Identification of Lipophagy-Related Gene Signature for Diagnosis and Risk Prediction of Alzheimer’s Disease
Abstract
1. Introduction
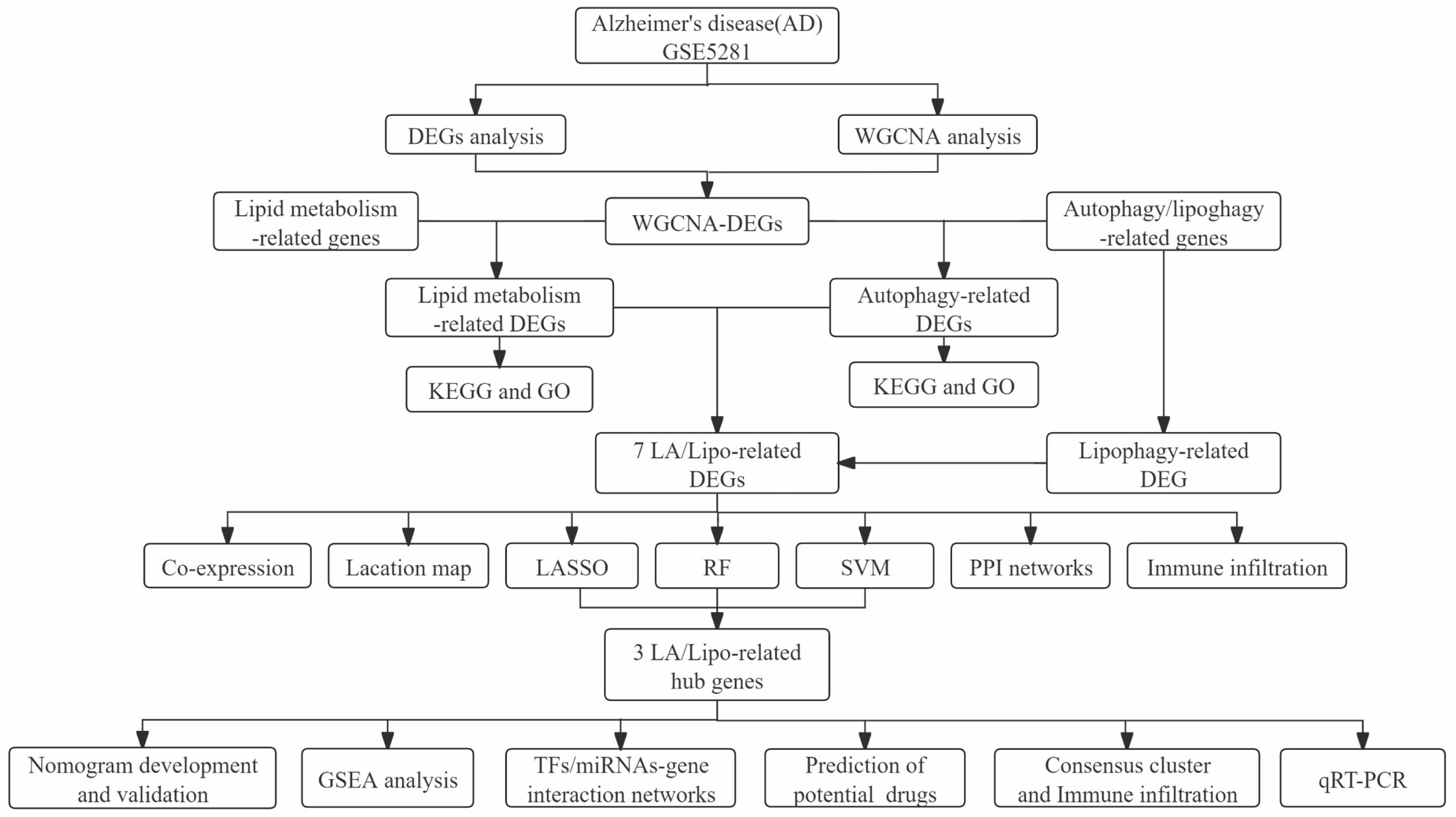
2. Materials and Methods
2.1. Data Acquisition
2.2. Identification of Differentially Expressed Genes (DEGs) and Weighted Gene Co-Expression Network Analysis (WGCNA)
2.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Functional Enrichment Analysis
