Circulating Gut Microbe-Derived Metabolites Are Associated with Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS)
2.3. Statistical Analysis
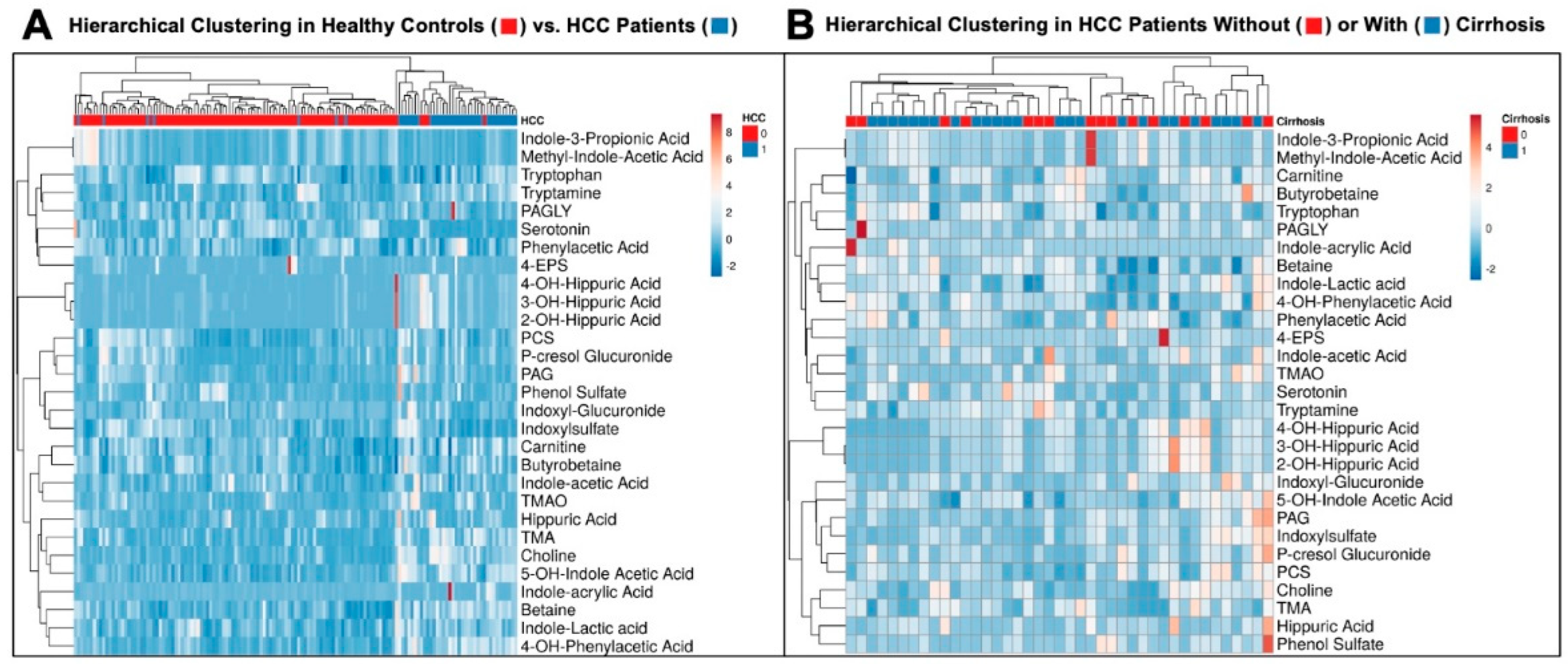
3. Results
3.1. Gut Microbe-Derived Metabolites Are Altered in HCC
3.2. Impact of Background Liver Morphology on Circulating Gut Microbe-Derived Metabolites
3.3. Hierarchical Clustering Clearly Distinguishes Healthy Controls from HCC Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marquardt, J.U.; Thorgeirsson, S.S. SnapShot: Hepatocellular carcinoma. Cancer Cell 2014, 25, 550.e1. [Google Scholar] [CrossRef] [PubMed]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of hepatocellular carcinoma. Hepatology 2021, 73 (Suppl. 1), 4–13. [Google Scholar] [CrossRef]
- Bruix, J.; Sherman, M. Management of hepatocellular carcinoma: An update. Hepatology 2011, 53, 1020–1022. [Google Scholar] [CrossRef]
- Temraz, S.; Nassar, F.; Kreidieh, F.; Mukherji, D.; Shamseddine, A.; Nasr, R. Hepatocellular carcinoma immunotherapy and the potential influence of gut microbiome. Int. J. Mol. Sci. 2021, 22, 7800. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Greten, T.F. Gut microbiome in HCC—Mechanisms, diagnosis and therapy. J. Hepatol. 2020, 72, 230–238. [Google Scholar] [CrossRef]
- Daniel, N.; Genua, F.; Jenab, M.; Mayén, A.L.; Chrysovalantou, C.A.; Keski-Rankonen, P.; Hughes, D.J. The role of the gut microbiome in the development of hepatobiliary cancers. Hepatology 2023. [Google Scholar] [CrossRef]
- Yang, J.; He, Q.; Lu, F.; Chen, K.; Ni, Z.; Wang, H.; Zhou, C.; Zhang, Y.; Chen, B.; Bo, Z.; et al. A distinct microbiota signature precedes the clinical diagnosis of hepatocellular carcinoma. Gut Microbes 2023, 15, 2201159. [Google Scholar] [CrossRef]
- Ren, Z.; Li, A.; Jiang, J.; Zhou, L.; Yu, Z.; Lu, H.; Xie, H.; Chen, X.; Shao, L.; Zhang, R.; et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019, 68, 1014–1023. [Google Scholar] [CrossRef]
- Yu, J.; Yang, X.; Zhang, B. Understanding gut dysbiosis for hepatocellular carcinoma and treatment. Trends Endocrinol. Metab. 2024, 24, 00163-2. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.; Wong, V.W.; Zhang, X.; Yu, J. Interplay between gut microbiome, host genetics and epigenetic modifications in MASLD and MASLD-related hepatocellular carcinoma. Gut 2024. [Google Scholar] [CrossRef] [PubMed]
- Elvevi, A.; Laffusa, A.; Gallo, C.; Invernizzi, P.; Massironi, S. Any role for microbiota in cholangiocarcinoma? A comprehensive review. Cells 2023, 12, 370. [Google Scholar] [CrossRef] [PubMed]
