Investigating the Relationship between Epigenetic Age and Cardiovascular Risk in a Population with Overweight/Obesity
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population, Personal Data, and Biological Samples
2.2. Sample Collection and DNA Extraction
2.3. Bisulphite Conversion
2.4. Determination of Epigenetic Age
2.5. Statistical Analysis
3. Results
3.1. Study Population
3.2. Cardiovascular Parameters and DNAm Age
3.3. DNAm Age Acceleration Is Associated with Cardiovascular Risk
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, P.L.; Carrera-Bastos, P.; Castillo-García, A.; Lieberman, D.E.; Santos-Lozano, A.; Lucia, A. Obesity and the Risk of Cardiometabolic Diseases. Nat. Rev. Cardiol. 2023, 20, 475–494. [Google Scholar] [CrossRef]
- Santos, A.L.; Sinha, S. Obesity and Aging: Molecular Mechanisms and Therapeutic Approaches. Ageing Res. Rev. 2021, 67, 101268. [Google Scholar] [CrossRef]
- Costa, E.; Santos-Silva, A.; Paúl, C.; González Gallego, J. Aging and Cardiovascular Risk. BioMed Res. Int. 2015, 2015, e871656. [Google Scholar] [CrossRef]
- Rutledge, J.; Oh, H.; Wyss-Coray, T. Measuring Biological Age Using Omics Data. Nat. Rev. Genet. 2022, 23, 715–727. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Yousefi, P.D.; Suderman, M.; Langdon, R.; Whitehurst, O.; Davey Smith, G.; Relton, C.L. DNA Methylation-Based Predictors of Health: Applications and Statistical Considerations. Nat. Rev. Genet. 2022, 23, 369–383. [Google Scholar] [CrossRef]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.-B.; Gao, Y.; et al. Genome-Wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.G.; Lowe, R.; Adams, P.D.; Baccarelli, A.A.; Beck, S.; Bell, J.T.; Christensen, B.C.; Gladyshev, V.N.; Heijmans, B.T.; Horvath, S.; et al. DNA Methylation Aging Clocks: Challenges and Recommendations. Genome Biol. 2019, 20, 249. [Google Scholar] [CrossRef]
- Zbieć-Piekarska, R.; Spólnicka, M.; Kupiec, T.; Parys-Proszek, A.; Makowska, Ż.; Pałeczka, A.; Kucharczyk, K.; Płoski, R.; Branicki, W. Development of a Forensically Useful Age Prediction Method Based on DNA Methylation Analysis. Forensic Sci. Int. Genet. 2015, 17, 173–179. [Google Scholar] [CrossRef]
- Daunay, A.; Baudrin, L.G.; Deleuze, J.-F.; How-Kit, A. Evaluation of Six Blood-Based Age Prediction Models Using DNA Methylation Analysis by Pyrosequencing. Sci. Rep. 2019, 9, 8862. [Google Scholar] [CrossRef] [PubMed]
- Spólnicka, M.; Pośpiech, E.; Pepłońska, B.; Zbieć-Piekarska, R.; Makowska, Ż.; Pięta, A.; Karłowska-Pik, J.; Ziemkiewicz, B.; Wężyk, M.; Gasperowicz, P.; et al. DNA Methylation in ELOVL2 and C1orf132 Correctly Predicted Chronological Age of Individuals from Three Disease Groups. Int. J. Leg. Med. 2018, 132, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, S.S.; Levy, D.; Vasan, R.S.; Wang, T.J. The Framingham Heart Study and the Epidemiology of Cardiovascular Disease: A Historical Perspective. Lancet 2014, 383, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Bollati, V.; Favero, C.; Albetti, B.; Tarantini, L.; Moroni, A.; Byun, H.-M.; Motta, V.; Conti, D.M.; Tirelli, A.S.; Vigna, L.; et al. Nutrients Intake Is Associated with DNA Methylation of Candidate Inflammatory Genes in a Population of Obese Subjects. Nutrients 2014, 6, 4625–4639. [Google Scholar] [CrossRef] [PubMed]
- Rubino, F.; Puhl, R.M.; Cummings, D.E.; Eckel, R.H.; Ryan, D.H.; Mechanick, J.I.; Nadglowski, J.; Ramos Salas, X.; Schauer, P.R.; Twenefour, D.; et al. Joint International Consensus Statement for Ending Stigma of Obesity. Nat. Med. 2020, 26, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Bollati, V.; Iodice, S.; Favero, C.; Angelici, L.; Albetti, B.; Cacace, R.; Cantone, L.; Carugno, M.; Cavalleri, T.; De Giorgio, B.; et al. Susceptibility to Particle Health Effects, miRNA and Exosomes: Rationale and Study Protocol of the SPHERE Study. BMC Public Health 2014, 14, 1137. [Google Scholar] [CrossRef] [PubMed]
- Rigamonti, A.E.; Bollati, V.; Favero, C.; Albetti, B.; Caroli, D.; Abbruzzese, L.; Cella, S.G.; Sartorio, A. Effect of a 3-Week Multidisciplinary Body Weight Reduction Program on the Epigenetic Age Acceleration in Obese Adults. J. Clin. Med. 2022, 11, 4677. [Google Scholar] [CrossRef] [PubMed]
- Methylation-Based Biological Age and Breast Cancer Risk-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/30794318/ (accessed on 26 February 2024).
