Fourier Transform Ion Cyclotron Resonance Mass Spectrometry Applications for Metabolomics
Abstract
1. Introduction
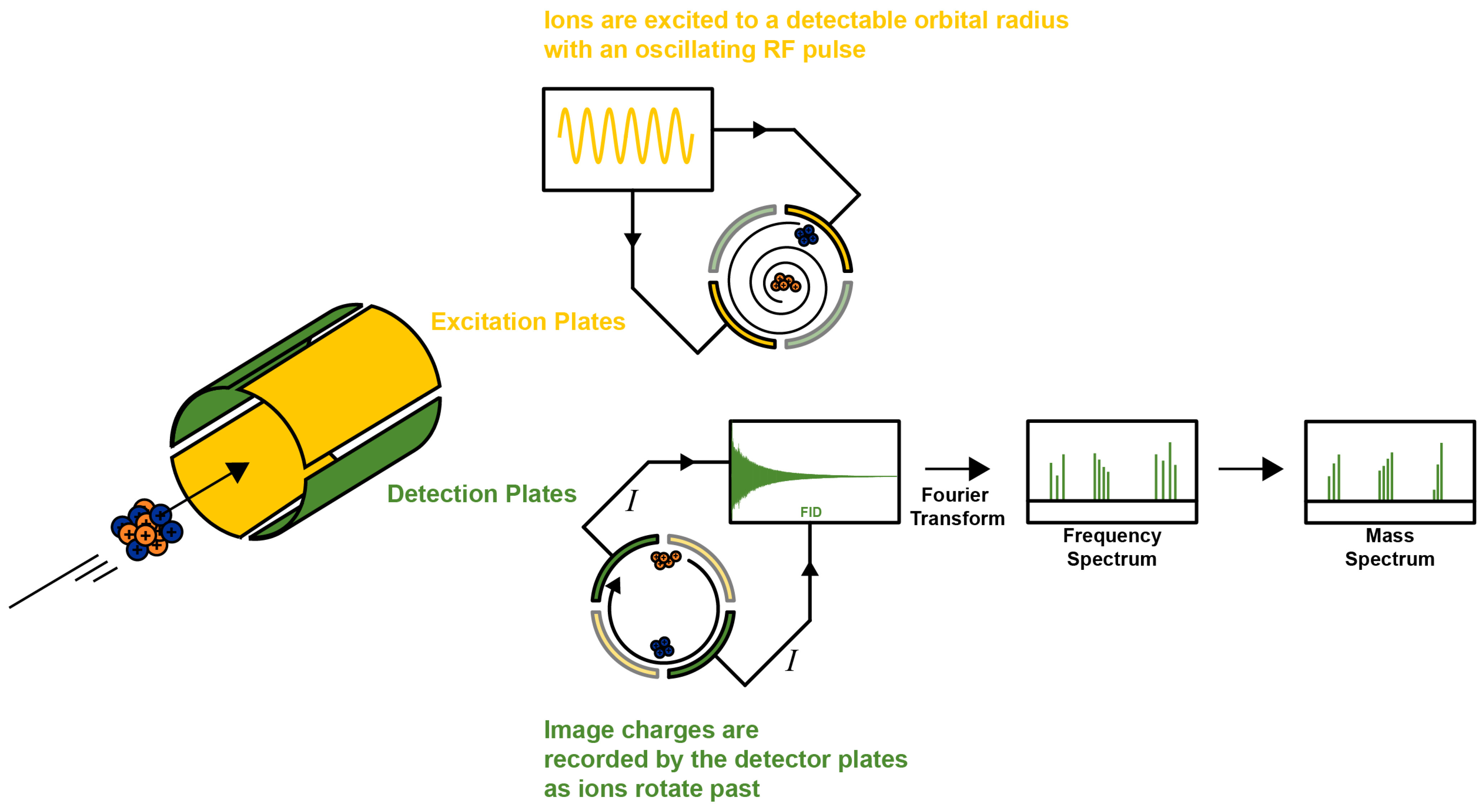
2. FT-ICR-MS Fundamentals
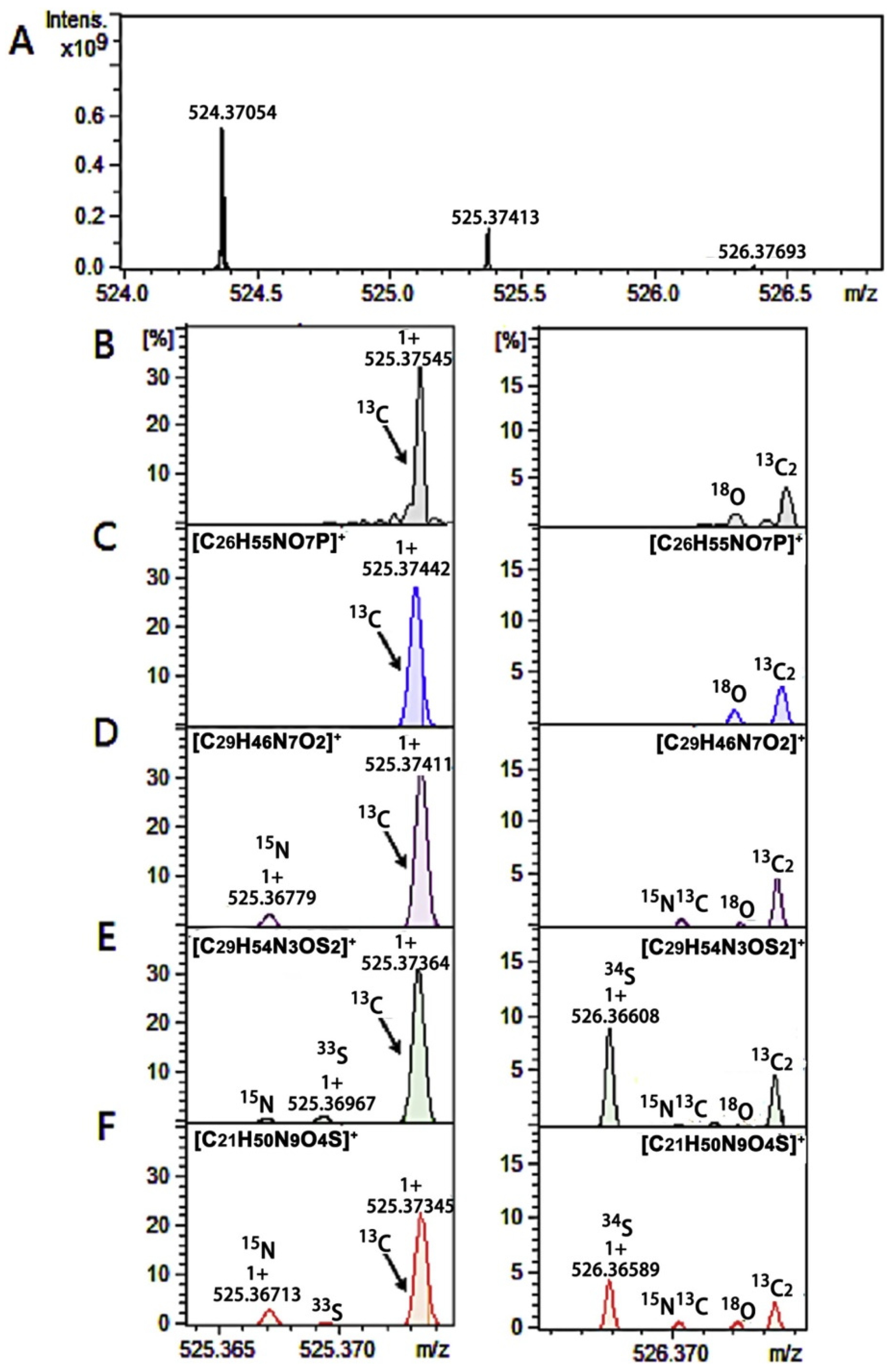
3. FT-ICR-MS in the Search for Clinical Biomarkers
4. FT-ICR-MS in the Search for Non-Human Biomarkers
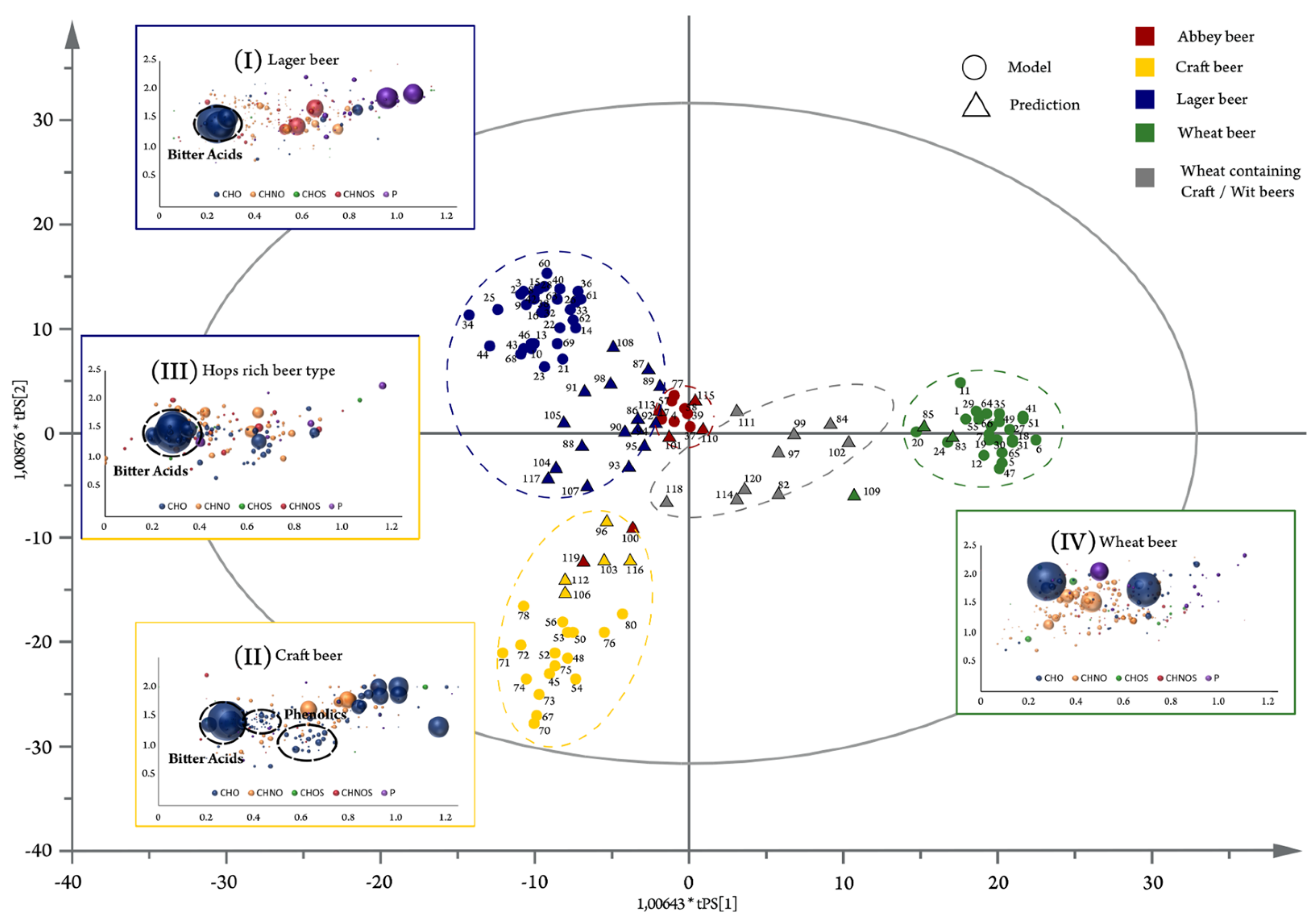
5. FT-ICR-MS for Food and Beverage Metabolomics
6. FT-ICR-MS in Environmental Applications
7. FT-ICR-MS High Resolution Imaging
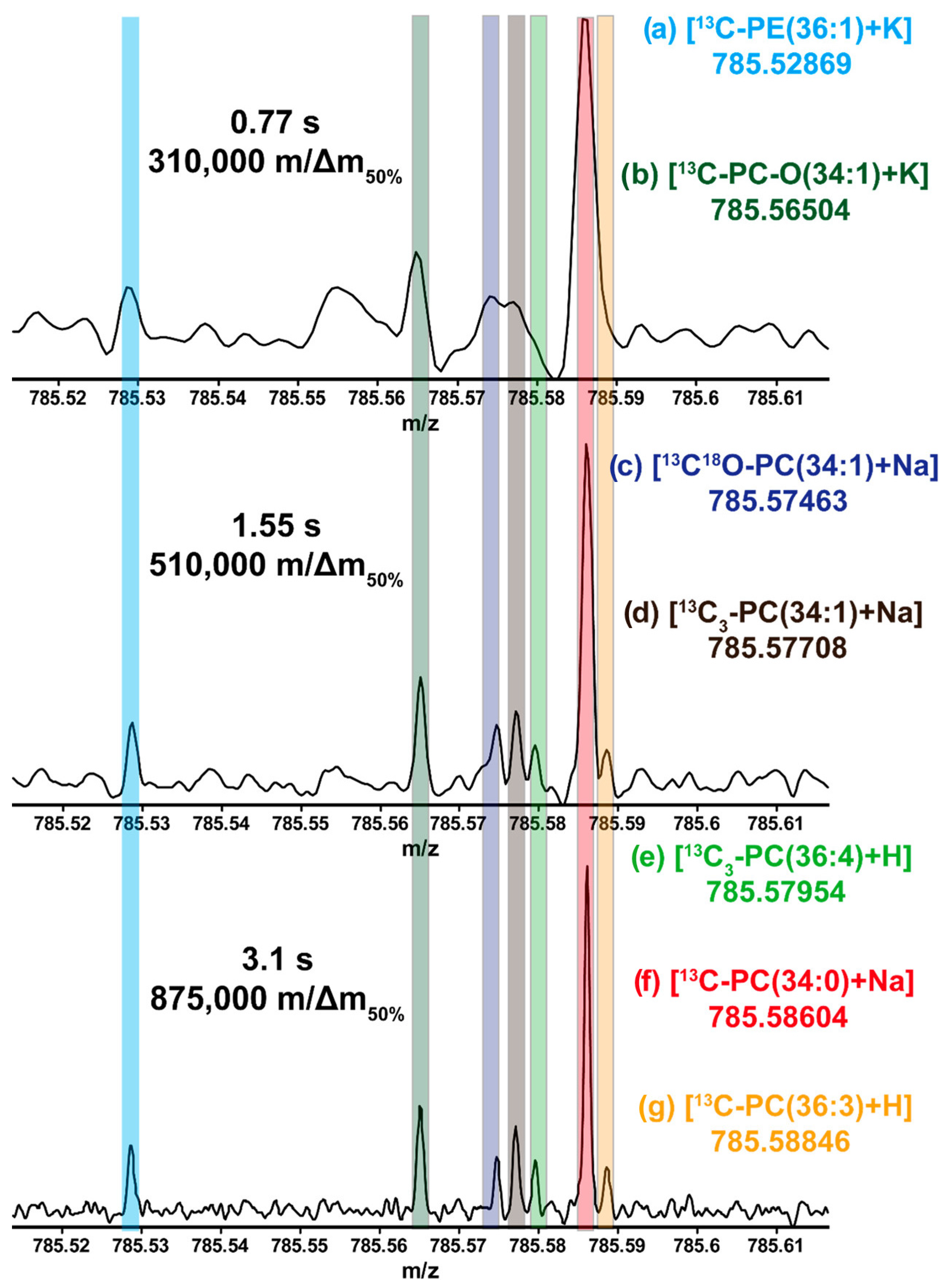
8. Practical Considerations for Implementing FT-ICR-MS
8.1. FT-ICR-MS Locations and Sample Submission Costs
8.2. FT-ICR-MS Sample Preparation Concerns
8.3. Sample Purity and Concentration
8.4. When Possible, Use Fresh Frozen Tissues for Mass Spectrometry Imaging
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
List of Abbreviations
| AD | Alzheimer’s disease |
| ANOVA | Analysis of variation |
| CID | Collision induced dissociation |
| CV | Coefficient of variation |
| DESI | Desorption electrospray ionization |
| DI-FT-ICR-MS | Direct injection FT-ICR-MS |
| DOM | Dissolved organic matter |
| ESI | Electrospray ionization |
| FF | Fresh frozen |
| FFPE | Formalin-fixed, paraffin-embedded |
| FID | Free induction decay |
| FIE | Flow injection electrospray |
| FT-ICR-MS | Fourier transform ion cyclotron resonance mass spectrometry |
| H/C | Hydrogen to carbon ratio |
| HMDB | Human metabolome database |
| HPLC | High performance liquid chromatography |
| HRMS | High resolution mass spectrometry |
| ICR | Ion cyclotron resonance |
| IFS | Isotopic fine structure |
| KMD | Kendrick mass defect |
| LC-MS | Liquid chromatography mass spectrometry |
| LPC | Lysophosphatidyl choline |
| MALDI | Matrix assisted laser desorption ionization |
| MS | Mass spectrometry |
| NMR | Nuclear magnetic resonance |
| NSO:C | Ratio of heteroatoms to carbon |
| O/C | Oxygen to carbon ratio |
| OPLS-DA | Orthogonal projections to latent structures discriminant analysis |
| PDS | Sodium persulfate |
| ppb | Parts per billion |
| QToF | Quadrupole time-of-flight |
| RCE | Rhodiola crenulate |
| RT | Retention time |
| SOM | Soil organic matter |
| T | Tesla |
| T2D | Type 2 diabetes |
| UK | United Kingdom |
| WAF | Water accommodated fraction |
| WSF | Water soluble fraction |
References
- Dhillon, B.K.; Smith, M.; Baghela, A.; Lee, A.H.Y.; Hancock, R.E.W. Systems Biology Approaches to Understanding the Human Immune System. Front. Immunol. 2020, 11, 1683. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, M.J.; Powers, R. Multiplatform untargeted metabolomics. Magn. Reson. Chem 2023, 61, 628–653. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Liebeke, M.; Sommer, U.; Viant, M.R.; Bundy, J.G.; Ebbels, T.M. Statistical Correlations between NMR Spectroscopy and Direct Infusion FT-ICR Mass Spectrometry Aid Annotation of Unknowns in Metabolomics. Anal. Chem. 2016, 88, 2583–2589. [Google Scholar] [CrossRef] [PubMed]
- Plumb, R.S.; Gethings, L.A.; Rainville, P.D.; Isaac, G.; Trengove, R.; King, A.M.; Wilson, I.D. Advances in high throughput LC/MS based metabolomics: A review. TrAC Trends Anal. Chem. 2023, 160, 116954. [Google Scholar] [CrossRef]
- Wishart, D.S.; Cheng, L.L.; Copie, V.; Edison, A.S.; Eghbalnia, H.R.; Hoch, J.C.; Gouveia, G.J.; Pathmasiri, W.; Powers, R.; Schock, T.B.; et al. NMR and Metabolomics-A Roadmap for the Future. Metabolites 2022, 12, 678. [Google Scholar] [CrossRef] [PubMed]
