SNHG1: Redefining the Landscape of Hepatocellular Carcinoma through Long Noncoding RNAs
Abstract
:1. Introduction
2. SNHG1 and Liver Carcinogenesis
2.1. The SNHG Family
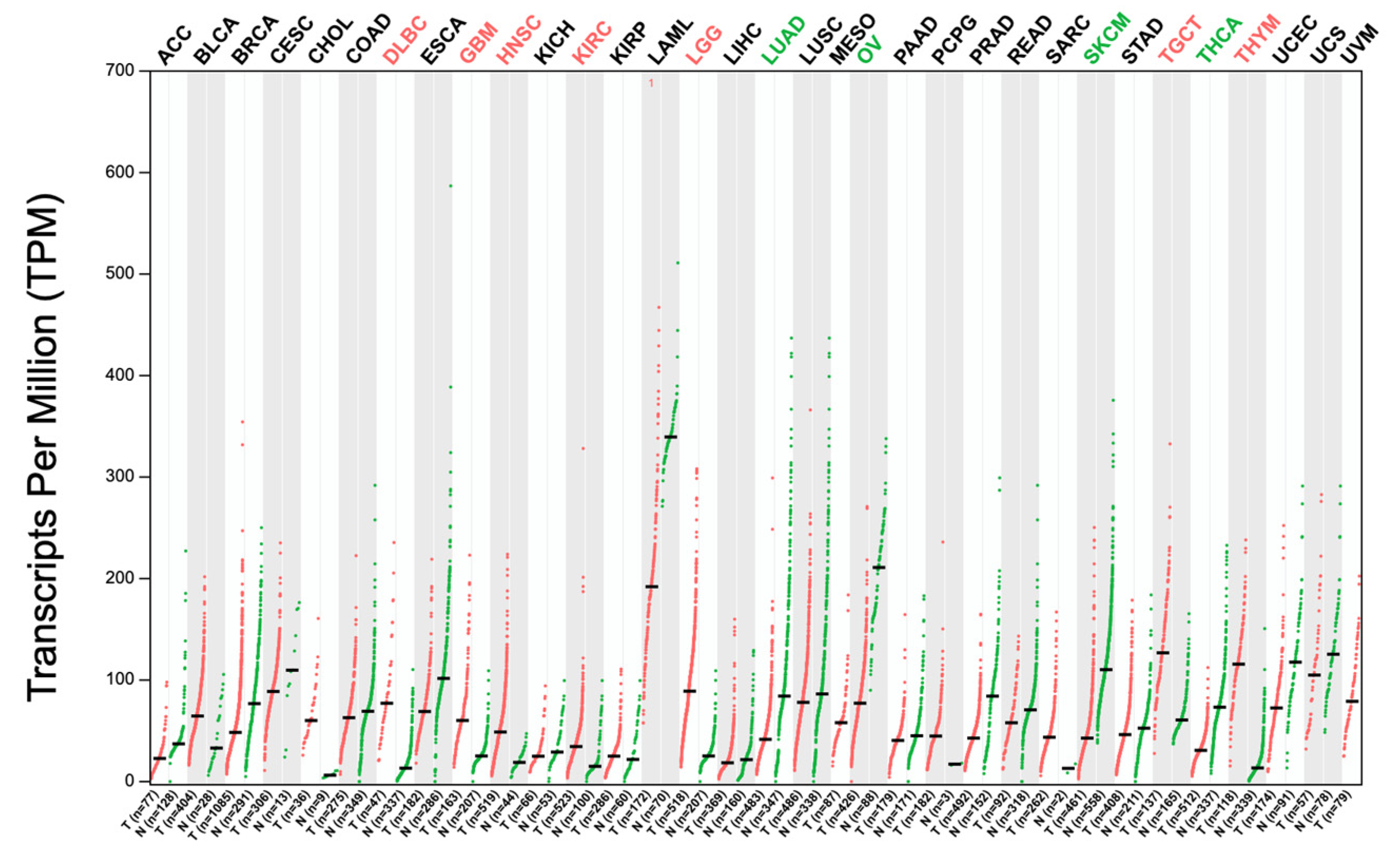
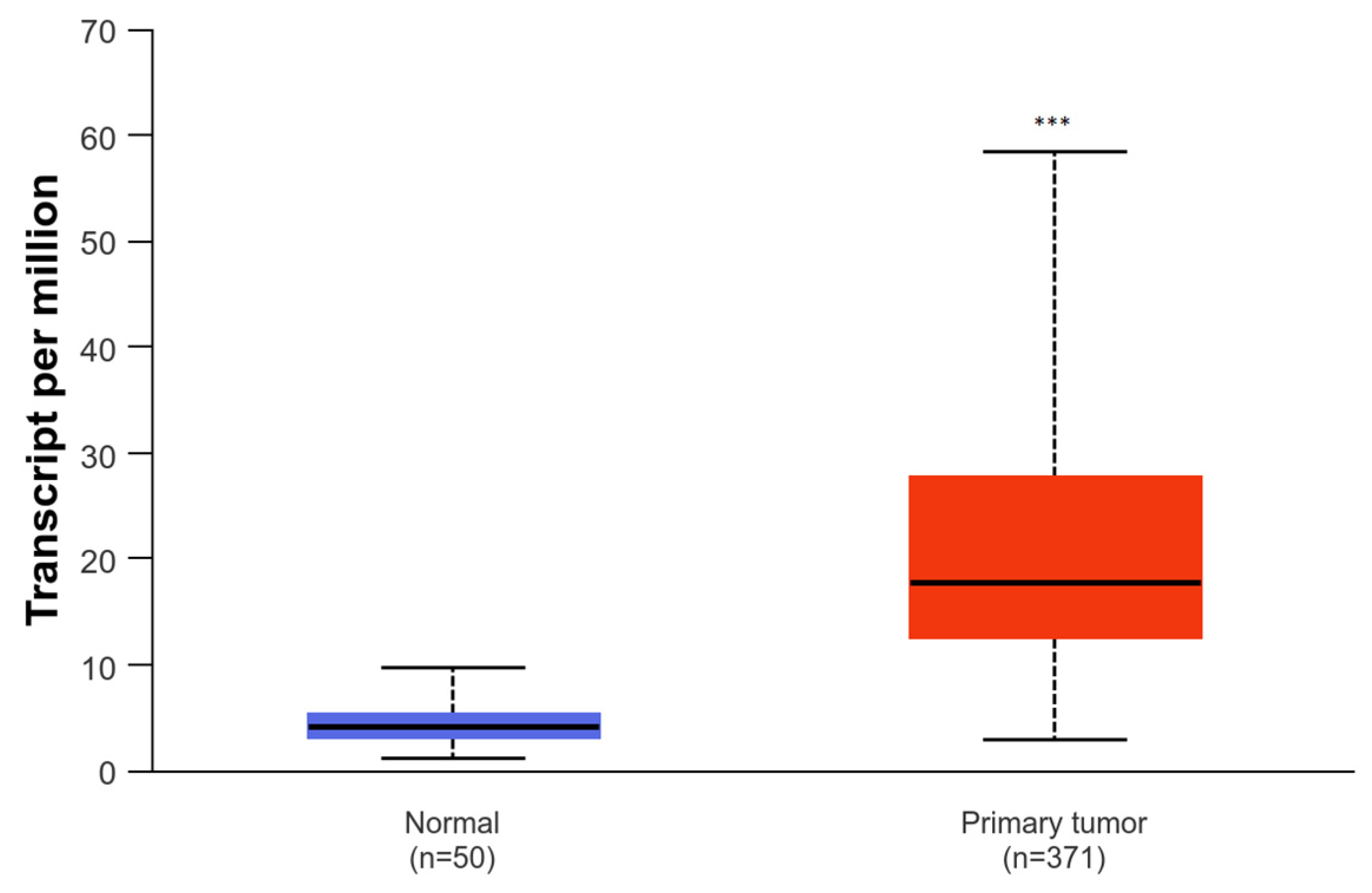
2.2. SNHG1
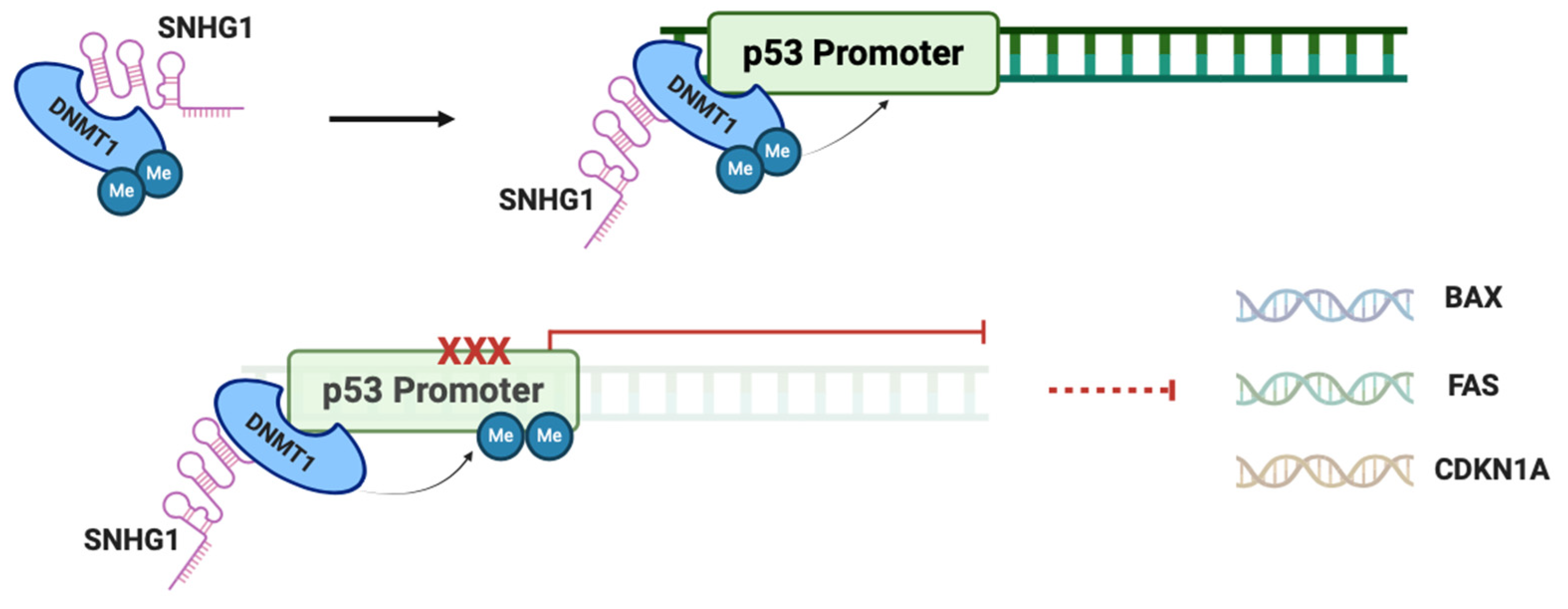
2.3. SNHG1 and p53 in HCC
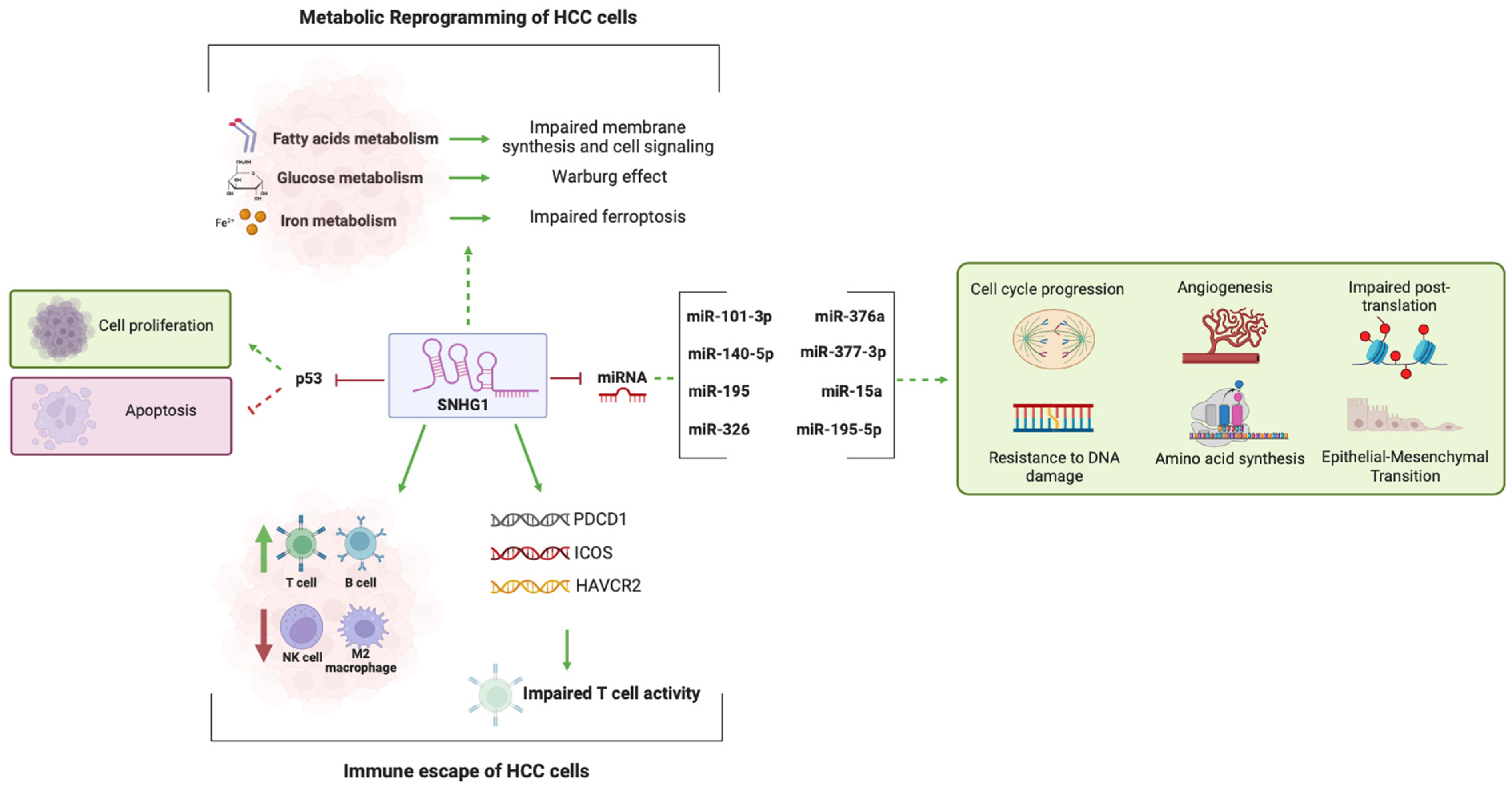
2.4. SNHG1 and miRNA Activity
2.5. SNHG1 and Metabolic Reprogramming in HCC
2.6. SNHG1 and the Immune Landscape of HCC
2.7. SNHG1 and Treatment Resistance
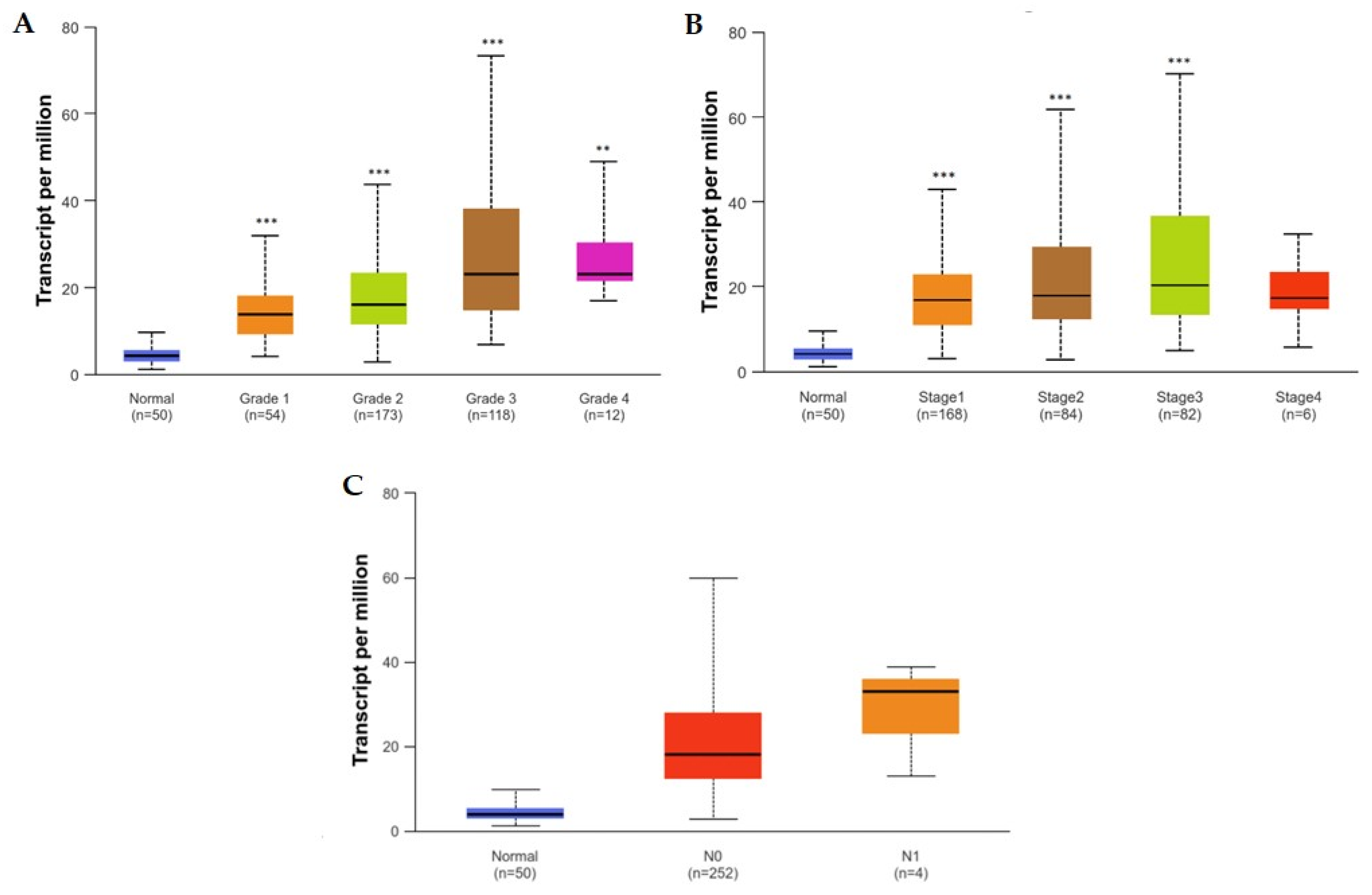
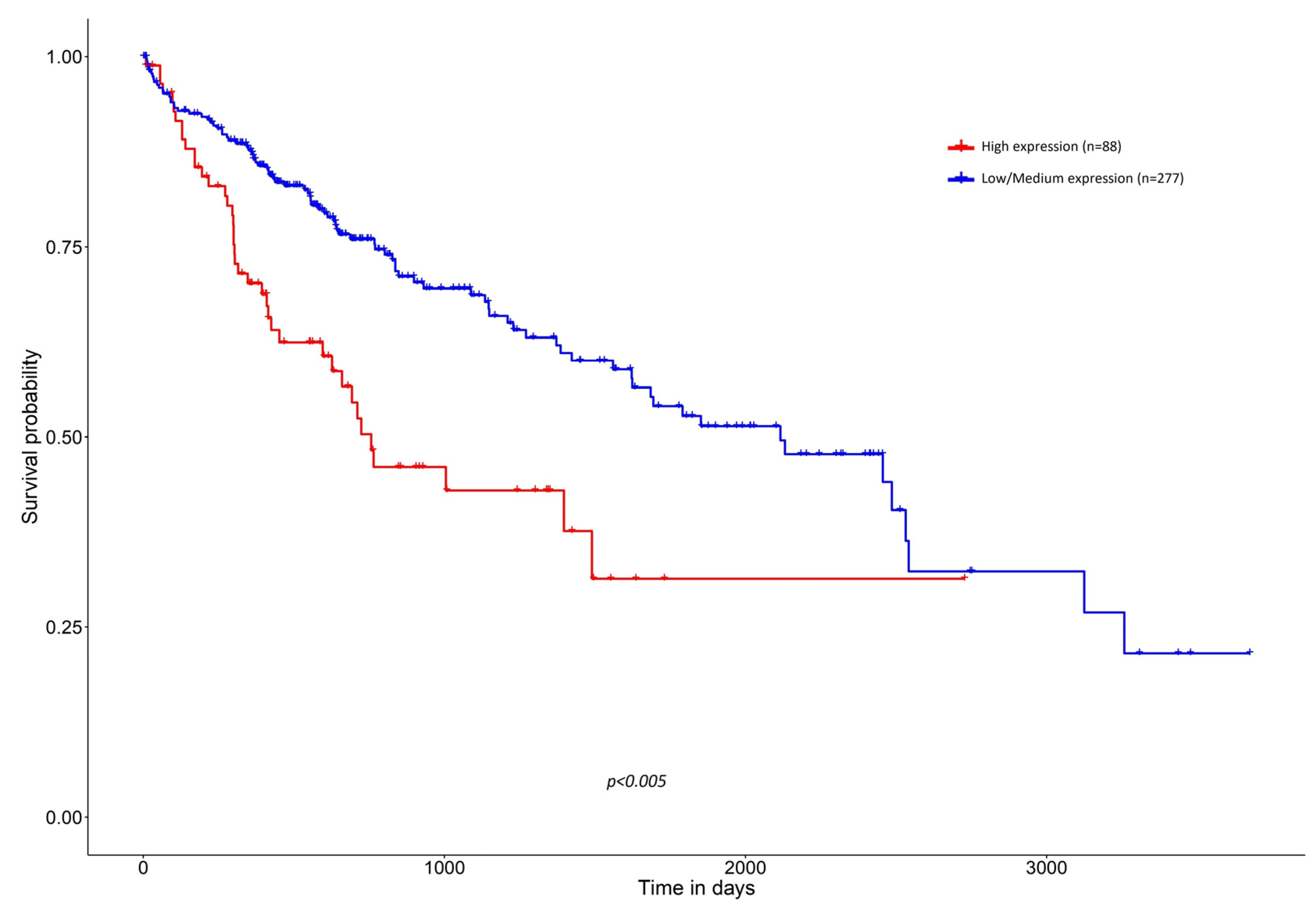
3. Clinicopathological Associations between High Expression of SNHG1 and HCC
| Population (n) | Age | Tumor Diameter (cm) | Differentiation | LN Metastasis | Distant Metastasis | TNM Stage | AFP (ng/mL) | Ref. |
|---|---|---|---|---|---|---|---|---|
| 82 | >55 vs. ≤55 | <5 vs. ≥5 | High/moderate vs. Low | - | - | BCLC A vs. B-C | >20 vs. ≤20 | [14] |
| 233 | >60 vs. ≤60 | - | G1 + G2 vs. G3 + G4 | N0 vs. N1 | M0 vs. M1 | AJCC I-II vs. III-IV | - | [27] |
| 150 | ≥46 vs. <46 | ≥7 vs. <7 | High/moderate vs. Low | N0 vs. N1/N2 | - | AJCC I-II vs. III-IV | - | [29] |
| 358 | >60 vs. ≤60 | - | G1 + G2 vs. G3 + G4 | N0 vs. N1 vs. Nx | M0 vs. M1 | AJCC I-II vs. III-IV | ≤400 vs. >400 | [55] |
| 576 | - | - | G1 vs. G2 vs. G3 vs. G4 | - | - | AJCC I vs. II vs. III vs. IV | - | [54] |
| 72 | <50 vs. ≥50 | <5 vs. ≥5 | - | - | - | AJCC I-II vs. III-IV | <200 vs. ≥200 | [76] |
| 389 | >60 vs. ≤60 | - | G1 vs. G2 vs. G3 vs. G4 | - | - | AJCC I vs. II vs. III vs. IV | - | [78] |
4. Future Directions
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACSBG1 | Acyl-CoA Synthetase Bubblegum Family Member 1 |
| AEG-1 | Astrocyte Elevated Gene-1 |
| AFP | Alpha-Fetoprotein |
| AJCC | American Joint Committee on Cancer |
| ATG5 | Autophagy related 5 |
| AUC | Area under the curve |
| BAX | Bcl-2 Associated X-protein |
| Bcl-2 | B-cell lymphoma 2 |
| BCLC | Barcelona Clinic Liver Cancer |
| BMSCs | Bone marrow stromal cells |
| BUB1 | Budding Uninhibited By Benzimidazole 1 |
| CCNA2 | Cyclin A2 |
| CCNB1 | Cyclin B1 |
| CD98 | Cluster of Differentiation 98 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B |
| CDK4 | Cyclin-dependent kinase 4 |
| ceRNA | Competitive endogenous RNA |
| CT | Computed tomography |
| DNA | Desoxyribonucleic acid |
| DNMT1 | DNA Methyltransferase 1 |
| EMT | Epithelial-to-mesenchymal transition |
| EZH2 | Histone-lysine N-methyltransferase EZH2 |
| FA | Fatty acid |
| FANCE | Fanconi Anemia Complementation Group E |
| FAS | Fas Cell Surface Death Receptor |
| FOXK1 | Forkhead Box K1 |
| GPX4 | Glutathione Peroxidase 4 |
| HAVCR2 | Hepatitis A Virus Cellular Receptor 2 |
| HCC | Hepatocellular carcinoma |
| HLC | Hepatocyte-like cell |
| hnRNCP | Heterogeneous Nuclear Ribonucleoprotein C1/C2 |
| ICOS | Inducible T-Cell Costimulator |