- White, A.J.; Kresovich, J.K.; Xu, Z.; Sandler, D.P.; Taylor, J.A. Shift Work, DNA Methylation and Epigenetic Age. Int. J. Epidemiol. 2019, 48, 1536–1544. [Google Scholar] [CrossRef] [PubMed]
- Sartorio, A.; Lafortuna, C.L.; Vangeli, V.; Tavani, A.; Bosetti, C.; La Vecchia, C. Short-Term Changes of Cardiovascular Risk Factors after a Non-Pharmacological Body Weight Reduction Program. Eur. J. Clin. Nutr. 2001, 55, 865–869. [Google Scholar] [CrossRef]
- Huang, R.-C.; Lillycrop, K.A.; Beilin, L.J.; Godfrey, K.M.; Anderson, D.; Mori, T.A.; Rauschert, S.; Craig, J.M.; Oddy, W.H.; Ayonrinde, O.T.; et al. Epigenetic Age Acceleration in Adolescence Associates with BMI, Inflammation, and Risk Score for Middle Age Cardiovascular Disease. J. Clin. Endocrinol. Metab. 2019, 104, 3012–3024. [Google Scholar] [CrossRef]
- Nevalainen, T.; Kananen, L.; Marttila, S.; Jylhävä, J.; Mononen, N.; Kähönen, M.; Raitakari, O.T.; Hervonen, A.; Jylhä, M.; Lehtimäki, T.; et al. Obesity Accelerates Epigenetic Aging in Middle-Aged but Not in Elderly Individuals. Clin. Epigenetics 2017, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhang, T.; Su, S.; Hao, G.; Chen, T.; Li, Q.-Z.; Bazzano, L.; He, J.; Wang, X.; Li, S.; et al. Body Mass Index Drives Changes in DNA Methylation. Circ. Res. 2019, 125, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Tekola-Ayele, F. Invited Commentary: Epigenetic Clocks and Obesity-Towards the Next Frontier Using Integrative Approaches and Early-Life Models. Am. J. Epidemiol. 2021, 190, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Franzago, M.; Pilenzi, L.; Di Rado, S.; Vitacolonna, E.; Stuppia, L. The Epigenetic Aging, Obesity, and Lifestyle. Front. Cell Dev. Biol. 2022, 10, 985274. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.J.; Chandra, T. Epigenetic Age Prediction. Aging Cell 2021, 20, e13452. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Mueller, A.; English, B.; Arena, A.; Vera, D.; Kane, A.E.; Sinclair, D.A. Novel Feature Selection Methods for Construction of Accurate Epigenetic Clocks. PLoS Comput. Biol. 2022, 18, e1009938. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Chao, D.L.; Rocha, L.; Kolar, M.; Nguyen Huu, V.A.; Krawczyk, M.; Dasyani, M.; Wang, T.; Jafari, M.; Jabari, M.; et al. The Lipid Elongation Enzyme ELOVL2 Is a Molecular Regulator of Aging in the Retina. Aging Cell 2020, 19, e13100. [Google Scholar] [CrossRef]
- Garagnani, P.; Bacalini, M.G.; Pirazzini, C.; Gori, D.; Giuliani, C.; Mari, D.; Di Blasio, A.M.; Gentilini, D.; Vitale, G.; Collino, S.; et al. Methylation of ELOVL2 Gene as a New Epigenetic Marker of Age. Aging Cell 2012, 11, 1132–1134. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Watanabe, M.; Hatakeyama, S. TRIM59 Interacts with ECSIT and Negatively Regulates NF-κB and IRF-3/7-Mediated Signal Pathways. Biochem. Biophys. Res. Commun. 2012, 422, 501–507. [Google Scholar] [CrossRef]