- Moritz, F.; Hemmler, D.; Kanawati, B.; Schnitzler, J.-P.; Schmitt-Kopplin, P. Chapter 12—Mass differences in metabolome analyses of untargeted direct infusion ultra-high resolution MS data. In Fundamentals and Applications of Fourier Transform Mass Spectrometry; Kanawati, B., Schmitt-Kopplin, P., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 357–405. [Google Scholar]
- Ohta, D.; Kanaya, S.; Suzuki, H. Application of Fourier-transform ion cyclotron resonance mass spectrometry to metabolic profiling and metabolite identification. Curr. Opin. Biotechnol. 2010, 21, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Ghaste, M.; Mistrik, R.; Shulaev, V. Applications of Fourier Transform Ion Cyclotron Resonance (FT-ICR) and Orbitrap Based High Resolution Mass Spectrometry in Metabolomics and Lipidomics. Int. J. Mol. Sci. 2016, 17, 816. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, D.; Jung, M.-J.; Kim, S. Analysis of environmental organic matters by Ultrahigh-Resolution mass spectrometry—A review on the development of analytical methods. Mass Spectrom. Rev. 2022, 41, 352–369. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chu, S.; Tan, S.; Yin, X.; Jiang, Y.; Dai, X.; Gong, X.; Fang, X.; Tian, D. Towards Higher Sensitivity of Mass Spectrometry: A Perspective From the Mass Analyzers. Front. Chem. 2021, 9, 813359. [Google Scholar] [CrossRef] [PubMed]
- Nikolaev, E.N.; Kostyukevich, Y.I.; Vladimirov, G.N. Fourier transform ion cyclotron resonance (FT ICR) mass spectrometry: Theory and simulations. Mass Spectrom. Rev. 2016, 35, 219–258. [Google Scholar] [CrossRef] [PubMed]
- Marshall, A.G.; Hendrickson, C.L.; Jackson, G.S. Fourier transform ion cyclotron resonance mass spectrometry: A primer. Mass Spectrom. Rev. 1998, 17, 1–35. [Google Scholar] [CrossRef]
- Gross, J.H. Mass Spectrometry: A Textbook; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Shaw, J.B.; Lin, T.Y.; Leach, F.E., 3rd; Tolmachev, A.V.; Tolic, N.; Robinson, E.W.; Koppenaal, D.W.; Pasa-Tolic, L. 21 Tesla Fourier Transform Ion Cyclotron Resonance Mass Spectrometer Greatly Expands Mass Spectrometry Toolbox. J. Am. Soc. Mass Spectrom. 2016, 27, 1929–1936. [Google Scholar] [CrossRef] [PubMed]
- Bowman, A.P.; Blakney, G.T.; Hendrickson, C.L.; Ellis, S.R.; Heeren, R.M.A.; Smith, D.F. Ultra-High Mass Resolving Power, Mass Accuracy, and Dynamic Range MALDI Mass Spectrometry Imaging by 21-T FT-ICR MS. Anal. Chem. 2020, 92, 3133–3142. [Google Scholar] [CrossRef] [PubMed]
- Bahureksa, W.; Borch, T.; Young, R.B.; Weisbrod, C.R.; Blakney, G.T.; McKenna, A.M. Improved Dynamic Range, Resolving Power, and Sensitivity Achievable with FT-ICR Mass Spectrometry at 21 T Reveals the Hidden Complexity of Natural Organic Matter. Anal. Chem. 2022, 94, 11382–11389. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.J.; Witt, M.; Forcisi, S.; Moritz, F.; Kessler, N.; Laukien, F.H.; Schmitt-Kopplin, P. An Enhanced Isotopic Fine Structure Method for Exact Mass Analysis in Discovery Metabolomics: FIA-CASI-FTMS. J. Am. Soc. Mass Spectrom. 2020, 31, 2025–2034. [Google Scholar] [CrossRef] [PubMed]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Hansson, O. Biomarkers for neurodegenerative diseases. Nat. Med. 2021, 27, 954–963. [Google Scholar] [CrossRef]
- Kim, M.; Legido-Quigley, C. Small molecule biomarkers in Alzheimer’s disease. Ocl 2018, 25, D404. [Google Scholar] [CrossRef]