| KIF11 | Kinesin Family Member 11 |
| KIF2C | Kinesin Family Member 2c |
| LMNB2 | Lamin B2 |
| LncRNA | Long noncoding RNA |
| M2 | M2 Macrophage |
| miRNA | microRNA |
| MRI | Magnet resonance imaging |
| mRNA | Messenger RNA |
| NCOA4 | Nuclear Receptor Coactivator 4 |
| NCAPG | Non-SMC Condensin I Complex Subunit G |
| NK | Natural killer |
| NRF2 | Nuclear Factor Erythroid 2-Related Factor |
| OS | Overall survival |
| PDCD1 | Programmed Cell Death Protein 1 |
| PDCD4 | Programmed Cell Death Protein 4 |
| PDGFR | Platelet-Derived Growth Factor Receptor |
| PKM2 | Pyruvate Kinase M1/2 |
| SAP30 | Sin3-Associated Protein 30 |
| SLC3A2 | Solute Carrier Family 3 Member 2 |
| SMURF1 | Smad Ubiquitination Regulatory Factor 1 |
| SNHG1 | Small Nucleolar RNA Host Gene 1 |
| SOX9 | SRY-Box Transcription Factor 9 |
| TKTL1 | Transketolase-Like-1 |
| TNB | Tumor Neoantigen Burden |
| TOP2A | DNA Topoisomerase II Alpha |
| VEGFR | Vascular Endothelial Growth Factor Receptor |
| Wnt5a | Wnt Family Member 5A |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Geller, D.A.; Goss, J.A.; Busuttil, R.W.; Tsung, A. Liver. In Schwartz’s Principles of Surgery, 11th ed.; Brunicardi, F.C., Andersen, D.K., Billiar, T.R., Dunn, D.L., Kao, L.S., Hunter, J.G., Matthews, J.B., Pollock, R.E., Eds.; McGraw-Hill Education: New York, NY, USA, 2019; Available online: http://accessmedicine.mhmedical.com/content.aspx?aid=1164318762 (accessed on 26 March 2024).
- Saffroy, R.; Pham, P.; Reffas, M.; Takka, M.; Lemoine, A.; Debuire, B. New perspectives and strategy research biomarkers for hepatocellular carcinoma. Clin. Chem. Lab. Med. 2007, 45, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Hu, X.; Zhao, X.; Zhang, L. Focally amplified lnc RNA genes in cancer. Oncoscience 2015, 2, 205–206. [Google Scholar] [CrossRef]
- Carlevaro-Fita, J.; Lanzós, A.; Feuerbach, L.; Hong, C.; Mas-Ponte, D.; Pedersen, J.S.; PCAWG Drivers and Functional Interpretation Group; Johnson, R.; PCAWG Consortium. Cancer LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. Commun. Biol. 2020, 3, 56. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Feng, X.; Liu, M.; Gong, H.; Zhou, X. SnoRNA and lncSNHG: Advances of nucleolar small RNA host gene transcripts in anti-tumor immunity. Front. Immunol. 2023, 14, 1143980. [Google Scholar] [CrossRef]
- Zimta, A.-A.; Tigu, A.B.; Braicu, C.; Stefan, C.; Ionescu, C.; Berindan-Neagoe, I. An Emerging Class of Long Non-coding RNA With Oncogenic Role Arises From the snoRNA Host Genes. Front. Oncol. 2020, 10, 389. [Google Scholar] [CrossRef]
- Frey, M.R.; Wu, W.; Dunn, J.M.; Matera, A.G. The U22 host gene (UHG): Chromosomal localization of UHG and distribution of U22 small nucleolar RNA. Histochem. Cell Biol. 1997, 108, 365–370. [Google Scholar] [CrossRef]