- Koch, C.M.; Wagner, W. Epigenetic-Aging-Signature to Determine Age in Different Tissues. Aging 2011, 3, 1018–1027. [Google Scholar] [CrossRef]
- Akash, M.S.H.; Rasheed, S.; Rehman, K.; Ibrahim, M.; Imran, M.; Assiri, M.A. Biochemical Activation and Regulatory Functions of Trans-Regulatory KLF14 and Its Association with Genetic Polymorphisms. Metabolites 2023, 13, 199. [Google Scholar] [CrossRef] [PubMed]
- Carugno, M.; Maggioni, C.; Ruggiero, V.; Crespi, E.; Monti, P.; Ferrari, L.; Pesatori, A.C. Can Night Shift Work Affect Biological Age? Hints from a Cross-Sectional Study on Hospital Female Nurses. Int. J. Environ. Res. Public Health 2021, 18, 10639. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y.; et al. An Epigenetic Biomarker of Aging for Lifespan and Healthspan. Aging 2018, 10, 573–591. [Google Scholar] [CrossRef] [PubMed]
- Belsky, D.W.; Caspi, A.; Arseneault, L.; Baccarelli, A.; Corcoran, D.L.; Gao, X.; Hannon, E.; Harrington, H.L.; Rasmussen, L.J.; Houts, R.; et al. Quantification of the Pace of Biological Aging in Humans through a Blood Test, the DunedinPoAm DNA Methylation Algorithm. eLife 2020, 9, e54870. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.E. Assessment of Epigenetic Clocks as Biomarkers of Aging in Basic and Population Research. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 463–465. [Google Scholar] [CrossRef]
- Horvath, S.; Ritz, B.R. Increased Epigenetic Age and Granulocyte Counts in the Blood of Parkinson’s Disease Patients. Aging 2015, 7, 1130–1142. [Google Scholar] [CrossRef] [PubMed]
- Boutari, C.; Mantzoros, C.S. Inflammation: A Key Player Linking Obesity with Malignancies. Metabolism 2018, 81, A3–A6. [Google Scholar] [CrossRef] [PubMed]
- Kirk, E.P.; Klein, S. Pathogenesis and Pathophysiology of the Cardiometabolic Syndrome. J. Clin. Hypertens. 2009, 11, 761–765. [Google Scholar] [CrossRef]
- Moqri, M.; Herzog, C.; Poganik, J.R.; Justice, J.; Belsky, D.W.; Higgins-Chen, A.; Moskalev, A.; Fuellen, G.; Cohen, A.A.; Bautmans, I.; et al. Biomarkers of Aging for the Identification and Evaluation of Longevity Interventions. Cell 2023, 186, 3758–3775. [Google Scholar] [CrossRef]
- Niccoli, T.; Partridge, L.; Isaacs, A.M. Ageing as a Risk Factor for ALS/FTD. Hum. Mol. Genet. 2017, 26, R105–R113. [Google Scholar] [CrossRef]
- Schmid-Schönbein, G.W. 2008 Landis Award Lecture. Inflammation and the Autodigestion Hypothesis. Microcirculation 2009, 16, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Nannini, D.R.; Joyce, B.T.; Zheng, Y.; Gao, T.; Liu, L.; Yoon, G.; Huan, T.; Ma, J.; Jacobs, D.R.; Wilkins, J.T.; et al. Epigenetic Age Acceleration and Metabolic Syndrome in the Coronary Artery Risk Development in Young Adults Study. Clin. Epigenetics 2019, 11, 160. [Google Scholar] [CrossRef] [PubMed]
- A Systematic Review of Biological, Social and Environmental Factors Associated with Epigenetic Clock Acceleration-ScienceDirect. Available online: https://www.sciencedirect.com/science/article/pii/S1568163721000957 (accessed on 23 February 2024).