- Alzheimer’s Association. 2023 Alzheimer’s disease facts and figures. Alzheimers Dement. 2023, 19, 1598–1695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiang, X.; Wang, X.; Zhao, Y.; Jia, L.; Chen, F.; Yin, R.; Han, F. A metabolomic study based on accurate mass and isotopic fine structures by dual mode combined-FT-ICR-MS to explore the effects of Rhodiola crenulata extract on Alzheimer disease in rats. J. Pharm. Biomed. Anal. 2019, 166, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Qu, Z.Q.; Zeng, Y.S.; Lin, Y.K.; Li, Y.; Chung, P.; Wong, R.; Hagg, U. Neuroprotective effect of preadministration with Ganoderma lucidum spore on rat hippocampus. Exp. Toxicol. Pathol. 2012, 64, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, Q.; Cheng, Q.; Ding, F. Protective effect of salidroside against H2O2-induced cell apoptosis in primary culture of rat hippocampal neurons. Mol. Cell. Biochem. 2009, 332, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Qu, Z.Q.; Wu, J.L.; Chung, P.; Zeng, Y.S. Mitochondrial protective and anti-apoptotic effects of Rhodiola crenulata extract on hippocampal neurons in a rat model of Alzheimer’s disease. Neural. Regen. Res. 2017, 12, 2025–2034. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wancewicz, B.; Schaid, M.; Tiambeng, T.N.; Wenger, K.; Jin, Y.; Heyman, H.; Thompson, C.J.; Barsch, A.; Cox, E.D.; et al. Ultrahigh-Resolution Mass Spectrometry-Based Platform for Plasma Metabolomics Applied to Type 2 Diabetes Research. J. Proteome Res. 2021, 20, 463–473. [Google Scholar] [CrossRef]
- Maia, M.; Figueiredo, A.; Cordeiro, C.; Sousa Silva, M. FT-ICR-MS-based metabolomics: A deep dive into plant metabolism. Mass Spectrom. Rev. 2023, 42, 1535–1556. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Fernie, A.R.; Luo, J. Exploring the Diversity of Plant Metabolism. Trends Plant Sci. 2019, 24, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.; McConville, M.; Loukopoulos, P. Metabolomics in the study of spontaneous animal diseases. J. Vet. Diagn. Investig. 2020, 32, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Vercauteren, M.; Ampe, B.; Devriese, L.; Moons, C.P.H.; Decostere, A.; Aerts, J.; Chiers, K. Explorative study on scale cortisol accumulation in wild caught common dab (Limanda limanda). BMC Vet. Res. 2022, 18, 324. [Google Scholar] [CrossRef] [PubMed]
- Mirbahai, L.; Southam, A.D.; Sommer, U.; Williams, T.D.; Bignell, J.P.; Lyons, B.P.; Viant, M.R.; Chipman, J.K. Disruption of DNA methylation via S-adenosylhomocysteine is a key process in high incidence liver carcinogenesis in fish. J. Proteome Res. 2013, 12, 2895–2904. [Google Scholar] [CrossRef] [PubMed]
- Stentiford, G.D.; Viant, M.R.; Johnson, P.J.; Martin, A.; Wenbin, W.; Cooper, H.J.; Lyons, B.P.; Feist, S.W. Liver tumors in wild flatfish: A histopathological, proteomic, and metabolomic study. OMICS 2005, 9, 281–299. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Wang, X. Evaluation Technologies for Food Quality; Woodhead Publishing: Sawston, UK, 2019. [Google Scholar]
- Kew, W.; Goodall, I.; Clarke, D.; Uhrin, D. Chemical Diversity and Complexity of Scotch Whisky as Revealed by High-Resolution Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2017, 28, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Kew, W.; Mackay, C.L.; Goodall, I.; Clarke, D.J.; Uhrin, D. Complementary Ionization Techniques for the Analysis of Scotch Whisky by High Resolution Mass Spectrometry. Anal. Chem. 2018, 90, 11265–11272. [Google Scholar] [CrossRef] [PubMed]
