Sloan-Heggen, C.M.; Kolbe, D.; Casavant, T.; et al. Genomic Landscape and Mutational Signatures of Deafness-Associated Genes. Am. J. Hum. Genet. 2018, 103, 484–497. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jian, X.; Boerwinkle, E. dbNSFP: A Lightweight Database of Human Nonsynonymous SNPs and Their Functional Predictions. Hum. Mutat. 2011, 32, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jian, X.; Boerwinkle, E. dbNSFP v2.0: A Database of Human Non-Synonymous SNVs and Their Functional Predictions and Annotations. Hum. Mutat. 2013, 34, E2393–E2402. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, C.; Li, C.; Boerwinkle, E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Non-Synonymous and Splice Site SNVs. Hum. Mutat. 2016, 37, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Steinhaus, R.; Proft, S.; Schuelke, M.; Cooper, D.N.; Schwarz, J.M.; Seelow, D. MutationTaster2021. Nucleic Acids Res. 2021, 49, W446–W451. [Google Scholar] [CrossRef]
- Quang, D.; Chen, Y.; Xie, X. DANN: A Deep Learning Approach for Annotating the Pathogenicity of Genetic Variants. Bioinformatics 2015, 31, 761–763. [Google Scholar] [CrossRef]
- Clark, J.G. Uses and Abuses of Hearing Loss Classification. ASHA 1981, 23, 493–500. [Google Scholar] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.Y.; Lee, C.; Han, J.H.; Kim, M.Y.; Park, H.-R.; Kim, N.; Park, W.-Y.; Oh, D.Y.; Choi, B.Y. Expansion of Phenotypic Spectrum of MYO15A Pathogenic Variants to Include Postlingual Onset of Progressive Partial Deafness. BMC Med. Genet. 2018, 19, 29. [Google Scholar] [CrossRef]
- Vona, B.; Müller, T.; Nanda, I.; Neuner, C.; Hofrichter, M.A.H.; Schröder, J.; Bartsch, O.; Läßig, A.; Keilmann, A.; Schraven, S.; et al. Targeted Next-Generation Sequencing of Deafness Genes in Hearing-Impaired Individuals Uncovers Informative Mutations. Genet. Med. 2014, 16, 945–953. [Google Scholar] [CrossRef]
- Schraders, M.; Ruiz-Palmero, L.; Kalay, E.; Oostrik, J.; del Castillo, F.J.; Sezgin, O.; Beynon, A.J.; Strom, T.M.; Pennings, R.J.E.; Seco, C.Z.; et al. Mutations of the Gene Encoding Otogelin Are a Cause of Autosomal-Recessive Nonsyndromic Moderate Hearing Impairment. Am. J. Hum. Genet. 2012, 91, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Ganaha, A.; Kaname, T.; Yanagi, K.; Tono, T.; Higa, T.; Suzuki, M. Clinical Characteristics with Long-Term Follow-up of Four Okinawan Families with Moderate Hearing Loss Caused by an OTOG Variant. Hum. Genome Var. 2019, 6, 37. [Google Scholar] [CrossRef]
- Chen, D.-Y.; Liu, X.-F.; Lin, X.-J.; Zhang, D.; Chai, Y.-C.; Yu, D.-H.; Sun, C.-L.; Wang, X.-L.; Zhu, W.-D.; Chen, Y.; et al. A Dominant Variant in DMXL2 Is Linked to Nonsyndromic Hearing Loss. Genet. Med. 2017, 19, 553–558. [Google Scholar] [CrossRef]
- Snoeckx, R.L.; Huygen, P.L.M.; Feldmann, D.; Marlin, S.; Denoyelle, F.; Waligora, J.; Mueller-Malesinska, M.; Pollak, A.; Ploski, R.; Murgia, A.; et al. GJB2 Mutations and Degree of Hearing Loss: A Multicenter Study. Am. J. Hum. Genet. 2005, 77, 945–957. [Google Scholar] [CrossRef]
- Lindsey, S.; Brewer, C.; Stakhovskaya, O.; Kim, H.J.; Zalewski, C.; Bryant, J.; King, K.A.; Naggert, J.K.; Gahl, W.A.; Marshall, J.D.; et al. Auditory and Otologic Profile of Alström Syndrome: Comprehensive Single Center Data on 38 Patients. Am. J. Med. Genet. A 2017, 173, 2210–2218. [Google Scholar] [CrossRef]