- Bellver-Sanchis, A.; Pallàs, M.; Griñán-Ferré, C. The Contribution of Epigenetic Inheritance Processes on Age-Related Cognitive Decline and Alzheimer’s Disease. Epigenomes 2021, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Gonzalez-Latapi, P.; Marras, C.; Visanji, N.P.; Yang, W.; Sato, C.; Lang, A.E.; Rogaeva, E.; Zhang, M. Epigenetic Clock Acceleration Is Linked to Age at Onset of Parkinson’s Disease. Mov. Disord. 2022, 37, 1831–1840. [Google Scholar] [CrossRef]
- Ammous, F.; Zhao, W.; Ratliff, S.M.; Mosley, T.H.; Bielak, L.F.; Zhou, X.; Peyser, P.A.; Kardia, S.L.R.; Smith, J.A. Epigenetic Age Acceleration Is Associated with Cardiometabolic Risk Factors and Clinical Cardiovascular Disease Risk Scores in African Americans. Clin. Epigenetics 2021, 13, 55. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.D.; Vittinghoff, E.; Lu, A.T.; Alonso, A.; Wang, B.; Sitlani, C.M.; Mohammadi-Shemirani, P.; Fornage, M.; Kornej, J.; Brody, J.A.; et al. Epigenetic Age and the Risk of Incident Atrial Fibrillation. Circulation 2021, 144, 1899–1911. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D.; et al. DNA Methylation GrimAge Strongly Predicts Lifespan and Healthspan. Aging 2019, 11, 303–327. [Google Scholar] [CrossRef] [PubMed]
- Systematic Review and Meta-Analysis of Environmental, Lifestyle, and Health Factors Associated with DNA Methylation Age|The Journals of Gerontology: Series A|Oxford Academic. Available online: https://academic.oup.com/biomedgerontology/article/75/3/481/5475146 (accessed on 26 February 2024).
- Fransquet, P.D.; Wrigglesworth, J.; Woods, R.L.; Ernst, M.E.; Ryan, J. The Epigenetic Clock as a Predictor of Disease and Mortality Risk: A Systematic Review and Meta-Analysis. Clin. Epigenetics 2019, 11, 62. [Google Scholar] [CrossRef] [PubMed]
- DNA Methylation-Based Biomarkers of Aging Were Slowed down in a Two-Year Diet and Physical Activity Intervention Trial: The DAMA Study-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/34535961/ (accessed on 9 July 2024).