2.4. Gene Expression Patterns and Protein–Protein Interaction Analyses of LA/Lipo-Related DEGs
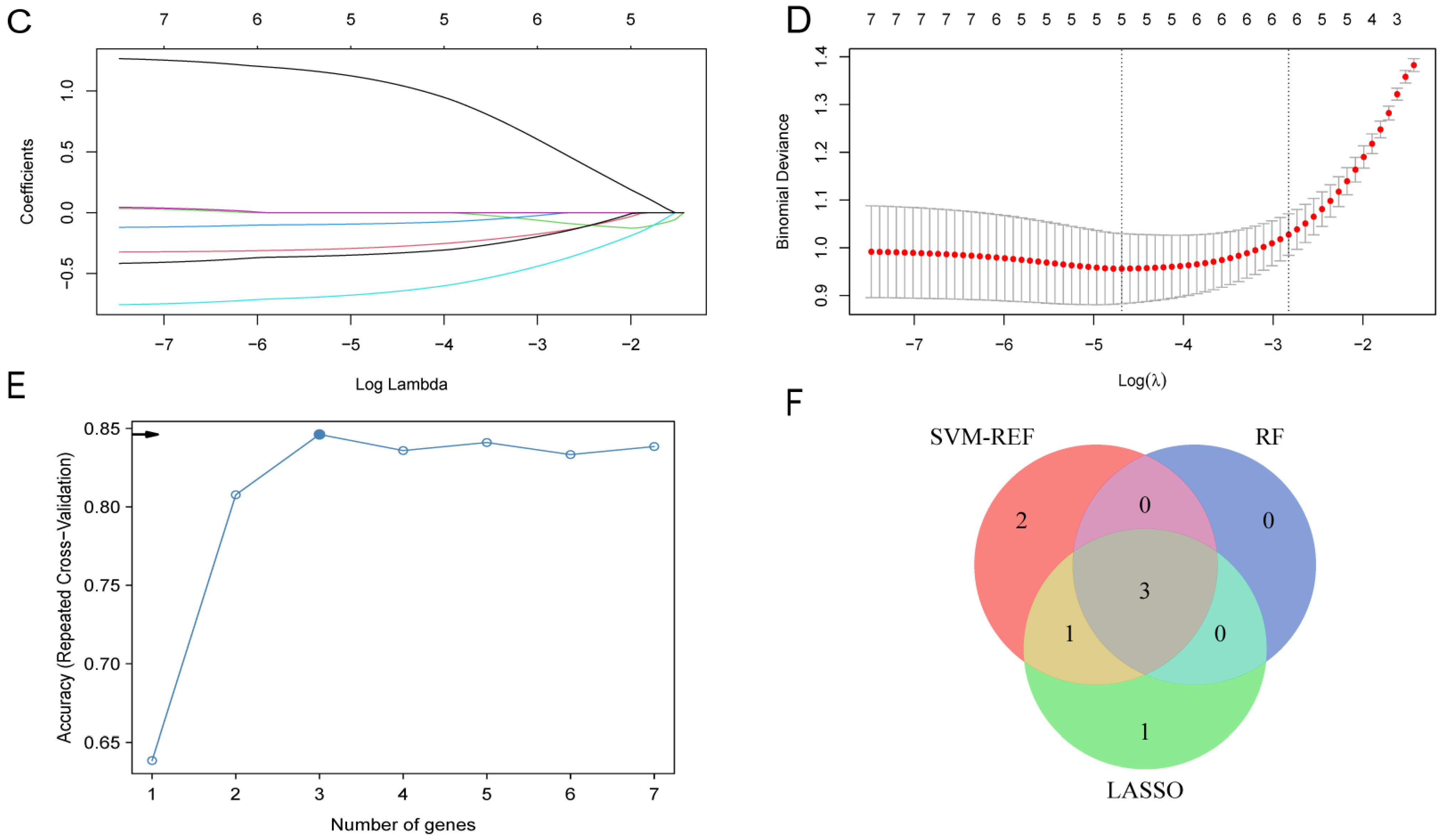
2.5. Identification of LA/Lipo-Related Hub Genes Based on Machine Learning Algorithms
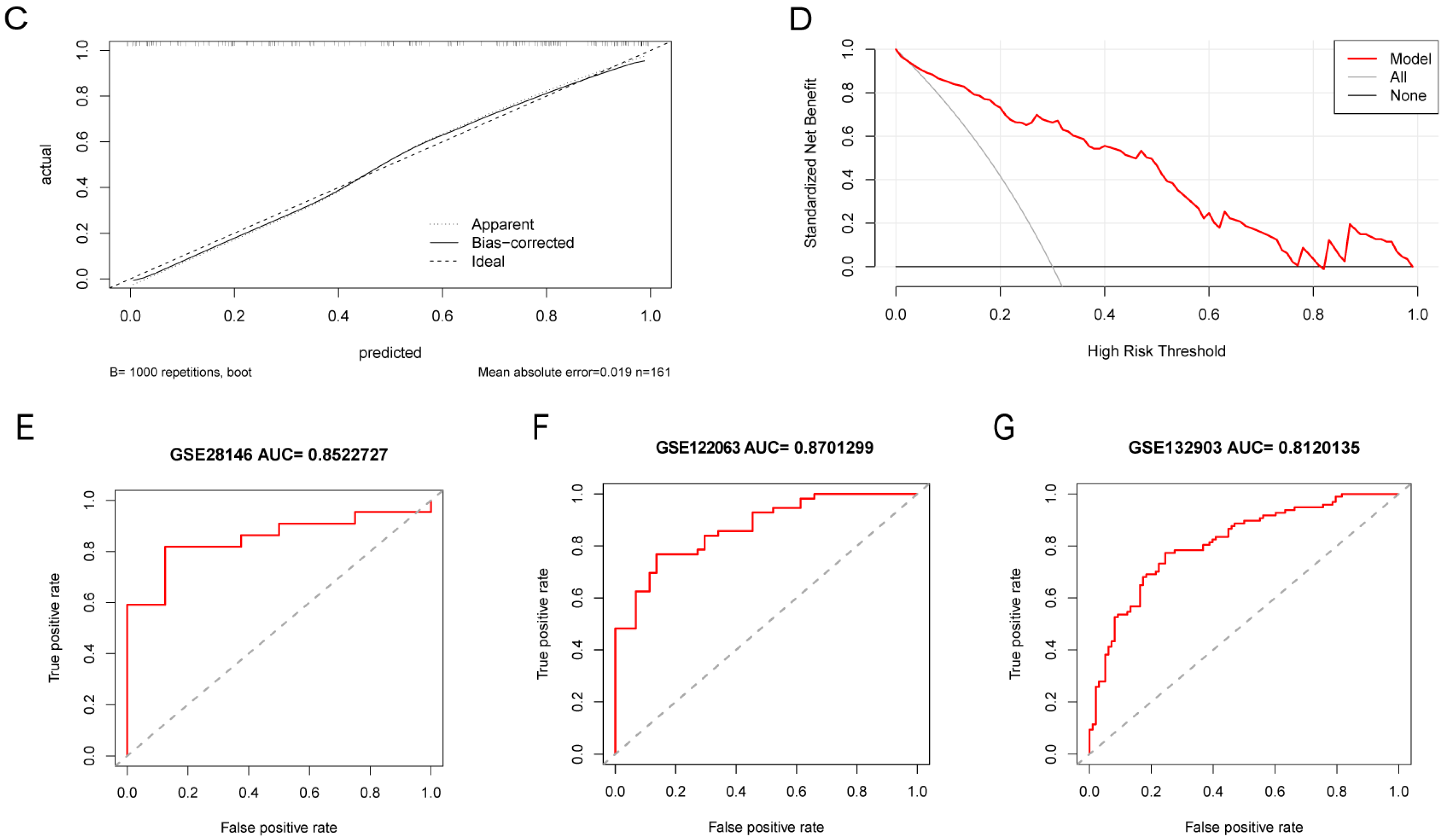
2.6. Construction and Validation of a Nomogram Model for AD Risk Prediction
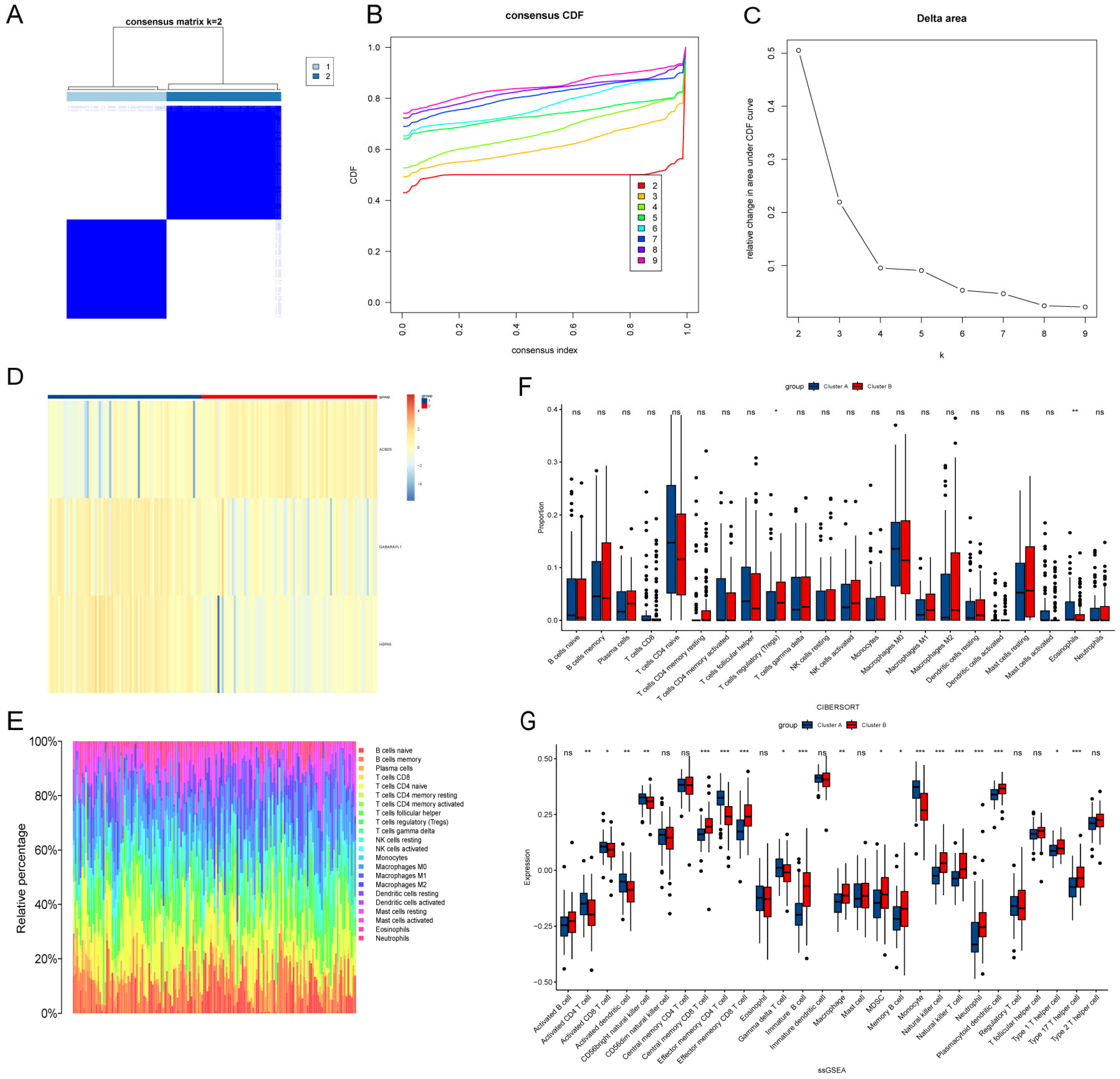