- Weston, M.D.; Luijendijk, M.W.J.; Humphrey, K.D.; Möller, C.; Kimberling, W.J. Mutations in the VLGR1 Gene Implicate G-Protein Signaling in the Pathogenesis of Usher Syndrome Type II. Am. J. Hum. Genet. 2004, 74, 357–366. [Google Scholar] [CrossRef]
- Santos-Cortez, R.L.P.; Lee, K.; Azeem, Z.; Antonellis, P.J.; Pollock, L.M.; Khan, S.; Irfanullah; Andrade-Elizondo, P.B.; Chiu, I.; Adams, M.D.; et al. Mutations in KARS, Encoding Lysyl-tRNA Synthetase, Cause Autosomal-Recessive Nonsyndromic Hearing Impairment DFNB89. Am. J. Hum. Genet. 2013, 93, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Jaijo, T.; Aller, E.; Beneyto, M.; Najera, C.; Graziano, C.; Turchetti, D.; Seri, M.; Ayuso, C.; Baiget, M.; Moreno, F.; et al. MYO7A Mutation Screening in Usher Syndrome Type I Patients from Diverse Origins. J. Med. Genet. 2007, 44, e71. [Google Scholar] [CrossRef]
- Kros, C.J.; Marcotti, W.; van Netten, S.M.; Self, T.J.; Libby, R.T.; Brown, S.D.M.; Richardson, G.P.; Steel, K.P. Reduced Climbing and Increased Slipping Adaptation in Cochlear Hair Cells of Mice with Myo7a Mutations. Nat. Neurosci. 2002, 5, 41–47. [Google Scholar] [CrossRef]
- Longoni, M.; Kantarci, S.; Donnai, D.; Pober, B.R. Donnai-Barrow Syndrome. In GeneReviews®; Adam, M.P., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J., Gripp, K.W., Amemiya, A., Eds.; University of Washington, Seattle: Seattle, WA, USA, 1993. [Google Scholar]
- Schowalter, D.B.; Pagon, R.A.; Kalina, R.E.; McDonald, R. Facio-Oculo-Acoustico-Renal (FOAR) Syndrome: Case Report and Review. Am. J. Med. Genet. 1997, 69, 45–49. [Google Scholar] [CrossRef]
- Faridi, R.; Yousaf, R.; Gu, S.; Inagaki, S.; Turriff, A.E.; Pelstring, K.; Guan, B.; Naik, A.; Griffith, A.J.; Adadey, S.M.; et al. Variants of LRP2, Encoding a Multifunctional Cell-Surface Endocytic Receptor, Associated with Hearing Loss and Retinal Dystrophy. Clin. Genet. 2023, 103, 699–703. [Google Scholar] [CrossRef]
- Tanaka, A.J.; Okumoto, K.; Tamura, S.; Abe, Y.; Hirsch, Y.; Deng, L.; Ekstein, J.; Chung, W.K.; Fujiki, Y. A Newly Identified Mutation in the PEX26 Gene Is Associated with a Milder Form of Zellweger Spectrum Disorder. Cold Spring Harb. Mol. Case Stud. 2019, 5, a003483. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yergeau, C.; Kawai, K.; Braverman, N.; Géléoc, G.S.G. A Retrospective Study of Hearing Loss in Patients Diagnosed with Peroxisome Biogenesis Disorders in the Zellweger Spectrum. Ear Hear. 2022, 43, 582–591. [Google Scholar] [CrossRef]
- Wolf, B.; Spencer, R.; Gleason, T. Hearing Loss Is a Common Feature of Symptomatic Children with Profound Biotinidase Deficiency. J. Pediatr. 2002, 140, 242–246. [Google Scholar] [CrossRef]
- Smith, R.J.; Berlin, C.I.; Hejtmancik, J.F.; Keats, B.J.; Kimberling, W.J.; Lewis, R.A.; Möller, C.G.; Pelias, M.Z.; Tranebjaerg, L. Clinical Diagnosis of the Usher Syndromes. Usher Syndrome Consortium. Am. J. Med. Genet. 1994, 50, 32–38. [Google Scholar] [CrossRef]
- Sadeghi, A.M.; Cohn, E.S.; Kimberling, W.J.; Halvarsson, G.; Möller, C. Expressivity of Hearing Loss in Cases with Usher Syndrome Type IIA. Int. J. Audiol. 2013, 52, 832–837. [Google Scholar] [CrossRef]