- Lederer, A.K.; Rasel, H.; Kohnert, E.; Kreutz, C.; Huber, R.; Badr, M.T.; Dellweg, P.K.E.; Bartsch, F.; Lang, H. Gut microbiota in diagnosis, therapy and prognosis of cholangiocarcinoma and gallbladder carcinoma—A scoping review. Microorganisms 2023, 11, 2363. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, C.E.; Hershberger, C.E.; Rodarte, A.I.; Siddiqi, S.; Moro, A.; Acevedo-Moreno, L.A.; Brown, J.M.; Allende, D.S.; Aucejo, F.; Rotroff, D.M. Salivary Metabolites are Promising Non-Invasive Biomarkers of Hepatocellular Carcinoma and Chronic Liver Disease. Liver Cancer Int. 2021, 2, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Acharya, C.; Sahingur, S.E.; Bajaj, J.S. Microbiota, cirrhosis, and the emerging oral-gut-liver axis. JCI Insight 2017, 2, e94416. [Google Scholar] [CrossRef]
- Brown, J.M.; Hazen, S.L. The gut microbial endocrine organ: Bacterially derived signals driving cardiometabolic disease. Annu. Rev. Med. 2015, 66, 343–359. [Google Scholar] [CrossRef]
- Donia, M.S.; Fischbach, M.A. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science 2015, 349, 1254766. [Google Scholar] [CrossRef]
- Nemet, I.; Li, X.S.; Haghikia, A.; Li, L.; Wilcox, J.; Romano, K.A.; Buffa, J.A.; Witkowski, M.; Demuth, I.; König, M.; et al. Atlas of gut microbe-derived products from aromatic amino acids and risk of cardiovascular morbidity and mortality. Eur. Heart J. 2023, 44, 3085–3096. [Google Scholar] [CrossRef]
- Wang, Z.; Bergeron, N.; Levison, B.S.; Xi, X.S.; Chiu, S.; Jia, X.; Koeth, R.A.; Li, L.; Wu, Y.; Tang, W.H.W.; et al. Impact of chronic dietary red meat, white meat, or non-meat protein on trimethylamine N-oxide metabolism and renal excretion in healthy men and women. Eur. Heart J. 2019, 40, 583–594. [Google Scholar] [CrossRef]
- Haynes, W. Bonferroni Correction. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; p. 154. [Google Scholar]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Sharpton, S.R.; Schnabl, B.; Knight, R.; Loomba, R. Current concepts, opportunities, and challenges of gut microbiome-based personalized medicine in nonalcoholic fatty liver disease. Cell Metab. 2021, 33, 21–32. [Google Scholar] [CrossRef]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Loomba, R.; Schnabl, B.; Knight, R. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Adolph, T.E.; Dudek, M.; Knolle, P. Non-alcoholic fatty liver disease: The interplay between metabolism, microbes and immunity. Nat. Metab. 2021, 3, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Guo, Y.; Liao, Y.; Liu, J. Mechanism-guided fine-tuned microbiome potentiates anti-tumor immunity in HCC. Front. Immunol. 2023, 14, 1333864. [Google Scholar] [CrossRef] [PubMed]
- Gok Yavuz, B.; Datar, S.; Chamseddine, S.; Mohamed, Y.I.; LaPelusa, M.; Lee, S.S.; Hu, Z.I.; Koay, E.J.; Tran Cao, H.S.; Jalal, P.K.; et al. The gut microbiome as a biomarker and therapeutic target in hepatocellular carcinoma. Cancers 2023, 15, 4875. [Google Scholar] [CrossRef] [PubMed]