- Ozaki, T.; Nakagawara, A. Role of p53 in Cell Death and Human Cancers. Cancers 2011, 3, 994. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-J.; Wang, L.; Sun, Z.-X.; Sun, S.-J.; Gao, J.; Ma, R.-L. LncRNA SNHG1 promotes liver cancer development through inhibiting p53 expression via binding to DNMT1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2768–2776. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, W.; Li, T.; Yu, X.; Zhu, Y.; Ding, F.; Li, D.; Yang, T. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed. Pharmacother. 2016, 80, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Niu, Z.; Liu, J.; Dang, X.; Feng, H.; Sun, J.; Pan, L.; Peng, Z. LncRNA SNHG1 promotes sepsis-induced myocardial injury by inhibiting Bcl-2 expression via DNMT1. J. Cell. Mol. Med. 2022, 26, 3648–3658. [Google Scholar] [CrossRef]
- Hu, Y.; Ma, Z.; He, Y.; Liu, W.; Su, Y.; Tang, Z. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem. Biophys. Res. Commun. 2017, 491, 926–931. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, Q.; Huang, W.; He, C.; Chang, C.; Zhou, J.; Ning, Y. Epigenetic silencing of ZCCHC10 by the lncRNA SNHG1 promotes progression and venetoclax resistance of acute myeloid leukemia. Int. J. Oncol. 2023, 62, 64. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Thin, K.Z.; Tu, J.C.; Raveendran, S. Long non-coding SNHG1 in cancer. Clin. Chim. Acta 2019, 494, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Liang, X.; Li, X.; Zhang, Y.; Sun, Z.; Liu, Y.; Wang, J. MicroRNA-195: A review of its role in cancers. OncoTargets Ther. 2018, 11, 7109–7123. [Google Scholar] [CrossRef]
- Xu, T.; Zhu, Y.; Xiong, Y.; Ge, Y.-Y.; Yun, J.-P.; Zhuang, S.-M. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology 2009, 50, 113–121. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, N.; Li, S.; Fang, J.-H.; Chen, M.-X.; Yang, J.; Jia, W.-H.; Yuan, Y.; Zhuang, S.-M. MicroRNA-195 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology 2013, 58, 642–653. [Google Scholar] [CrossRef]
- Xu, H.; Hu, Y.-W.; Zhao, J.-Y.; Hu, X.-M.; Li, S.-F.; Wang, Y.-C.; Gao, J.-J.; Sha, Y.-H.; Kang, C.-M.; Lin, L.; et al. MicroRNA-195-5p acts as an anti-oncogene by targeting PHF19 in hepatocellular carcinoma. Oncol. Rep. 2015, 34, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Song, X. Long non-coding RNA SNHG1 promotes cell proliferation and invasion of hepatocellular carcinoma by acting as a molecular sponge to modulate miR-195. Arch. Med. Sci. 2020, 16, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Wang, X. The role of MTDH/AEG-1 in the progression of cancer. Int. J. Clin. Exp. Med. 2015, 8, 4795–4807. [Google Scholar] [PubMed]