- Roullier-Gall, C.; Signoret, J.; Hemmler, D.; Witting, M.A.; Kanawati, B.; Schafer, B.; Gougeon, R.D.; Schmitt-Kopplin, P. Usage of FT-ICR-MS Metabolomics for Characterizing the Chemical Signatures of Barrel-Aged Whisky. Front. Chem. 2018, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Laszakovits, J.R.; MacKay, A.A. Data-Based Chemical Class Regions for Van Krevelen Diagrams. J. Am. Soc. Mass Spectrom. 2022, 33, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Pieczonka, S.A.; Lucio, M.; Rychlik, M.; Schmitt-Kopplin, P. Decomposing the molecular complexity of brewing. NPJ Sci. Food 2020, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Pieczonka, S.A.; Hemmler, D.; Moritz, F.; Lucio, M.; Zarnkow, M.; Jacob, F.; Rychlik, M.; Schmitt-Kopplin, P. Hidden in its color: A molecular-level analysis of the beer’s Maillard reaction network. Food Chem. 2021, 361, 130112. [Google Scholar] [CrossRef] [PubMed]
- Pieczonka, S.A.; Paravicini, S.; Rychlik, M.; Schmitt-Kopplin, P. On the Trail of the German Purity Law: Distinguishing the Metabolic Signatures of Wheat, Corn and Rice in Beer. Front. Chem. 2021, 9, 715372. [Google Scholar] [CrossRef]
- Pieczonka, S.A.; Zarnkow, M.; Ampenberger, F.; Gastl, M.; Rychlik, M.; Schmitt-Kopplin, P. FT-ICR-MS reveals the molecular imprints of the brewing process. Front. Nutr. 2023, 10, 1243503. [Google Scholar] [CrossRef] [PubMed]
- Sousa Silva, M.; Soeiro, M.; Cordeiro, C. From the grapevine to the glass: A wine metabolomics tale by FT-ICR-MS. J. Mass Spectrom. 2024, 59, e5019. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, M.; Di Vita, G.; Monaco, L. Exploring environmental consciousness and consumer preferences for organic wines without sulfites. J. Clean. Prod. 2016, 120, 64–71. [Google Scholar] [CrossRef]
- Roullier-Gall, C.; Hemmler, D.; Gonsior, M.; Li, Y.; Nikolantonaki, M.; Aron, A.; Coelho, C.; Gougeon, R.D.; Schmitt-Kopplin, P. Sulfites and the wine metabolome. Food Chem. 2017, 237, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Zacs, D.; Perkons, I.; Bartkevics, V. Evaluation of analytical performance of gas chromatography coupled with atmospheric pressure chemical ionization Fourier transform ion cyclotron resonance mass spectrometry (GC-APCI-FT-ICR-MS) in the target and non-targeted analysis of brominated and chlorinated flame retardants in food. Chemosphere 2019, 225, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Rodgers, R.P.; Marshall, A.G. Characterization of Vegetable Oils: Detailed Compositional Fingerprints Derived from Electrospray Ionization Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. J. Agric. Food Chem. 2004, 52, 5322–5328. [Google Scholar] [CrossRef] [PubMed]
- Costa, H.B.; Souza, L.M.; Soprani, L.C.; Oliveira, B.G.; Ogawa, E.M.; Korres, A.M.; Ventura, J.A.; Romao, W. Monitoring the physicochemical degradation of coconut water using ESI-FT-ICR MS. Food Chem. 2015, 174, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Kuhnert, N.; D’Souza, R.N.; Behrends, B.; Ullrich, M.S.; Witt, M. Investigating time dependent cocoa bean fermentation by ESI-FT-ICR mass spectrometry. Food Res. Int. 2020, 133, 109209. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.K.; He, W.; Guo, H.; Karanfil, T.; Hur, J. Effects of organic additives on spectroscopic and molecular-level features of photo-induced dissolved organic matter from microplastics. Water Res. 2023, 242, 120272. [Google Scholar] [CrossRef] [PubMed]