- Boeckhaus, J.; Strenzke, N.; Storz, C.; Gross, O. Characterization of Sensorineural Hearing Loss in Children with Alport Syndrome. Life 2020, 10, 360. [Google Scholar] [CrossRef]
- Kurima, K.; Peters, L.M.; Yang, Y.; Riazuddin, S.; Ahmed, Z.M.; Naz, S.; Arnaud, D.; Drury, S.; Mo, J.; Makishima, T.; et al. Dominant and Recessive Deafness Caused by Mutations of a Novel Gene, TMC1, Required for Cochlear Hair-Cell Function. Nat. Genet. 2002, 30, 277–284. [Google Scholar] [CrossRef]
- Nishio, S.; Usami, S. Prevalence and Clinical Features of Autosomal Dominant and Recessive TMC1-Associated Hearing Loss. Hum. Genet. 2022, 141, 929–937. [Google Scholar] [CrossRef]
- Azaiez, H.; Yang, T.; Prasad, S.; Sorensen, J.L.; Nishimura, C.J.; Kimberling, W.J.; Smith, R.J.H. Genotype–Phenotype Correlations for SLC26A4-Related Deafness. Hum. Genet. 2007, 122, 451–457. [Google Scholar] [CrossRef]
- Mustapha, M.; Weil, D.; Chardenoux, S.; Elias, S.; El-Zir, E.; Beckmann, J.S.; Loiselet, J.; Petit, C. An Alpha-Tectorin Gene Defect Causes a Newly Identified Autosomal Recessive Form of Sensorineural Pre-Lingual Non-Syndromic Deafness, DFNB21. Hum. Mol. Genet. 1999, 8, 409–412. [Google Scholar] [CrossRef][Green Version]
- Naz, S. Distinctive Audiometric Profile Associated with DFNB21 Alleles of TECTA. J. Med. Genet. 2003, 40, 360–363. [Google Scholar] [CrossRef]
- Liu, X.; Newton, V.; Read, A. Hearing Loss and Pigmentary Disturbances in Waardenburg Syndrome with Reference to WS Type II. J. Laryngol. Otol. 1995, 109, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Aldè, M.; Cantarella, G.; Zanetti, D.; Pignataro, L.; La Mantia, I.; Maiolino, L.; Ferlito, S.; Di Mauro, P.; Cocuzza, S.; Lechien, J.R.; et al. Autosomal Dominant Non-Syndromic Hearing Loss (DFNA): A Comprehensive Narrative Review. Biomedicines 2023, 11, 1616. [Google Scholar] [CrossRef]
- Deng, Y.; Sang, S.; Wen, J.; Liu, Y.; Ling, J.; Chen, H.; Cai, X.; Mei, L.; Chen, X.; Li, M.; et al. Reproductive Guidance through Prenatal Diagnosis and Genetic Counseling for Recessive Hereditary Hearing Loss in High-Risk Families. Int. J. Pediatr. Otorhinolaryngol. 2018, 115, 114–119. [Google Scholar] [CrossRef]
- Sharma, S.D.; Cushing, S.L.; Papsin, B.C.; Gordon, K.A. Hearing and Speech Benefits of Cochlear Implantation in Children: A Review of the Literature. Int. J. Pediatr. Otorhinolaryngol. 2020, 133, 109984. [Google Scholar] [CrossRef] [PubMed]
- Bruijnzeel, H.; Ziylan, F.; Stegeman, I.; Topsakal, V.; Grolman, W. A Systematic Review to Define the Speech and Language Benefit of Early (<12 Months) Pediatric Cochlear Implantation. Audiol. Neurootol. 2016, 21, 113–126. [Google Scholar] [CrossRef]
- Cejas, I.; Barker, D.H.; Petruzzello, E.; Sarangoulis, C.M.; Quittner, A.L. Cochlear Implantation and Educational and Quality-of-Life Outcomes in Adolescence. JAMA Otolaryngol. Head. Neck Surg. 2023, 149, 708–715. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, D.; He, Y.; Shu, Y. Advances in Gene Therapy Hold Promise for Treating Hereditary Hearing Loss. Mol. Ther. 2023, 31, 934–950. [Google Scholar] [CrossRef]