2.7. Consensus Cluster Analysis
2.8. Immune Cell Infiltration and Gene Set Enrichment Analysis (GSEA)
2.9. TF/miRNA-Gene Regulatory Networks and Drug–Gene Interaction Predication
2.10. Animals
2.11. Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
2.12. Statistical Analysis
3. Results
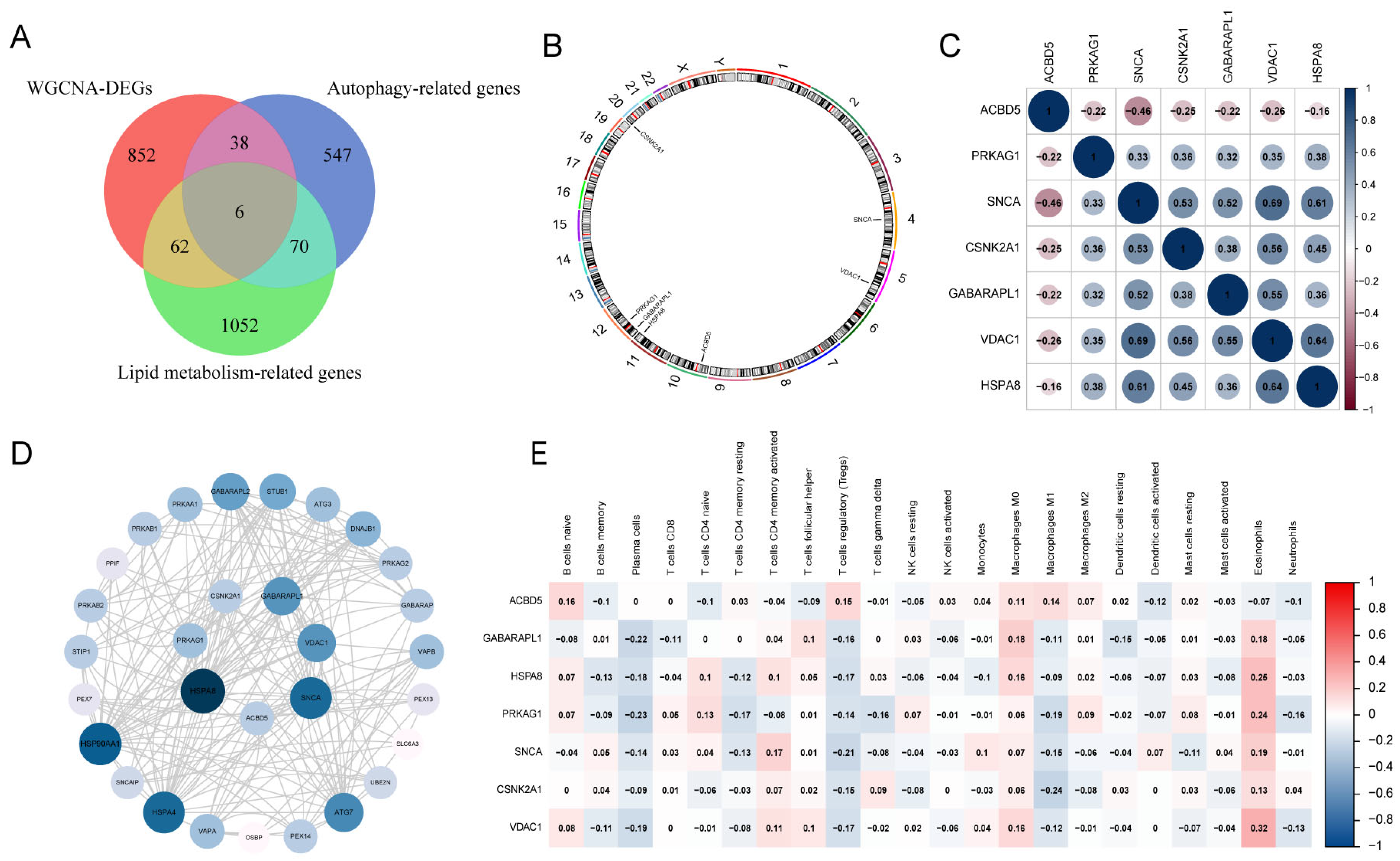
3.1. Identification of DEGs and Construction of Weighted Gene Co-Expression Networks