- Ranjbarian, T.; Schnabl, B. Gut microbiome-centered therapies for alcohol-associated liver disease. Semin. Liver Dis. 2023, 43, 311–322. [Google Scholar] [CrossRef]
- Ladd, A.D.; Duarte, S.; Sahin, I.; Zarrinpar, A. Mechanisms of drug resistance in HCC. Hepatology 2023, 79, 926–940. [Google Scholar] [CrossRef]
- Greten, T.F.; Villanueva, A.; Korangy, F.; Ruf, B.; Yarchoan, M.; Ma, L.; Ruppin, E.; Wang, X.W. Biomarkers for immunotherapy of hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2023, 20, 780–798. [Google Scholar] [CrossRef]
- Muscolino, P.; Granata, B.; Omero, F.; De Pasquale, C.; Campana, S.; Calabrò, A.; D’Anna, F.; Drommi, F.; Pezzino, G.; Cavaliere, R.; et al. Potential predictive role of gut microbiota to immunotherapy in HCC patients: A brief review. Front. Oncol. 2023, 13, 1247614. [Google Scholar] [CrossRef]
- Craciun, S.; Marks, J.A.; Balskus, E.P. Characterization of choline trimethylamine-lyase expands the chemistry of glycyl radical enzymes. ACS Chem. Biol. 2014, 9, 1408–1413. [Google Scholar] [CrossRef]
- Rajakovich, L.J.; Fu, B.; Bollenbach, M.; Balskus, E.P. Elucidation of an anaerobic pathway for metabolism of L-carnitine-derived g-butyrobetaine to trimethylamine in human gut bacteria. Proc. Natl. Acad. Sci. USA 2021, 118, e2101498118. [Google Scholar] [CrossRef]
- Koeth, R.A.; Levison, B.S.; Culley, M.K.; Buffa, J.A.; Wang, Z.; Gregory, J.C.; Org, E.; Wu, Y.; Li, L.; Smith, J.D.; et al. γ-butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab. 2014, 20, 799–812. [Google Scholar] [CrossRef] [PubMed]
- Buffa, J.A.; Romano, K.A.; Copeland, M.F.; Cody, D.B.; Zhu, W.; Galvez, R.; Fu, X.; Ward, K.; Ferrell, M.; Dai, H.J.; et al. The microbial gbu gene cluster links cardiovascular disease risk associated with red meat consumption to microbiota L-carnitine catabolism. Nat. Microbiol. 2022, 7, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Jameson, E.; Crosatti, M.; Schäfer, H.; Rajakumar, K.; Bugg, T.D.; Chen, Y. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc. Natl. Acad. Sci. USA 2014, 111, 4268–4273. [Google Scholar] [CrossRef] [PubMed]
- Rossner, R.; Kaeberlein, M.; Leiser, S.F. Flavin-containing monooxygenases in aging and disease: Emerging roles for ancient enzymes. J. Biol. Chem. 2017, 292, 11138–11146. [Google Scholar] [CrossRef]