- Schoenaker, D.A.J.M.; Jackson, C.A.; Rowlands, J.V.; Mishra, G.D. Socioeconomic Position, Lifestyle Factors and Age at Natural Menopause: A Systematic Review and Meta-Analyses of Studies across Six Continents. Int. J. Epidemiol. 2014, 43, 1542–1562. [Google Scholar] [CrossRef]
- Levine, M.E.; Lu, A.T.; Chen, B.H.; Hernandez, D.G.; Singleton, A.B.; Ferrucci, L.; Bandinelli, S.; Salfati, E.; Manson, J.E.; Quach, A.; et al. Menopause Accelerates Biological Aging. Proc. Natl. Acad. Sci. USA 2016, 113, 9327–9332. [Google Scholar] [CrossRef]
- Opoku, A.A.; Abushama, M.; Konje, J.C. Obesity and Menopause. Best. Pract. Res. Clin. Obstet. Gynaecol. 2023, 88, 102348. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Chung, H.-F.; Dobson, A.J.; Pandeya, N.; Giles, G.G.; Bruinsma, F.; Brunner, E.J.; Kuh, D.; Hardy, R.; Avis, N.E.; et al. Age at Natural Menopause and Risk of Incident Cardiovascular Disease: A Pooled Analysis of Individual Patient Data. Lancet Public Health 2019, 4, e553–e564. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.; Minhas, A.S.; Kazzi, B.; Varma, B.; Choi, E.; Thakkar, A.; Michos, E.D. Sex-Specific Differences in Cardiovascular Risk Factors and Implications for Cardiovascular Disease Prevention in Women. Atherosclerosis 2023, 384, 117269. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, C.P.; Ogunmoroti, O.; Minhas, A.S.; Vaidya, D.; Kazzi, B.; Osibogun, O.; Whelton, S.; Kovell, L.C.; Harrington, C.M.; Honigberg, M.C.; et al. Female-Specific Risk Factors of Parity and Menopause Age and Risk of Carotid Plaque: The Multi-Ethnic Study of Atherosclerosis. Am. J. Cardiovasc. Dis. 2023, 13, 222–234. [Google Scholar] [PubMed]
- Prabakaran, S.; Schwartz, A.; Lundberg, G. Cardiovascular Risk in Menopausal Women and Our Evolving Understanding of Menopausal Hormone Therapy: Risks, Benefits, and Current Guidelines for Use. Ther. Adv. Endocrinol. Metab. 2021, 12, 20420188211013917. [Google Scholar] [CrossRef]