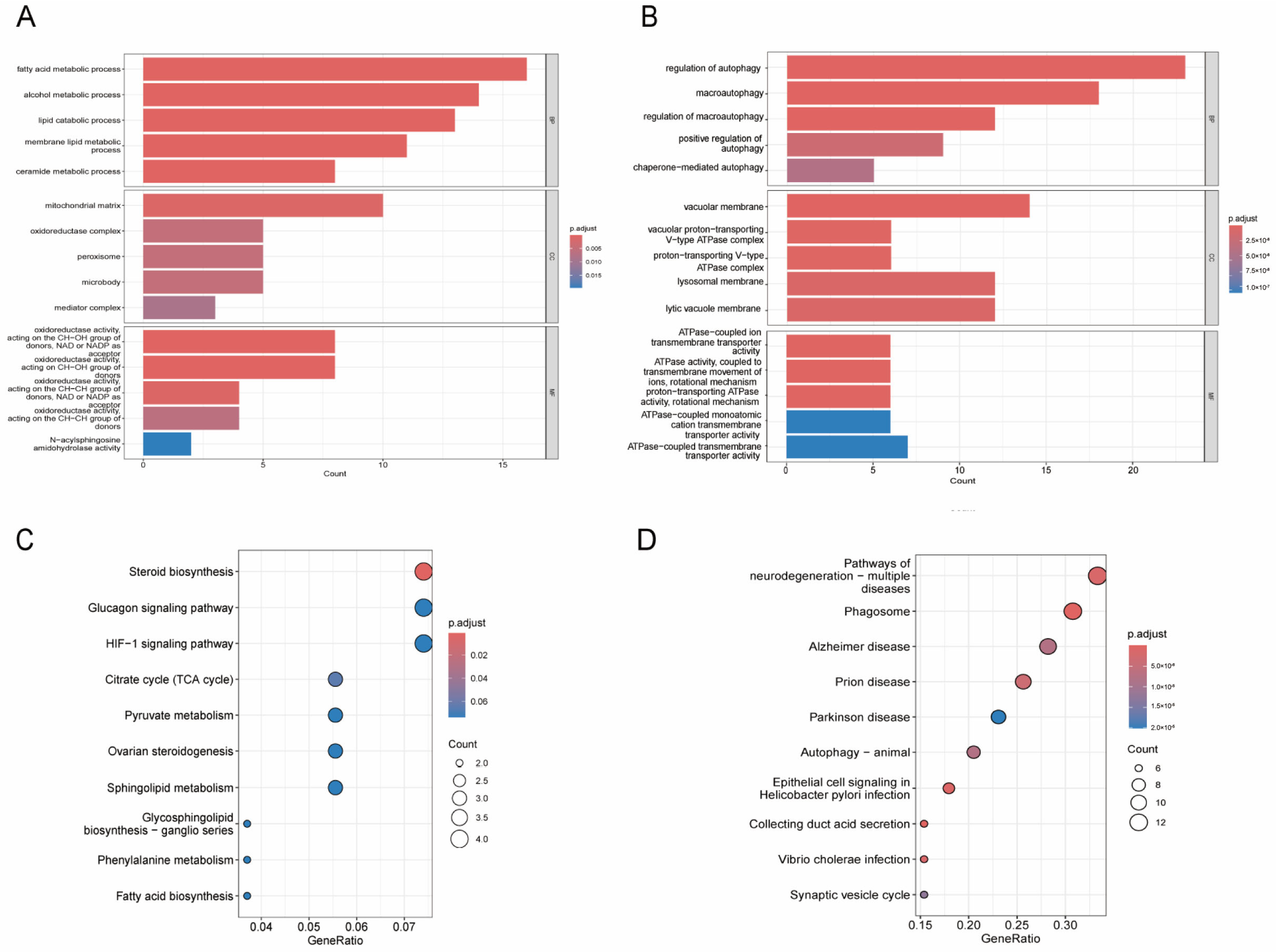
3.2. GO and KEGG Functional Enrichment Analysis
3.3. Gene Expression Patterns and PPI Network of LA/Lipo-Related DEGs
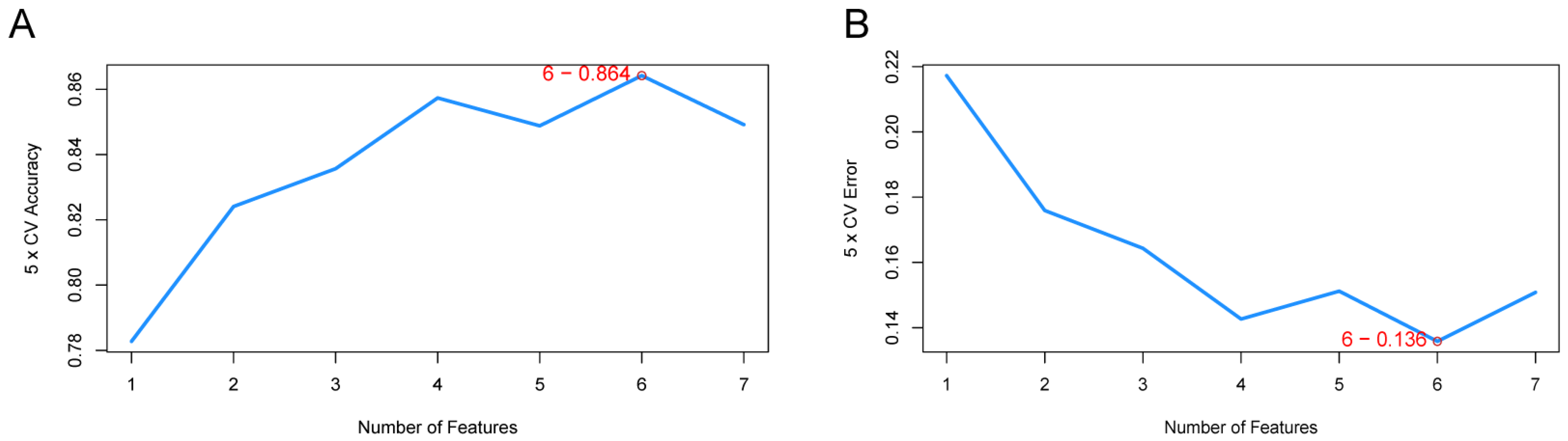
3.4. Identification of LA/Lipo-Related Hub Genes via Machine Learning Algorithm
3.5. Performance of LA/Lipo-Related Hub Genes