- Warrier, M.; Shih, D.M.; Burrows, A.C.; Ferguson, D.; Gromovsky, A.D.; Brown, A.L.; Marshall, S.; McDaniel, A.; Schugar, R.C.; Wang, Z.; et al. The TMAO-generating enzyme flavin monooxygenase 3 is a central regulator of cholesterol balance. Cell Rep. 2015, 10, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Ling, A.V.; Manthena, P.V.; Gearing, M.E.; Graham, M.J.; Crooke, R.M.; Croce, K.J.; Esquejo, R.M.; Clish, C.B.; Morbid Obesity Study Group; et al. Flavin-containing monooxygenase 3 as a potential player in diabetes-associated atherosclerosis. Nat. Comm. 2015, 6, 6498. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef]
- Zhou, D.; Zhang, J.; Xiao, C.; Mo, C.; Ding, B.S. Trimethylamine-N-oxide (TMAO) mediates the crosstalk between the gut microbiota and hepatic vascular niche to alleviate liver fibrosis in nonalcoholic steatohepatitis. Front. Immunol. 2022, 13, 964477. [Google Scholar] [CrossRef]
- Liu, Z.-Y.; Tan, X.Y.; Li, Q.J.; Liao, G.C.; Fang, A.P.; Zhang, D.M.; Chen, P.Y.; Wang, X.Y.; Luo, Y.; Long, J.A.; et al. Trimethylamine N-oxide, a gut microbiota-dependent metabolite of choline, is positively associated with the risk of primary liver cancer: A case-control study. Nutr. Metab. 2018, 15, 81. [Google Scholar] [CrossRef] [PubMed]
- Flores-Guerrero, J.L.; Post, A.; van Dijk, P.R.; Connelly, M.A.; Garcia, E.; Navis, G.; Bakker, S.J.L.; Dullaart, R.P.F. Circulating trimethylamine-N-oxide is associated with all-cause mortality in subjects with nonalcoholic fatty liver disease. Liver Int. 2021, 41, 2371–2382. [Google Scholar] [CrossRef]
- Tan, X.; Liu, Y.; Long, J.; Chen, S.; Liao, G.; Wu, S.; Li, C.; Wang, L.; Ling, W.; Zhu, H. Trimethylamine N-oxide aggravates liver steatosis through modulation of bile acid metabolism and inhibition of farnesoid X receptor signaling in nonalcoholic fatty liver disease. Mol. Nutr. Food Res. 2019, 63, e1900257. [Google Scholar] [CrossRef] [PubMed]
- Theofilis, P.; Vordoni, A.; Kalaitzidis, R.G. Trimethylamine N-oxide levels in non-alcoholic fatty liver disease. Metabolites 2022, 12, 1243. [Google Scholar] [CrossRef] [PubMed]
- Helsley, R.N.; Miyata, T.; Kadam, A.; Varadharajan, V.; Sangwan, N.; Huang, E.C.; Banerjee, R.; Brown, A.L.; Fung, K.K.; Massey, W.J.; et al. Gut microbial trimethylamine is elevated in alcohol-associated hepatitis and contributes to ethanol-induced liver injury in mice. eLife 2022, 11, e76554. [Google Scholar] [CrossRef] [PubMed]
- Blachier, F.; Andriamihaja, M. Effects of the L-tyrosine-derived bacterial metabolite p-cresol on colonic and peripheral cells. Amino Acids 2022, 54, 325–338. [Google Scholar] [CrossRef]
- Fu, H.Y.; Xu, J.M.; Ai, X.; Dang, F.T.; Tan, X.; Yu, H.Y.; Feng, J.; Yang, W.X.; Ma, H.T.; Tu, R.F.; et al. The clostridium metabolite p-cresol sulfate relieves inflammation of primary biliary cholangitis by regulating Kupffer cells. Cells 2022, 11, 3782. [Google Scholar] [CrossRef]