| Characteristics | |
|---|---|
| Age, years | 51.7 ± 18.1 |
| Gender | |
| Males | 47 (24.7%) |
| Females | 143 (75.6%) |
| BMI, kg/m2 | 33 ± 4.6 |
| BMI < 30 | 50 (26.3%) |
| BMI 30–35 | 80 (42.1%) |
| BMI ≥ 35 | 60 (31.6%) |
| Smoking status | |
| Non-smoker | 88 (46.3%) |
| Smoker | 102 (53.7%) |
| Alcohol consumption | |
| Yes | 80 (42.2%) |
| No | 55 (28.9%) |
| Missing | 55 (28.9%) |
| Physical activity levels | |
| Sedentary | 108 (56.8%) |
| Active | 59 (31.0%) |
| Sporty | 11 (5.8%) |
| Active and sporty | 6 (3.2%) |
| Missing | 6 (3.2%) |
| Clinical Characteristics | |
|---|---|
| Menopause (only for 143 women) | |
| Yes | 84 (58.8%) |
| No | 55 (38.5%) |
| Missing | 4 (2.7%) |
| Metabolic syndrome | |
| Yes | 79 (41.6%) |
| No | 111 (58.4%) |
| Blood pressure, mmHg | |
| Systolic | 124.2 ± 17.9 |
| Diastolic | 77.0 ± 9.6 |
| Antihypertensive medications | |
| Yes | 71 (37.4%) |
| No | 119 (62.6%) |
| Glucose, mg/dL | 92 [85, 103] |
| Glycated hemoglobin, mmol/mol | 39.8 [36.6, 43] |
| Insulin level, U/mL | 12.0 [8.8, 17.9] |
| Diabetes | |
| Yes | 40 (21.0%) |
| No | 150 (79.0%) |
| Diabetes medications | |
| Yes | 24 (12.6%) |
| No | 166 (87.4%) |
| Triglycerides, mg/dL | 100 [75, 145] |
| Total cholesterol, mg/dL | 204.1 ± 41.2 |
| HDL, mg/dL | 58.6 ± 15.8 |
| LDL, mg/dL | 125.2 ± 36.7 |
| Lipid-lowering medications | |
| Yes | 26 (13.7%) |
| No | 164 (86.3%) |
| Heart rate, bpm | 66.8 ± 10.2 |
| Fibrinogen, mg/dL | 334 ± 63.6 |
| C-reactive protein, mg/L | 0.30 [0.15, 0.49] |
| Serum creatinine, mg/dL | 0.8 ± 0.2 |
| AST, U/L | 20 [17, 24] |
| ALT, U/L | 20 [15, 30] |
| Gamma-flutamyltransferase, U/L | 17 [12, 28] |
| TSH, U/mL | 1.9 [1.2, 2.6] |
| Neutrophils, % | 58.5 ± 7.9 |
| Eosinophils, % | 2.6 ± 1.6 |
| Lymphocytes, % | 30.5 ± 7.7 |
| Monocytes, % | 7.7 ± 1.9 |
| Basophils, % | 0.5 ± 0.3 |
| Granulocytes, % | 61.7 ± 7.6 |
| Framingham risk score, % | 5.8 [2.1, 12.2] |
| β | SE | p-Value | ||