- Viant, M.R.; Sommer, U. Mass spectrometry based environmental metabolomics: A primer and review. Metabolomics 2012, 9, 144–158. [Google Scholar] [CrossRef]
- Bedia, C. Metabolomics in environmental toxicology: Applications and challenges. Trends Environ. Anal. Chem. 2022, 34, e00161. [Google Scholar] [CrossRef]
- Peters, K.; Worrich, A.; Weinhold, A.; Alka, O.; Balcke, G.; Birkemeyer, C.; Bruelheide, H.; Calf, O.W.; Dietz, S.; Duhrkop, K.; et al. Current Challenges in Plant Eco-Metabolomics. Int. J. Mol. Sci. 2018, 19, 1385. [Google Scholar] [CrossRef] [PubMed]
- Nebbioso, A.; Piccolo, A. Molecular characterization of dissolved organic matter (DOM): A critical review. Anal. Bioanal. Chem. 2013, 405, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.; Guo, P.; Wu, Y.; Zhao, Y. A Novel Method for Unraveling the Black Box of Dissolved Organic Matter in Soils by FT-ICR-MS Coupled with Induction-Based Nanospray Ionization. Environ. Sci. Technol. Lett. 2021, 8, 356–361. [Google Scholar] [CrossRef]
- Wojnarovits, L.; Homlok, R.; Kovacs, K.; Tegze, A.; Takacs, E. Wastewater Characterization: Chemical Oxygen Demand or Total Organic Carbon Content Measurement? Molecules 2024, 29, 405. [Google Scholar] [CrossRef] [PubMed]
- Bercovici, S.K.; Dittmar, T.; Niggemann, J. The detection of bacterial exometabolites in marine dissolved organic matter through ultrahigh-resolution mass spectrometry. Limnol. Oceanogr. Methods 2022, 20, 350–360. [Google Scholar] [CrossRef]
- Lechtenfeld, O.J.; Kaesler, J.; Jennings, E.K.; Koch, B.P. Direct Analysis of Marine Dissolved Organic Matter Using LC-FT-ICR MS. Environ. Sci. Technol. 2024, 58, 4637–4647. [Google Scholar] [CrossRef] [PubMed]
- Ruan, M.; Wu, F.; Sun, F.; Song, F.; Li, T.; He, C.; Jiang, J. Molecular-level exploration of properties of dissolved organic matter in natural and engineered water systems: A critical review of FTICR-MS application. Crit. Rev. Environ. Sci. Technol. 2023, 53, 1534–1562. [Google Scholar] [CrossRef]
- Xu, W.; Gao, Q.; He, C.; Shi, Q.; Hou, Z.Q.; Zhao, H.Z. Using ESI FT-ICR MS to Characterize Dissolved Organic Matter in Salt Lakes with Different Salinity. Environ. Sci. Technol. 2020, 54, 12929–12937. [Google Scholar] [CrossRef] [PubMed]
- Geng, W.; Wei, Y.; Ke, Y.; Qin, J.; Yu, X.; Guo, X.; Long, M. Unveiling Molecular Transformations of Soil Organic Matter after Remediation by Chemical Oxidation Based on ESI-FT-ICR-MS analysis. ACS EST Eng. 2023, 3, 831–840. [Google Scholar] [CrossRef]
- Liu, Y.; Kujawinski, E.B. Chemical Composition and Potential Environmental Impacts of Water-Soluble Polar Crude Oil Components Inferred from ESI FT-ICR MS. PLoS ONE 2015, 10, e0136376. [Google Scholar] [CrossRef] [PubMed]
- Ansell, D.V.; Dicks, B.; Guenette, C.C.; Moller, T.H.; Santner, R.S.; White, I.C. A review of the problems posed by spills of heavy fuel oils. In Proceedings of the 2001 International Oil Spill Conference, Tampa, FL, USA, 6–29 March 2001. [Google Scholar]
- Marshall, A.G.; Rodgers, R.P. Petroleomics: Chemistry of the Underworld. Proc. Natl. Acad. Sci. USA 2008, 105, 18090–19095. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, L.; Traunspurger, W.; Roeleveld, K.; Lynen, F.; Moens, T. Direct toxicity of the water-soluble fractions of a crude and a diesel-motor oil on the survival of free-living nematodes. Ecol. Indic. 2018, 93, 13–23. [Google Scholar] [CrossRef]