- Wypych, T.P.; Pattaroni, C.; Perdijk, O.; Yap, C.; Trompette, A.; Anderson, D.; Creek, D.J.; Harris, N.L.; Marsland, B.J. Microbial metabolism of L-tyrosine protects against allergic airway inflammation. Nat. Immunol. 2021, 22, 279–286. [Google Scholar] [CrossRef]
- Khanmohammadi, S.; Kuchay, M.S. Toll-like receptors and metabolic (dysfunction)-associated fatty liver disease. Pharmacol. Res. 2022, 185, 106507. [Google Scholar] [CrossRef] [PubMed]
- Roh, Y.S.; Seki, E. Toll-like receptors in alcoholic liver disease, non-alcoholic steatohepatitis and carcinogenesis. J. Gastroenterol. Hepatol. 2013, 28 (Suppl. 1), 38–42. [Google Scholar] [CrossRef]
- Tang, Y.L.; Zhu, L.; Tao, Y.; Lu, W.; Cheng, H. Role of targeting TLR4 signaling axis in liver-related disease. Pathol. Res. Pract. 2023, 244, 154410. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Treuren, W.V.; Fischer, C.R.; Merrill, B.D.; DeFelice, B.C.; Sanchez, J.M.; Higginbottom, S.K.; Guthrie, L.; Fall, L.A.; Dodd, D.; et al. A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature 2021, 595, 415–420. [Google Scholar] [CrossRef]
- Ferrell, M.; Bazeley, P.; Wang, Z.; Levison, B.S.; Li, X.S.; Jia, X.; Krauss, R.M.; Knight, R.; Lusis, A.J.; Garcia-Garcia, J.C.; et al. Fecal microbiome composition does not accurately predict diet-induced TMAO production in healthy adults. J. Am. Heart Assoc. 2021, 10, e021934. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Hazen, S.L. Targeting of microbe-derived metabolites to improve human health. J. Biol. Chem. 2017, 292, 8560–8568. [Google Scholar] [CrossRef] [PubMed]
- Woo, A.Y.M.; Aguilar Ramos, M.A.; Narayan, R.; Richards-Corke, K.C.; Wang, M.L.; Sandoval-Espinola, W.J.; Balskus, E.P. Targeting the human gut microbiome with small-molecule inhibitors. Nat. Rev. Chem. 2023, 7, 319–339. [Google Scholar] [CrossRef]
- Oellgaard, J.; Winther, S.A.; Hansen, T.S.; Rossing, P.; von Scholten, B.J. Trimethylamine N-oxide (TMAO) as a New Potential Therapeutic Target for Insulin Resistance and Cancer. Curr. Pharm. Des. 2017, 23, 3699–3712. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hong, J.; Wang, Y.; Pei, M.; Wang, K.; Gong, Z. Trimethylamine-N-Oxide Pathway: A Potential Target for the Treatment of MAFLD. Front. Mol. Biosci. 2021, 8, 733507. [Google Scholar] [CrossRef]


| (a) | |||
| Total (n = 96) | |||
| Average Age | 50.38 | ||
| Male Sex | 44 | ||
| Race | |||
| White | 32 (72.72%) | ||
| Black | 11 (25.0%) | ||
| Asian | 1 (2.27%) | ||
| Average BMI (kg/m2) | 28.06 | ||
| Female Sex | 50 | ||
| Race | |||
| White | 45 (90.0%) | ||
| Black | 5 (10.0%) | ||
| Average BMI (kg/m2) | 27.46 | ||
| (b) | |||
| Total HCC (n = 41) | HCC + Cirrhosis (n = 25) | HCC Alone (n = 16) | |