|---|---|---|---|---|
| Gender | ||||
| Female vs. male | 0.997 | 0.791 | 0.209 | |
| BMI, kg/m2 | 0.009 | 0.073 | 0.898 | |
| BMI 30;35 vs. BMI < 30 | 1.053 | 0.833 | 0.208 | 0.523 |
| BMI ≥ 35 vs. BMI < 30 | −0.287 | 0.884 | 0.746 | |
| Smoking habits | ||||
| Smoker vs. non-smoker | −0.708 | 0.675 | 0.269 | |
| Alcohol consumption | ||||
| Yes vs. No | −0.198 | 0.752 | 0.793 | |
| Physical activity levels | ||||
| Active vs. sedentary behavior | 0.968 | 0.746 | 0.196 | |
| Sporty vs. sedentary behavior | −0.927 | 1.475 | 0.531 | 0.619 |
| Active and sporty vs. sedentary behavior | 1.811 | 1.869 | 0.334 | |
| Menopause (only women) | ||||
| Yes vs. no | 2.219 | 0.767 | 0.005 | |
| Metabolic syndrome | ||||
| Yes vs. no | 0.875 | 0.688 | 0.205 | |
| Blood pressure, mmHg | ||||
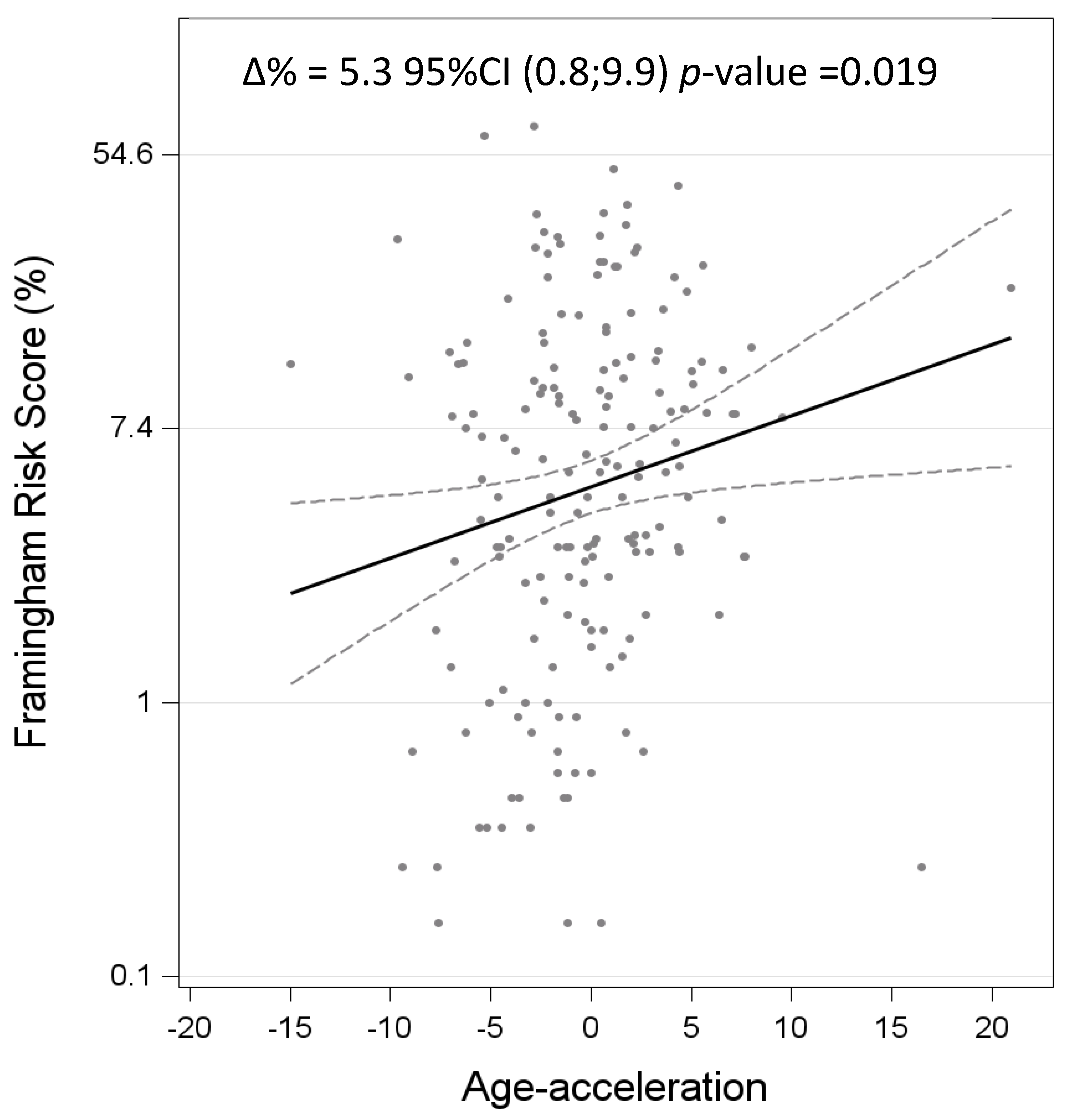
| Systolic | 0.045 | 0.019 | 0.019 | |
| Diastolic | 0.036 | 0.035 | 0.303 | |
| Antihypertensive medications | ||||
| Yes vs. no | 0.336 | 0.703 | 0.633 | |
| Glucose, mg/dL | 0.025 | 0.012 | 0.030 | |
| Glycated hemoglobin, mmol/mol | 0.105 | 0.042 | 0.013 | |
| Insulin level, U/mL | 0.033 | 0.038 | 0.389 | |
| Diabetes | ||||
| Yes vs. No | 2.247 | 0.841 | 0.008 | |
| Diabetes medications | ||||
| Yes vs. no | 1.145 | 1.042 | 0.273 | |
| Triglycerides, mg/dL | 0.001 | 0.003 | 0.640 | |
| Total cholesterol, mg/dL | 0.015 | 0.008 | 0.069 | |
| HDL, mg/dL | 0.006 | 0.021 | 0.774 | |
| LDL, mg/dL | 0.013 | 0.009 | 0.173 | |
| Lipid-lowering medications | ||||
| Yes vs. no | 0.896 | 1.022 | 0.382 | |
| Heart rate, bpm | 0.096 | 0.032 | 0.003 | |
| Fibrinogen, mg/dL | −0.001 | 0.005 | 0.904 | |
| C-reactive protein, mg/L | 0.175 | 0.492 | 0.723 | |
| Serum creatinine, mg/dL | −0.327 | 1.526 | 0.831 | |
| AST, U/L | −0.013 | 0.035 | 0.713 | |
| ALT, U/L | −0.011 | 0.014 | 0.434 | |
| Gamma-glutamyltransferase, U/L | 0.002 | 0.016 | 0.922 | |
| TSH, U/mL | 0.295 | 0.255 | 0.249 | |
| Neutrophils, % | 0.100 | 0.042 | 0.018 | |