- Huang, D.; Wei, Y.; Zhu, J.; Wang, F. Long non-coding RNA SNHG1 functions as a competitive endogenous RNA to regulate PDCD4 expression by sponging miR-195-5p in hepatocellular carcinoma. Gene 2019, 714, 143994. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, Q.; Deng, H.; Ou, S.; Liang, T.; Zhou, J. The SNHG1-Centered ceRNA Network Regulates Cell Cycle and Is a Potential Prognostic Biomarker for Hepatocellular Carcinoma. Tohoku J. Exp. Med. 2022, 258, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-F.; Liu, A.-D.; Li, S.-L. SNHG1 promotes proliferation, invasion and EMT of prostate cancer cells through miR-195-5p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 9880–9888. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Liu, J.; Lu, T.; Zang, L.; Wang, J.; He, Q.; Zhou, A. SNHG1 knockdown upregulates miR-376a and downregulates FOXK1/Snail axis to prevent tumor growth and metastasis in HCC. Mol. Ther.-Oncolytics 2021, 21, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Yin, L.; Chen, H.; Yang, S.; Pan, C.; Lu, S.; Miao, M.; Jiao, B. miR-376a suppresses proliferation and induces apoptosis in hepatocellular carcinoma. FEBS Lett. 2012, 586, 2396–2403. [Google Scholar] [CrossRef]
- Olmeda, D.; Moreno-Bueno, G.; Flores, J.M.; Fabra, A.; Portillo, F.; Cano, A. SNAI1 is required for tumor growth and lymph node metastasis of human breast carcinoma MDA-MB-231 cells. Cancer Res. 2007, 67, 11721–11731. [Google Scholar] [CrossRef]
- Wencong, M.; Jinghan, W.; Yong, Y.; Jianyang, A.; Bin, L.; Qingbao, C.; Chen, L.; Xiaoqing, J. FOXK1 Promotes Proliferation and Metastasis of Gallbladder Cancer by Activating AKT/mTOR Signaling Pathway. Front. Oncol. 2020, 10, 545. [Google Scholar] [CrossRef]
- Zhang, J.; Lou, W. A Key mRNA-miRNA-lncRNA Competing Endogenous RNA Triple Sub-network Linked to Diagnosis and Prognosis of Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 340. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Shan, X.; Chen, K.; Liu, Y.; Yu, G.; Chen, Q.; Zeng, T.; Zhu, L.; Dang, H.; Chen, F.; et al. LINC00052/miR-101-3p axis inhibits cell proliferation and metastasis by targeting SOX9 in hepatocellular carcinoma. Gene 2018, 679, 138–149. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, H.; Chen, L.; Zhang, T.; Xu, N.; Huang, Z. Identification of Hypoxia-Related Prognostic Signature and Competing Endogenous RNA Regulatory Axes in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2022, 23, 13590. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, J.-H.; Guo, C.-K.; Cai, P.-C. Gene silencing of TKTL1 by RNAi inhibits cell proliferation in human hepatoma cells. Cancer Lett. 2007, 253, 108–114. [Google Scholar] [CrossRef]
- Sun, W.; Liu, Y.; Glazer, C.A.; Shao, C.; Bhan, S.; Demokan, S.; Zhao, M.; Rudek, M.A.; Ha, P.K.; Califano, J.A. TKTL1 is activated by promoter hypomethylation and contributes to head and neck squamous cell carcinoma carcinogenesis through increased aerobic glycolysis and HIF1alpha stabilization. Clin. Cancer Res. 2010, 16, 857–866. [Google Scholar] [CrossRef]