| Median Age (IQR) | 68 (62.5–76.5) | 66 (62–72) | 73 (62.5–79.3) |
| Male Sex | 13 | 7 (28%) | 6 (37.5%) |
| Race | |||
| White | 34 (82.9%) | 19 (76%) | 15 (93.7%) |
| Black | 4 (9.8%) | 4 (16%) | - |
| Asian | 3 (7.3%) | 2 (8%) | 1 (6.3%) |
| BMI (kg/m2) | 27.3 (24–30.7) | 27.3 (24–31.3) | 27.6 (25.7–30.7) |
| Etiology of Cirrhosis | - | ||
| NASH | 4 (9.8%) | 4 (16%) | |
| Hepatitis B | 4 (9.8%) | 4 (16%) | |
| Hepatitis C | 14 (34.1%) | 14 (56%) | |
| Idiopathic | 2 (4.9%) | 2 (8%) | |
| Hemochromatosis | 1 (2.4%) | 1 (4%) | |
| Diabetes | 11 (26.8%) | 8 (32%) | 3 (18.8%) |
| No. Lesions Median (IQR) | 1 (1–2) | 1 (1–2) | 1 (1–3) |
| Size Largest Lesion Median cm (IQR) | 4 (2.5–6) | 3 (2–4) | 10 (4.3–14.3) |
| Treatment Pre-Sample Collection | |||
| Y90 | 3 (7.3%) | 2 (8%) | 1 (6.3%) |
| TACE | 4 (9.8%) | 2 (8%) | 2 (12.6%) |
| None | 34 (82.9%) | 21 (84%) | 13 (81.1%) |
| Grade of Differentiation | |||
| Well | 4 (9.8%) | - | 4 (25%) |
| Moderate | 34 (82.9%) | 24 (96%) | 10 (62.5%) |
| Poor | 3 (7.3%) | 1 (4%) | 2 (12.6%) |
| Vascular Invasion | 30 (73.2%) | 20 (80%) | 10 (62.5%) |
| Recurrence | 15 (36.6%) | 9 (36%) | 6 (37.5%) |
| HCC (n = 41) | Healthy Control (n = 96) | p-Value | |
|---|---|---|---|
| Choline | 18.10 (8.54) | 8.49 (2.75) | <0.001 |
| Betaine | 46.0 (14.5) | 31.70 (14.1) | <0.001 |
| Carnitine | 29.5 (9.2) | 25.60 (6.2) | 0.007 |
| g-butyrobetaine | 0.75 (0.33) | 0.67 (0.22) | 0.029 |
| 5-OH-indole acetic acid | 0.064 (0.025) | 0.033 (0.010) | <0.001 |
| Hippuric acid | 9.61 (12.2) | 6.27 (5.65) | 0.124 |
| 4-OH-hippuric acid | 0.89 (0.037) | 0.40 (1.68) | 0.106 |
| 3-OH-hippuric acid | 0.84 (0.98) | 0.36 (1.48) | <0.001 |
| 2-OH-hippuric acid | 1.09 (1.32) | 0.43 (1.91) | 0.108 |
| Indole-3-propionic acid | 0.63 (0.83) | 1.03 (1.08) | 0.028 |
| Methyl-indole-acetic acid | 0.18 (0.23) | 0.29 (0.30) | 0.030 |
| Indole-lactic acid | 1.06 (0.36) | 0.91 (0.42) | 0.038 |
| Phenylacetic acid | 2.59 (1.19) | 2.36 (0.68) | 0.105 |
| 4-OH-Phenyllactic acid | 1.41 (0.70) | 0.625 (0.36) | <0.001 |
| p-Cresol glucuronide | 26.9 (28.1) | 15.8 (20.3) | 0.032 |
| Tryptophan | 86.7 (22.3) | 94.9 (20.7) | 0.056 |
| Serotonin | 0.015 (0.014) | 0.18 (0.15) | <0.001 |
| Indole-acetic acid | 1.79 (1.22) | 1.58 (0.81) | 0.177 |
| Phenol Sulfate | 4.69 (7.09) | 3.94 (4.66) | 0.508 |
| Trimethylamine N-oxide (TMAO) | 7.11 (7.72) | 3.99 (3.26) | <0.001 |
| Phenylacetylglutamine (PAG) | 3.66 (3.73) | 2.11 (1.50) | 0.002 |
| Indoxyl sulfate | 3.80 (3.65) | 4.03 (2.20) | 0.748 |
| p-Cresol sulfate | 30.7 (23.9) | 24.70 (19.0) | 0.224 |
| Phenylacetylglycine (PAGLY) | 0.011 (0.014) | 0.0096 (0.0064) | 0.281 |
| 4-ethylphenyl sulfate (4-EPS) | 0.88 (2.33) | 1.25 (3.78) | 0.487 |