- Chan, D.K.; Chang, K.W. GJB2-Associated Hearing Loss: Systematic Review of Worldwide Prevalence, Genotype, and Auditory Phenotype. Laryngoscope 2014, 124, E34–E53. [Google Scholar] [CrossRef]
- del Castillo, I.; Morín, M.; Domínguez-Ruiz, M.; Moreno-Pelayo, M.A. Genetic Etiology of Non-Syndromic Hearing Loss in Europe. Hum. Genet. 2022, 141, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Tamagawa, Y.; Ishikawa, K.; Ishikawa, K.; Ishida, T.; Kitamura, K.; Makino, S.; Tsuru, T.; Ichimura, K. Phenotype of DFNA11: A Nonsyndromic Hearing Loss Caused by a Myosin VIIA Mutation. Laryngoscope 2002, 112, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Gooch, C.; Rudy, N.; Smith, R.J.; Robin, N.H. Genetic Testing Hearing Loss: The Challenge of Non Syndromic Mimics. Int. J. Pediatr. Otorhinolaryngol. 2021, 150, 110872. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, R.K.; Walls, W.D.; Corrigan, R.; Schaefer, A.; Wang, K.; Huygen, P.; Casavant, T.L.; Smith, R.J.H. AudioGene: Refining the Natural History of KCNQ4, GSDME, WFS1, and COCH-Associated Hearing Loss. Hum. Genet. 2022, 141, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Tropitzsch, A.; Schade-Mann, T.; Gamerdinger, P.; Dofek, S.; Schulte, B.; Schulze, M.; Fehr, S.; Biskup, S.; Haack, T.B.; Stöbe, P.; et al. Variability in Cochlear Implantation Outcomes in a Large German Cohort With a Genetic Etiology of Hearing Loss. Ear Hear. 2023, 44, 1464–1484. [Google Scholar] [CrossRef] [PubMed]
- Ajay, E.; Gunewardene, N.; Richardson, R. Emerging Therapies for Human Hearing Loss. Expert. Opin. Biol. Ther. 2022, 22, 689–705. [Google Scholar] [CrossRef] [PubMed]
- Moreland, Z.G.; Bird, J.E. Myosin Motors in Sensory Hair Bundle Assembly. Curr. Opin. Cell Biol. 2022, 79, 102132. [Google Scholar] [CrossRef] [PubMed]
- Kuppa, A.; Sergeev, Y.V. Homology Modeling and Global Computational Mutagenesis of Human Myosin VIIa. J. Anal. Pharm. Res. 2021, 10, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, M.S.; Morín, M.; Meyer, N.C.; Mayo, F.; Modamio-Hoybjor, S.; Mencía, A.; Olavarrieta, L.; Morales-Angulo, C.; Nishimura, C.J.; Workman, H.; et al. DFNA8/12 Caused by TECTA Mutations Is the Most Identified Subtype of Nonsyndromic Autosomal Dominant Hearing Loss. Hum. Mutat. 2011, 32, 825–834. [Google Scholar] [CrossRef]
- Santos, R.L.P.; Wajid, M.; Khan, M.N.; McArthur, N.; Pham, T.L.; Bhatti, A.; Lee, K.; Irshad, S.; Mir, A.; Yan, K.; et al. Novel Sequence Variants in the TMC1 Gene in Pakistani Families with Autosomal Recessive Hearing Impairment. Hum. Mutat. 2005, 26, 396. [Google Scholar] [CrossRef]
- Bairoch, A. PROSITE: A Dictionary of Sites and Patterns in Proteins. Nucleic Acids Res. 1991, 19, 2241–2245. [Google Scholar] [CrossRef] [PubMed]
- Bennett-Lovsey, R.M.; Herbert, A.D.; Sternberg, M.J.E.; Kelley, L.A. Exploring the Extremes of Sequence/Structure Space with Ensemble Fold Recognition in the Program Phyre. Proteins 2008, 70, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein Domains Identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
| ID | Age 1 | Sex | Ethnicity | Other Clinical Findings 2 | Temporal Bone Imaging | Panel |
|---|---|---|---|---|---|---|
| 1 | 0.6 | M | NA | None | MRI: Normal | Invitae |
| 2 | 0.8 | F | Hispanic | None | CT and MRI: Normal | Invitae |