- Chen, Z.; Zou, Y.; Zhang, Y.; Chen, Z.; Wu, F.; Shi, N.; Jin, H. A novel prognostic signature based on four glycolysis-related genes predicts survival and clinical risk of hepatocellular carcinoma. J. Clin. Lab. Anal. 2021, 35, e24005. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Zhang, J.; Chen, J. lncRNA SNHG1 negatively regulates miRNA-101-3p to enhance the expression of ROCK1 and promote cell proliferation, migration and invasion in osteosarcoma. Int. J. Mol. Med. 2018, 43, 1157–1166. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Guo, L.; Liu, Y.; Yang, H.; Ning, S.; Lv, G. Long Noncoding RNA SNHG1 Regulates LMNB2 Expression by Sponging miR-326 and Promotes Cancer Growth in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 784067. [Google Scholar] [CrossRef]
- Zheng, C.; Yu, S. Expression and gene regulatory network of SNHG1 in hepatocellular carcinoma. BMC Med. Genom. 2021, 14, 28. [Google Scholar] [CrossRef]
- Qu, A.; Yang, Q. LncRNA SNHG1 promotes cell progression and metastasis via sponging miR-377-3p in hepatocellular carcinoma. Neoplasma 2020, 67, 557–566. [Google Scholar] [CrossRef]
- Li, B.; Li, A.; You, Z.; Xu, J.; Zhu, S. Epigenetic silencing of CDKN1A and CDKN2B by SNHG1 promotes the cell cycle, migration and epithelial-mesenchymal transition progression of hepatocellular carcinoma. Cell Death Dis. 2020, 11, 823. [Google Scholar] [CrossRef] [PubMed]
- Bogliotti, Y.; Roest, M.V.; Mattis, A.N.; Gish, R.G.; Peltz, G.; Anwyl, R.; Kivlighn, S.; Schuur, E.R. Clinical Application of Induced Hepatocyte-like Cells Produced from Mesenchymal Stromal Cells: A Literature Review. Cells 2022, 11, 1998. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Weng, W.; Qiao, Y.; Ma, L.; Xiao, W.; Yu, Y.; Pan, Q.; Sun, F. Impaired phosphorylation and ubiquitination by p70 S6 kinase (p70S6K) and Smad ubiquitination regulatory factor 1 (Smurf1) promote tribbles homolog 2 (TRIB2) stability and carcinogenic property in liver cancer. J. Biol. Chem. 2013, 288, 33667–33681. [Google Scholar] [CrossRef] [PubMed]
- Chaker, D.; Mouawad, C.; Azar, A.; Quilliot, D.; Achkar, I.; Fajloun, Z.; Makdissy, N. Inhibition of the RhoGTPase Cdc42 by ML141 enhances hepatocyte differentiation from human adipose-derived mesenchymal stem cells via the Wnt5a/PI3K/miR-122 pathway: Impact of the age of the donor. Stem Cell Res. Ther. 2018, 9, 167. [Google Scholar] [CrossRef]
- Sun, J.; Sun, X.; Hu, S.; Wang, M.; Ma, N.; Chen, J.; Duan, F. Long noncoding RNA SNHG1 silencing accelerates hepatocyte-like cell differentiation of bone marrow-derived mesenchymal stem cells to alleviate cirrhosis via the microRNA-15a/SMURF1/UVRAG axis. Cell Death Discov. 2022, 8, 77. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Han, J.; Xing, H.; Zhang, H.; Li, Z.; Liang, L.; Li, C.; Dai, S.; Wu, M.; Shen, F.; et al. Dysregulated fatty acid metabolism in hepatocellular carcinoma. Hepatic Oncol. 2016, 3, 241–251. [Google Scholar] [CrossRef]