- Faksness, L.G.; Altin, D.; Nordtug, T.; Daling, P.S.; Hansen, B.H. Chemical comparison and acute toxicity of water accommodated fraction (WAF) of source and field collected Macondo oils from the Deepwater Horizon spill. Mar. Pollut. Bull. 2015, 91, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Bünemann, E.K.; Bongiorno, G.; Bai, Z.; Creamer, R.E.; De Deyn, G.; de Goede, R.; Fleskens, L.; Geissen, V.; Kuyper, T.W.; Mäder, P.; et al. Soil quality—A critical review. Soil Biol. Biochem. 2018, 120, 105–125. [Google Scholar] [CrossRef]
- Lehmann, J.; Kleber, M. The contentious nature of soil organic matter. Nature 2015, 528, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Withers, E.; Hill, P.W.; Chadwick, D.R.; Jones, D.L. Use of untargeted metabolomics for assessing soil quality and microbial function. Soil Biol. Biochem. 2020, 143, 107758. [Google Scholar] [CrossRef]
- Lim, M.W.; Lau, E.V.; Poh, P.E. A comprehensive guide of remediation technologies for oil contaminated soil—Present works and future directions. Mar. Pollut. Bull. 2016, 109, 14–45. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Zeng, G.; Wu, H.; Zhang, C.; Liang, J.; Dai, J.; Liu, Z.; Xiong, W.; Wan, J.; Xu, P.; et al. Co-occurrence and interactions of pollutants, and their impacts on soil remediation—A review. Crit. Rev. Environ. Sci. Technol. 2017, 47, 1528–1553. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, X.; Xue, Y. Application of calcium peroxide in water and soil treatment: A review. J. Hazard Mater. 2017, 337, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Hughey, C.A.; Hendrickson, C.L.; Rodgers, R.P.; Qian, K.; Marshall, A.G. Kendrick Mass Defect Spectrum: A Compact Visual Analysis for Ultrahigh-Resolution Broadband Mass Spectra. Anal. Chem. 2001, 73, 4676–4681. [Google Scholar] [CrossRef] [PubMed]
- Merel, S. Critical assessment of the Kendrick mass defect analysis as an innovative approach to process high resolution mass spectrometry data for environmental applications. Chemosphere 2023, 313, 137443. [Google Scholar] [CrossRef] [PubMed]
- Thurman, E.M.; Ferrer, I. The isotopic mass defect: A tool for limiting molecular formulas by accurate mass. Anal. Bioanal. Chem. 2010, 397, 2807–2816. [Google Scholar] [CrossRef] [PubMed]
- Amato, P.; Joly, M.; Besaury, L.; Oudart, A.; Taib, N.; Mone, A.I.; Deguillaume, L.; Delort, A.M.; Debroas, D. Active microorganisms thrive among extremely diverse communities in cloud water. PLoS ONE 2017, 12, e0182869. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.; Deguillaume, L.; Chaumerliac, N.; Vaitilingom, M.; Wang, M.; Delort, A.M.; Bridoux, M.C. Effect of endogenous microbiota on the molecular composition of cloud water: A study by Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR MS). Sci. Rep. 2019, 9, 7663. [Google Scholar] [CrossRef]
- Kim, S.; Kramer, R.W.; Hatcher, P.G. Graphical Method for Analysis of Ultrahigh-Resolution Broadband Mass Spectra of Natural Organic Matter, the Van Krevelen Diagram. Anal. Chem. 2003, 75, 5336–5344. [Google Scholar] [CrossRef] [PubMed]
- El-Aneed, A.; Cohen, A.; Banoub, J. Mass Spectrometry, Review of the Basics: Electrospray, MALDI, and Commonly Used Mass Analyzers. Appl. Spectrosc. Rev. 2009, 44, 210–230. [Google Scholar] [CrossRef]
- Dreisewerd, K. Recent methodological advances in MALDI mass spectrometry. Anal. Bioanal. Chem. 2014, 406, 2261–2278. [Google Scholar] [CrossRef] [PubMed]