| 3 | 3 | M | NA | None | None | Invitae |
| 4 | 15 | M | White | Ocular | CT: Normal | Invitae |
| 5 | 6 | F | White | None | None | Invitae |
| 6 | 1.7 | M | Hispanic | None | MRI: Normal | GeneDx |
| ID | Fhx | NBHS | Onset | R Severity | R Audiogram Shape | R PTA 1 | L Severity 2 | L Audiogram Shape | L PTA | HI | Age at HI |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | None | F | Birth | Severe-to-profound | Flat | 92 | Severe-to-profound | Flat | 92 | CI | 8 mo |
| 2 | Brother | F | Birth | Moderately severe-to-profound | Sloping | 77 | Severe | Flat | 83 | CI | 10 mo |
| 3 | None | P | 3 yr | Mild-to-moderate | Sloping | 27 | Mild-to-moderate | Sloping | 32 | HA | 3 yr |
| 4 | None | F | 2 yr | Moderate-to-moderately severe | Cookie-bite | 52 | Moderate-to-moderately severe | Sloping | 50 | HA | 2 yr |
| 5 | None | P | 3 yr | Moderate-to-severe | Sloping | 52 | Mild-to-severe | Sloping | 55 | HA | 3 yr |
| 6 | Cousin | F | Birth | Profound | Cookie-bite | 65 | Profound | Cookie-bite | 65 | CI | 23 mo |
| ID | Phenotype | Genes | DFN | Variant(s) | Genotype | GPT | Analysis a | New Var | Gene- or Variant- Associated Phenotypes in Literature |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Bilateral severe-to-profound SNHL present at birth | MYO15A | DFNB3 | c.7124_7127del (p.Asp2375Valfs*41) | Het | P | P | No | Gene b: Congenital bilateral severe-to-profound SNHL [32]. Variant: Steeply sloping severe, progressive SNHL [33]; childhood-onset bilateral severe–profound AR SNHL [15]. |
| MYO15A | DFNB3 | c.9109G>T (p.Glu3037*) | Het | P | P | Yes c | |||
| OTOG | DFNB18B | c.4856C>T (p.(Ser1619Leu)) | Het | VUS | -- | Yes | Prelingual moderate AR SNHL [34,35]. | ||
| DMXL2 | DFNA73 | c.7543A>G (p.(Met2515Val)) | Het | VUS | P | Yes | Bilateral mild-to-moderate AD SNHL beginning in 20s, progressing to severe to profound in 60s [36]. | ||
| 2 | Right moderately severe-to-profound SNHL and left severe SNHL present at birth | GJB2 | DFNB1A | c.35delG (p.Gly12Valfs*2) | Hom | P | P | No | Congenital moderate-to-profound bilateral SNHL; severity is variant-dependent [37]. |
| ALMS1 | -- | c.11708G>A (p.(Arg3903Gln)) | Het | VUS | -- | Yes | Associated with AR Alström Syndrome; progressive bilateral moderate SNHL in childhood [38]. | ||
| ADGRV1 | -- | c.13757A>G (p.(Glu4586Gly)) | Het | VUS | -- | Yes | Associated with AR Usher Syndrome type IIC causing congenital moderate-to-severe SNHL [39]. | ||
| KARS1 | DFNB89 | c.1259G>A (p.(Arg420Gln)) | Het | VUS | -- | Yes | Bilateral, symmetric severe-to-profound or moderate-to-severe AR SNHL [40]. | ||
| MYO15A | DFNB3 | c.9620G>A (p.Arg3207His) | Het | P | -- | No | See patient 1; MYO15A. | ||
| 3 | Bilateral mild-to-moderate SNHL, onset at 3 years old | MYO7A | DFNB2/ DFNA11 | c.2543G>A (p.(Arg848Gln)) | Het | VUS | P | Yes | Severe bilateral SNHL [41,42]. |
| LRP2 | -- | c.2426G>A (p.(Ser809Asn)) | Het | VUS | -- | Yes | Associated with AR Donnai–Barrow syndrome (Facio-oculoacousticorenal syndrome) with congenital bilateral profound SNHL though moderate SNHL was also reported [43,44]. May present as non-syndromic bilateral moderate HL in childhood [45]. | ||
| ADGRV1 | -- | c.12052G>A (p.(Val4018Ile)) | Het | VUS | -- | Yes | See patient 2; ADGRV1. | ||