- Chen, E.; Yi, J.; Jiang, J.; Zou, Z.; Mo, Y.; Ren, Q.; Lin, Z.; Lu, Y.; Zhang, J.; Liu, J. Identification and validation of a fatty acid metabolism-related lncRNA signature as a predictor for prognosis and immunotherapy in patients with liver cancer. BMC Cancer 2022, 22, 1037. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, W.; Razak, S.R.A.; Han, T.; Ahmad, N.H.; Li, X. Ferroptosis as a potential target for cancer therapy. Cell Death Dis. 2023, 14, 460. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, Q.; Cheng, J.; Shen, X.; Li, J.; Chen, M.; Zhou, C.; Zhou, J. LncRNA SNHG1 upregulates FANCD2 and G6PD to suppress ferroptosis by sponging miR-199a-5p/3p in hepatocellular carcinoma. Drug Discov. Ther. 2023, 17, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, S.; Shen, L.; Li, J.; Zhang, D.; Yu, J.; Huang, L.; Liu, N.; Lu, H.; Xu, M. SNHG1, interacting with SND1, contributes to sorafenib resistance of liver cancer cells by increasing m6A-mediated SLC7A11 expression and promoting aerobic glycolysis. Environ. Toxicol. 2023, 39, 1269–1282. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, F.; Peng, Q.; Mei, K.; He, H.; Yang, Q. Long non-coding RNA SNHG1 activates glycolysis to promote hepatocellular cancer progression through the miR-326/PKM2 axis. J. Gene Med. 2022, 24, e3440. [Google Scholar] [CrossRef] [PubMed]
- Zahra, K.; Dey, T.; Ashish; Mishra, S.P.; Pandey, U. Pyruvate Kinase M2 and Cancer: The Role of PKM2 in Promoting Tumorigenesis. Front. Oncol. 2020, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yang, H.; Zhang, W.; Wang, J.; Sun, L.; Gao, J.; Zhao, H.; Wang, Z. Identification of m6A-Related lncRNA to Predict the Prognosis of Patients with Hepatocellular Carcinoma. BioMed Res. Int. 2022, 2022, 4169150. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.C.; Duffy, C.R.; Allison, J.P. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018, 8, 1069–1086. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.; Wang, X.; Li, H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int. J. Biol. Macromol. 2018, 118 Pt A, 24–30. [Google Scholar] [CrossRef]
- Tian, P.; Wei, J.; Li, J.; Ren, J.; Yang, J. LncRNA SNHG1 regulates immune escape of renal cell carcinoma by targeting miR-129-3p to activate STAT3 and PD-L1. Cell Biol. Int. 2021, 45, 1546–1560. [Google Scholar] [CrossRef]
- Vogel, A.; Martinelli, E.; Cervantes, A.; Chau, I.; Daniele, B.; Llovet, J.; Meyer, T.; Nault, J.-C.; Neumann, U.; Ricke, J.; et al. Updated treatment recommendations for hepatocellular carcinoma (HCC) from the ESMO Clinical Practice Guidelines. Ann. Oncol. 2021, 32, 801–805. [Google Scholar] [CrossRef]
- Mousa, A. Sorafenib in the Treatment of Advanced Hepatocellular Carcinoma. Saudi J. Gastroenterol. 2008, 14, 40–42. [Google Scholar] [CrossRef]
- Zhai, B.; Hu, F.; Jiang, X.; Xu, J.; Zhao, D.; Liu, B.; Pan, S.; Dong, X.; Tan, G.; Wei, Z.; et al. Inhibition of Akt reverses the acquired resistance to sorafenib by switching protective autophagy to autophagic cell death in hepatocellular carcinoma. Mol. Cancer Ther. 2014, 13, 1589–1598. [Google Scholar] [CrossRef]