- Caprioli, R.M.; Farmer, T.B.; Gile, J. Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal. Chem. 1997, 69, 4751–4760. [Google Scholar] [CrossRef] [PubMed]
- Aichler, M.; Walch, A. MALDI Imaging mass spectrometry: Current frontiers and perspectives in pathology research and practice. Lab. Investig. 2015, 95, 422–431. [Google Scholar] [CrossRef]
- Gessel, M.M.; Norris, J.L.; Caprioli, R.M. MALDI imaging mass spectrometry: Spatial molecular analysis to enable a new age of discovery. J. Proteom. 2014, 107, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Berry, K.A.; Hankin, J.A.; Barkley, R.M.; Spraggins, J.M.; Caprioli, R.M.; Murphy, R.C. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem. Rev. 2011, 111, 6491–6512. [Google Scholar] [CrossRef] [PubMed]
- Zemaitis, K.J.; Izydorczak, A.M.; Thompson, A.C.; Wood, T.D. Streamlined Multimodal DESI and MALDI Mass Spectrometry Imaging on a Singular Dual-Source FT-ICR Mass Spectrometer. Metabolites 2021, 11, 253. [Google Scholar] [CrossRef] [PubMed]
- Bhinderwala, F.; Lei, S.; Woods, J.; Rose, J.; Marshall, D.D.; Riekeberg, E.; Leite, A.L.; Morton, M.; Dodds, E.D.; Franco, R.; et al. Metabolomics Analyses from Tissues in Parkinson’s Disease. Methods Mol. Biol. 2019, 1996, 217–257. [Google Scholar] [CrossRef] [PubMed]
- Bhinderwala, F.; Powers, R. NMR Metabolomics Protocols for Drug Discovery. Methods Mol. Biol. 2019, 2037, 265–311. [Google Scholar] [CrossRef] [PubMed]
- Hohenester, U.M.; Barbier Saint-Hilaire, P.; Fenaille, F.; Cole, R.B. Investigation of space charge effects and ion trapping capacity on direct introduction ultra-high-resolution mass spectrometry workflows for metabolomics. J. Mass Spectrom. 2020, 55, e4613. [Google Scholar] [CrossRef] [PubMed]
- Leach, F.E.; Kharchenko, A.; Vladimirov, G.; Aizikov, K.; O’Connor, P.B.; Nikolaev, E.; Heeren, R.M.A.; Amster, I.J. Analysis of phase dependent frequency shifts in simulated FTMS transients using the filter diagonalization method. Int. J. Mass Spectrom. 2012, 325–327, 19–24. [Google Scholar] [CrossRef]
- Mathieson, W.; Thomas, G. Using FFPE Tissue in Genomic Analyses: Advantages, Disadvantages and the Role of Biospecimen Science. Curr. Pathobiol. Rep. 2019, 7, 35–40. [Google Scholar] [CrossRef]
- Buck, A.; Ly, A.; Balluff, B.; Sun, N.; Gorzolka, K.; Feuchtinger, A.; Janssen, K.P.; Kuppen, P.J.; van de Velde, C.J.; Weirich, G.; et al. High-resolution MALDI-FT-ICR MS imaging for the analysis of metabolites from formalin-fixed, paraffin-embedded clinical tissue samples. J. Pathol. 2015, 237, 123–132. [Google Scholar] [CrossRef]
- Moore, J.L.; Charkoftaki, G. A Guide to MALDI Imaging Mass Spectrometry for Tissues. J. Proteome Res. 2023, 22, 3401–3417. [Google Scholar] [CrossRef] [PubMed]
- Ly, A.; Buck, A.; Balluff, B.; Sun, N.; Gorzolka, K.; Feuchtinger, A.; Janssen, K.P.; Kuppen, P.J.; van de Velde, C.J.; Weirich, G.; et al. High-mass-resolution MALDI mass spectrometry imaging of metabolites from formalin-fixed paraffin-embedded tissue. Nat. Protoc. 2016, 11, 1428–1443. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D. Interest is high in improving quality control for clinical metabolomics: Setting the path forward for community harmonization of quality control standards. Metabolomics 2018, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.B.; Bandukwala, A.; Bethan, B.; Broadhurst, D.; Clish, C.B.; Dasari, S.; Derr, L.; Evans, A.; Fischer, S.; et al. Towards quality assurance and quality control in untargeted metabolomics studies. Metabolomics 2019, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.M.; O’Donovan, C.; Playdon, M.; Beecher, C.; Beger, R.D.; Bowden, J.A.; Broadhurst, D.; Clish, C.B.; Dasari, S.; Dunn, W.B.; et al. Dissemination and analysis of the quality assurance (QA) and quality control (QC) practices of LC-MS based untargeted metabolomics practitioners. Metabolomics 2020, 16, 113. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, R.; Broadhurst, D.; Smilde, A.K.; Kristal, B.S.; Baker, J.D.; Beger, R.; Bessant, C.; Connor, S.; Capuani, G.; Craig, A.; et al. Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics 2007, 3, 231–241. [Google Scholar] [CrossRef]
- Kirwan, J.A.; Gika, H.; Beger, R.D.; Bearden, D.; Dunn, W.B.; Goodacre, R.; Theodoridis, G.; Witting, M.; Yu, L.R.; Wilson, I.D.; et al. Quality assurance and quality control reporting in untargeted metabolic phenotyping: mQACC recommendations for analytical quality management. Metabolomics 2022, 18, 70. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
| Analyzer | Accuracy | Resolution | m/z Range | Scan Speed | General Comments |
|---|---|---|---|---|---|
| Magnetic Sector | <5 ppm | 30,000 | 10,000 | 1 s | Large footprint |
| Quadrupole | 100 ppm | 4000 | 4000 | 1 s | Low cost |
| Ion Trap | 100 ppm | 4000 | 1000 | 1 s | Low cost, well suited for MSn |
| Time of Flight (ToF) | 200 ppm | 8000 | >300,000 | ms | Low cost |
| ToF Reflectron | 10 ppm | 15,000 | 10,000 | ms | Good accuracy, good resolution |
| Quadrupole-TOF | 10 ppm | 10,000 | 10,000 | 1 s | High sensitivity and accuracy when used for MS2 |
| FT-ICR-MS | 100 ppb | 106–7 | 10,000 | 1–10 s | Expensive, large footprint but has highest accuracy and resolving power |
| Orbitrap | 1 ppm | 105–6 | 10,000 | 1 s | Faster scan speeds allow for easier combination with LC systems |
| Facility | Location |
|---|---|
| National High Magnetic Field Laboratory (MagLab) at Florida State University | Tallahassee, FL |
| The Ohio State University | Columbus, OH |
| University of Alabama at Birmingham | Birmingham, AL |
| University of Nebraska-Lincoln | Lincoln, NE |
| University of Wisconsin-Madison | Madison, WI |
| Florida International University | Miami, FL |
| Old Dominion University | Norfolk, VA |
| University of Maryland School of Pharmacy | Baltimore, MD |
| Weill Cornell Medicine | New York, NY |
| Woods Hole Oceanographic Institution | Woods Hole, MA |
| European Network of FT-ICR-MS Centers | Various (www.eu-fticr-ms.eu, accessed on 1 August 2024) |
| University of Birmingham (UK) | Birmingham, UK |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cochran, D.; Powers, R. Fourier Transform Ion Cyclotron Resonance Mass Spectrometry Applications for Metabolomics. Biomedicines 2024, 12, 1786. https://doi.org/10.3390/biomedicines12081786
Cochran D, Powers R. Fourier Transform Ion Cyclotron Resonance Mass Spectrometry Applications for Metabolomics. Biomedicines. 2024; 12(8):1786. https://doi.org/10.3390/biomedicines12081786
Chicago/Turabian StyleCochran, Darcy, and Robert Powers. 2024. "Fourier Transform Ion Cyclotron Resonance Mass Spectrometry Applications for Metabolomics" Biomedicines 12, no. 8: 1786. https://doi.org/10.3390/biomedicines12081786
APA StyleCochran, D., & Powers, R. (2024). Fourier Transform Ion Cyclotron Resonance Mass Spectrometry Applications for Metabolomics. Biomedicines, 12(8), 1786. https://doi.org/10.3390/biomedicines12081786