| Trimethylamine (TMA) | 3.30 (2.42) | 1.17 (1.38) | <0.001 |
| Cirrhosis + HCC (n = 25) | HCC Alone (n = 16) | p-Value | |
|---|---|---|---|
| Choline | 17.3 (7.67) | 19.3 (9.90) | 0.321 |
| Betaine | 48.7 (14.7) | 41.9 (13.7) | 0.078 |
| Carnitine | 30.6 (9.54) | 27.6 (8.53) | 0.300 |
| g-butyrobetaine | 0.75 (0.30) | 0.75 (0.39) | 0.776 |
| 5-OH-indole acetic acid | 0.061 (0.019) | 0.067 (0.032) | 0.618 |
| Hippuric acid | 8.46 (9.70) | 11.4 (15.5) | 0.162 |
| 4-OH-hippuric acid | 0.84 (0.76) | 0.96 (1.09) | 0.744 |
| 3-OH-hippuric acid | 0.88 (1.00) | 0.74 (0.92) | 0.539 |
| 2-OH-hippuric acid | 0.866 (1.00) | 0.78 (0.94) | 0.804 |
| Indole-3-propionic acid | 0.699 (0.51) | 0.15 (0.32) | 0.645 |
| Methyl-indole-acetic acid | 0.32 (0.14) | 1.00 (1.24) | 0.620 |
| Indole-lactic acid | 1.15 (0.33) | 0.92 (0.38) | 0.033 |
| Phenylacetic acid | 2.50 (1.07) | 2.73 (1.37) | 0.546 |
| 4-OH-Phenyllactic acid | 1.54 (0.62) | 1.21 (0.79) | 0.079 |
| p-Cresol glucuronide | 28.6 (26.5) | 28.5 (31.2) | 0.507 |
| Tryptophan | 93.9 (19.4) | 75.5 (22.4) | 0.009 |
| Serotonin | 0.17 (0.16) | 0.011 (0.011) | 0.131 |
| Indole-acetic acid | 1.67 (0.90) | 2.00 (1.61) | 0.414 |
| Phenol Sulfate | 3.40 (2.61) | 6.71 (10.78) | 0.141 |
| Trimethylamine N-oxide (TMAO) | 7.7 (8.27) | 6.17 (6.93) | 0.412 |
| Phenylacetylglutamine (PAG) | 3.06 (3.31) | 4.61 (6.93) | 0.183 |
| Indoxyl sulfate | 3.56 (3.63) | 4.61 (4.26) | 0.652 |
| p-Cresol sulfate | 32.4 (27.2) | 28.1 (17.8) | 0.787 |
| Phenylacetylglycine (PAGLY) | 0.0085 (0.005) | 0.15 (0.21) | 0.136 |
| 4-ethylphenyl sulfate (4-EPS) | 0.97 (2.45) | 2.29 (6.03) | 0.372 |
| Trimethylamine (TMA) | 1.05 (2.66) | 0.611 (1.74) | 0.632 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banerjee, R.; Wehrle, C.J.; Wang, Z.; Wilcox, J.D.; Uppin, V.; Varadharajan, V.; Mrdjen, M.; Hershberger, C.; Reizes, O.; Yu, J.S.; et al. Circulating Gut Microbe-Derived Metabolites Are Associated with Hepatocellular Carcinoma. Biomedicines 2024, 12, 1946. https://doi.org/10.3390/biomedicines12091946
Banerjee R, Wehrle CJ, Wang Z, Wilcox JD, Uppin V, Varadharajan V, Mrdjen M, Hershberger C, Reizes O, Yu JS, et al. Circulating Gut Microbe-Derived Metabolites Are Associated with Hepatocellular Carcinoma. Biomedicines. 2024; 12(9):1946. https://doi.org/10.3390/biomedicines12091946
Chicago/Turabian StyleBanerjee, Rakhee, Chase J. Wehrle, Zeneng Wang, Jennifer D. Wilcox, Vinayak Uppin, Venkateshwari Varadharajan, Marko Mrdjen, Courtney Hershberger, Ofer Reizes, Jennifer S. Yu, and et al. 2024. "Circulating Gut Microbe-Derived Metabolites Are Associated with Hepatocellular Carcinoma" Biomedicines 12, no. 9: 1946. https://doi.org/10.3390/biomedicines12091946