- Nitulescu, G.M.; Van De Venter, M.; Ungurianu, A.; Juzenas, P.; Peng, Q.; Olaru, O.T.; Grădinaru, D.; Tsatsakis, A.; Tsoukalas, D.; Spandidos, D.A.; et al. The Akt pathway in oncology therapy and beyond (Review). Int. J. Oncol. 2018, 53, 2319–2331. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wei, G.; Luo, H.; Wu, W.; Skogerbø, G.; Luo, J.; Chen, R. The long noncoding RNA SNHG1 promotes tumor growth through regulating transcription of both local and distal genes. Oncogene 2017, 36, 6774–6783. [Google Scholar] [CrossRef]
- Li, W.; Dong, X.; He, C.; Tan, G.; Li, Z.; Zhai, B.; Feng, J.; Jiang, X.; Liu, C.; Jiang, H.; et al. LncRNA SNHG1 contributes to sorafenib resistance by activating the Akt pathway and is positively regulated by miR-21 in hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 2019, 38, 183. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Kon, N.; Li, T.; Wang, S.-J.; Su, T.; Hibshoosh, H.; Baer, R.; Gu, W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015, 520, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Bruix, J.; Chan, S.L.; Galle, P.R.; Rimassa, L.; Sangro, B. Systemic treatment of hepatocellular carcinoma: An EASL position paper. J. Hepatol. 2021, 75, 960–974. [Google Scholar] [CrossRef]
- Wang, P.; Chen, Y.; Wang, C. Beyond Tumor Mutation Burden: Tumor Neoantigen Burden as a Biomarker for Immunotherapy and Other Types of Therapy. Front. Oncol. 2021, 11, 672677. [Google Scholar] [CrossRef]
- Shen, Y.; Liu, S.; Fan, J.; Jin, Y.; Tian, B.; Zheng, X.; Fu, H. Nuclear retention of the lncRNA SNHG1 by doxorubicin attenuates hnRNPC–p53 protein interactions. EMBO Rep. 2017, 18, 536–548. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, B.; Liu, Z.; Jiang, L.; Wang, G.; Lv, M.; Li, D. Up-regulation of lncRNA SNHG1 indicates poor prognosis and promotes cell proliferation and metastasis of colorectal cancer by activation of the Wnt/β-catenin signaling pathway. Oncotarget 2017, 8, 111715–111727. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Qiu, R.; Qiu, X. SNHG1 promotes cell proliferation by acting as a sponge of miR-145 in colorectal cancer. Oncotarget 2018, 9, 2128–2139. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, F.; Zhu, C.; Geng, L.; Tian, T.; Liu, H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget 2017, 8, 17785–17794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-Y.; Yang, W.; Zheng, F.-S.; Wang, Y.-B.; Lu, J.-B. Long non-coding RNA SNHG1 regulates zinc finger E-box binding homeobox 1 expression by interacting with TAp63 and promotes cell metastasis and invasion in Lung squamous cell carcinoma. Biomed. Pharmacother. 2017, 90, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Xu, X.; Wang, Y.; Zhang, W.; Wang, X. Diagnostic utility of plasma lncRNA small nucleolar RNA host gene 1 in patients with hepatocellular carcinoma. Mol. Med. Rep. 2018, 18, 3305–3313. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Leng, Q.; Zhan, M.; Jiang, F. A Plasma Long Noncoding RNA Signature for Early Detection of Lung Cancer. Transl. Oncol. 2018, 11, 1225–1231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhuo, C.; Zhou, D.; Zhang, M.; Zhang, F.; Chen, M.; Xu, S.; Chen, Z. Small Nucleolar RNA Host Gene 1 (SNHG1) and Chromosome 2 Open Reading Frame 48 (C2orf48) as Potential Prognostic Signatures for Liver Cancer by Constructing Regulatory Networks. Med. Sci. Monit. 2020, 26, e920482-1. [Google Scholar] [CrossRef] [PubMed]
- Panzitt, K.; Tschernatsch, M.M.; Guelly, C.; Moustafa, T.; Stradner, M.; Strohmaier, H.M.; Buck, C.R.; Denk, H.; Schroeder, R.; Trauner, M.; et al. Characterization of HULC, a Novel Gene with Striking Up-Regulation in Hepatocellular Carcinoma, as Noncoding RNA. Gastroenterology 2007, 132, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Arita, T.; Ichikawa, D.; Konishi, H.; Komatsu, S.; Shiozaki, A.; Shoda, K.; Kawaguchi, T.; Hirajima, S.; Nagata, H.; Kubota, T.; et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013, 33, 3185–3193. [Google Scholar]
- Wang, X.; Tian, L.; Lu, J.; Ng, I.O.-L. Exosomes and cancer—Diagnostic and prognostic biomarkers and therapeutic vehicle. Oncogenesis 2022, 11, 54. [Google Scholar] [CrossRef]
- Tan, C.; Cao, J.; Chen, L.; Xi, X.; Wang, S.; Zhu, Y.; Yang, L.; Ma, L.; Wang, D.; Yin, J.; et al. Noncoding RNAs Serve as Diagnosis and Prognosis Biomarkers for Hepatocellular Carcinoma. Clin. Chem. 2019, 65, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Baek, G.O.; A Son, J.; Ahn, H.R.; Yoon, M.K.; Cho, H.J.; Yoon, J.H.; Nam, S.W.; Cheong, J.Y.; Eun, J.W. Early detection of hepatocellular carcinoma via liquid biopsy: Panel of small extracellular vesicle-derived long noncoding RNAs identified as markers. Mol. Oncol. 2021, 15, 2715–2731. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Gholipour, M.; Hussen, B.M.; Taheri, M. The Impact of Long Non-Coding RNAs in the Pathogenesis of Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 649107. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonseca, T.S.; Martins, R.M.; Rolo, A.P.; Palmeira, C.M. SNHG1: Redefining the Landscape of Hepatocellular Carcinoma through Long Noncoding RNAs. Biomedicines 2024, 12, 1696. https://doi.org/10.3390/biomedicines12081696
Fonseca TS, Martins RM, Rolo AP, Palmeira CM. SNHG1: Redefining the Landscape of Hepatocellular Carcinoma through Long Noncoding RNAs. Biomedicines. 2024; 12(8):1696. https://doi.org/10.3390/biomedicines12081696
Chicago/Turabian StyleFonseca, Tiago S., Rui Miguel Martins, Anabela P. Rolo, and Carlos M. Palmeira. 2024. "SNHG1: Redefining the Landscape of Hepatocellular Carcinoma through Long Noncoding RNAs" Biomedicines 12, no. 8: 1696. https://doi.org/10.3390/biomedicines12081696