| OTOG | DFNB18B | c.7817_7820dup (p.Tyr2608Serfs*76) | Het | P | -- | Yes | Prelingual bilateral moderate AR SNHL, stable throughout time [34,35]. | ||
| PEX26 | -- | c.98C>T (p.(Pro33Leu)) | Het | VUS | -- | Yes | Post-lingual bilateral moderate-to-severe SNHL [46]. AR Zellweger spectrum disorder results in moderately severe-to-severe SNHL [47]. | ||
| BTD | -- | c.1270G>C (p.Asp424His) | Het | P | -- | No | AR biotinidase deficiency may present with moderate-to-severe sloping SNHL within the 1st year [48]. No variant-specific hearing loss pattern reported previously. | ||
| 4 | Bilateral moderate-to-moderately severe SNHL, onset at 2 years old; retinitis pigmentosa at age 15 | USH2A | -- | c.13130C>A (p.Ser4377*) | Het | P | P | No | Usher Syndrome, type 2—moderate to severe congenital SNHL with retinitis pigmentosa presenting at age 20–30 years [49]. |
| USH2A | -- | c.2299del (p.Glu767Serfs*21) | Het | P | P | No | Variant: Variable severity of progressive HL according to variant [50]. | ||
| COL4A4 | -- | c.980A>G (p.(Glu327Gly)) | Het | VUS | -- | Yes | Associated with AR Alport Syndrome with progressive mild-to-moderate bilateral SNHL that affects mid-to-high frequencies [51]. | ||
| 5 d | Right moderate-to-severe SNHL and left mild-to-severe SNHL, onset at 3 years old | TMC1 | DFNA36/DFNB7/ DFNB11 | c.928A>G (p.(Thr310Ala)) | Het | VUS | P? | Yes | Congenital severe-to-profound SNHL if AR; bilateral, symmetric SNHL that begins at 5–10 years old and rapidly progresses to profound deafness within 10–15 years if AD [52,53]. |
| SLC26A4 | DFNB4 | c.1246A>C (p.Thr416Pro) | Het | P | -- | No | Bilateral fluctuating or progressive moderate-to-severe AR congenital SNHL [54]. | ||
| VCAN | -- | c.3917C>G (p.(Ala1306Gly)) | Het | VUS | -- | Yes | AD Wagner vitreoretinopathy. | ||
| 6 | Bilateral profound non-syndromic SNHL present at birth | TECTA | DFNA8/DFNA12 | c.2266 A>G (p.(Lys756Glu)) | Het | VUS | P | Yes | Moderate-to-severe AD SNHL, most pronounced in the mid-frequencies [55,56]. |
| MITF | -- | c.560-7T>A | Het | VUS | P | Yes | AD Waardenburg syndrome, type II, with congenital bilateral profound SNHL [57]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, N.K.; Uhler, K.M.; Yoon, P.J.; Santos-Cortez, R.L.P. Clinical Genetic Testing for Hearing Loss: Implications for Genetic Counseling and Gene-Based Therapies. Biomedicines 2024, 12, 1427. https://doi.org/10.3390/biomedicines12071427
Lee NK, Uhler KM, Yoon PJ, Santos-Cortez RLP. Clinical Genetic Testing for Hearing Loss: Implications for Genetic Counseling and Gene-Based Therapies. Biomedicines. 2024; 12(7):1427. https://doi.org/10.3390/biomedicines12071427
Chicago/Turabian StyleLee, Nam K., Kristin M. Uhler, Patricia J. Yoon, and Regie Lyn P. Santos-Cortez. 2024. "Clinical Genetic Testing for Hearing Loss: Implications for Genetic Counseling and Gene-Based Therapies" Biomedicines 12, no. 7: 1427. https://doi.org/10.3390/biomedicines12071427
APA StyleLee, N. K., Uhler, K. M., Yoon, P. J., & Santos-Cortez, R. L. P. (2024). Clinical Genetic Testing for Hearing Loss: Implications for Genetic Counseling and Gene-Based Therapies. Biomedicines, 12(7), 1427. https://doi.org/10.3390/biomedicines12071427