| Eosinophils, % | −0.252 | 0.207 | 0.225 | |
| Lymphocytes, % | −0.080 | 0.043 | 0.068 | |
| Monocytes, % | −0.150 | 0.183 | 0.416 | |
| Basophils, % | −0.483 | 1.149 | 0.674 | |
| Granulocytes, % | 0.095 | 0.044 | 0.033 | |
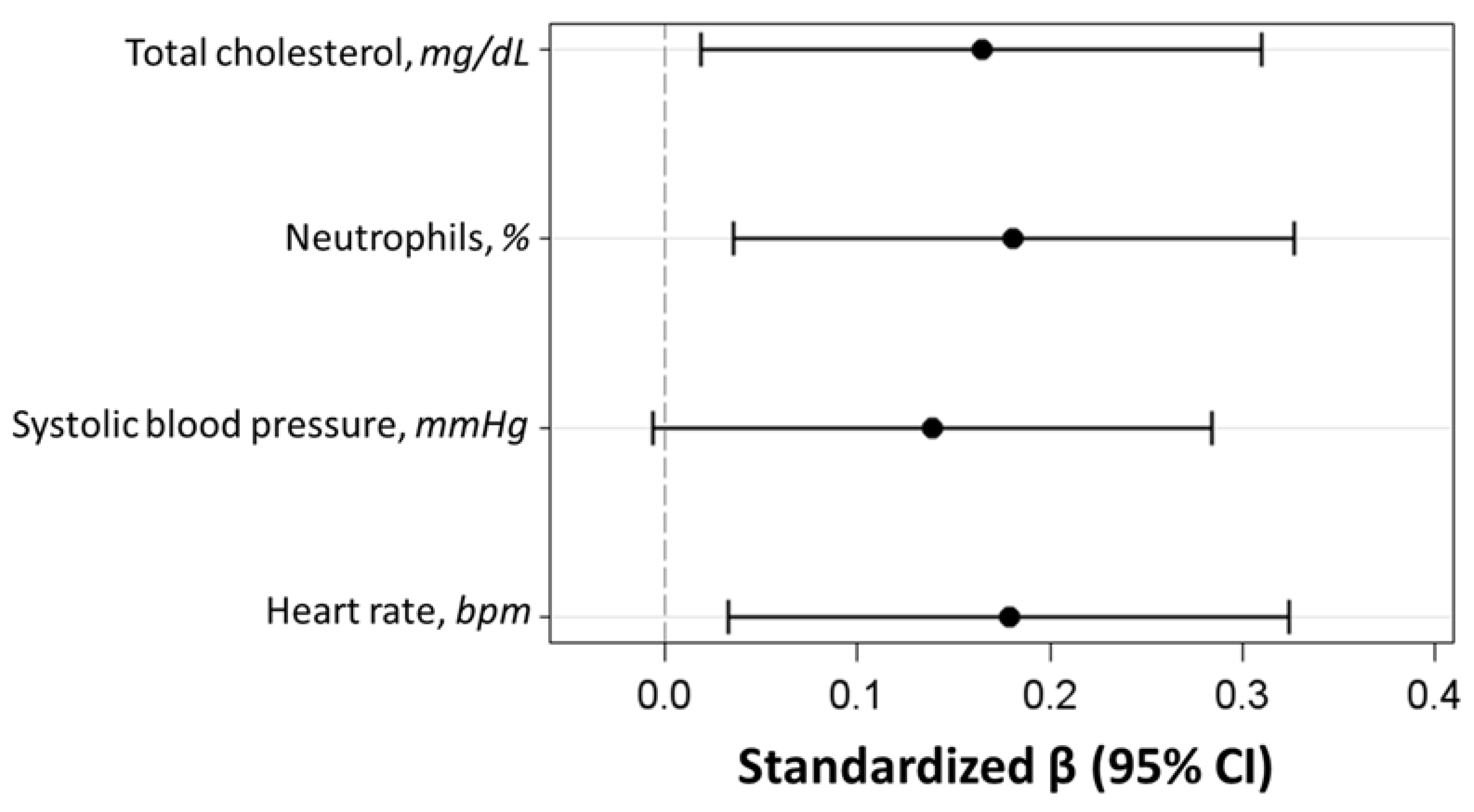
| β | SE | 95% CI | Partial Correlation Coefficient | p-Value | |
|---|---|---|---|---|---|
| Heart rate, bpm | 0.078 | 0.032 | (0.014; 0.141) | 0.183 | 0.016 |
| Systolic blood pressure, mmHg | 0.035 | 0.018 | (−0.002; 0.071) | 0.143 | 0.061 |
| Total cholesterol, mg/dL | 0.019 | 0.008 | (0.020; 0.185) | 0.168 | 0.028 |
| Neutrophils, % | 0.102 | 0.041 | (0.002; 0.035) | 0.185 | 0.015 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marinello, D.; Favero, C.; Albetti, B.; Barbuto, D.; Vigna, L.; Pesatori, A.C.; Bollati, V.; Ferrari, L. Investigating the Relationship between Epigenetic Age and Cardiovascular Risk in a Population with Overweight/Obesity. Biomedicines 2024, 12, 1631. https://doi.org/10.3390/biomedicines12081631
Marinello D, Favero C, Albetti B, Barbuto D, Vigna L, Pesatori AC, Bollati V, Ferrari L. Investigating the Relationship between Epigenetic Age and Cardiovascular Risk in a Population with Overweight/Obesity. Biomedicines. 2024; 12(8):1631. https://doi.org/10.3390/biomedicines12081631
Chicago/Turabian StyleMarinello, Davide, Chiara Favero, Benedetta Albetti, Davide Barbuto, Luisella Vigna, Angela Cecilia Pesatori, Valentina Bollati, and Luca Ferrari. 2024. "Investigating the Relationship between Epigenetic Age and Cardiovascular Risk in a Population with Overweight/Obesity" Biomedicines 12, no. 8: 1631. https://doi.org/10.3390/biomedicines12081631
APA StyleMarinello, D., Favero, C., Albetti, B., Barbuto, D., Vigna, L., Pesatori, A. C., Bollati, V., & Ferrari, L. (2024). Investigating the Relationship between Epigenetic Age and Cardiovascular Risk in a Population with Overweight/Obesity. Biomedicines, 12(8), 1631. https://doi.org/10.3390/biomedicines12081631