3.6. Identification LA/Lipo-Related Sub-Clusters and Differences in the Immune Cell Infiltration Between Sub-Clusters
3.7. GSEA Analysis
3.8. Construction of Regulatory Network and Predication of Drug–Gene Interaction
3.9. Validation of Hub Genes Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scheltens, P.; De Strooper, B.; Kivipelto, M.; Holstege, H.; Chételat, G.; Teunissen, C.; Cummings, J.; van der Flier, W. Alzheimer’s disease. Lancet 2021, 397, 1577–1590. [Google Scholar] [CrossRef] [PubMed]
- Jack, C. Advances in Alzheimer’s disease research over the past two decades. Lancet Neurol. 2022, 21, 866–869. [Google Scholar] [CrossRef]
- Rhodius-Meester, H.F.M.; Tijms, B.M.; Lemstra, A.W.; Prins, N.D.; Pijnenburg, Y.A.L.; Bouwman, F.; Scheltens, P.; van der Flier, W.M. Survival in memory clinic cohort is short, even in young-onset dementia. J. Neurol. Neurosurg. Psychiatry 2019, 90, 726–728. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer’s Association. 2023 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2023, 19, 1598–1695. [Google Scholar] [CrossRef] [PubMed]
- Migliore, L.; Coppedè, F. Gene-environment interactions in Alzheimer disease: The emerging role of epigenetics. Nat. Rev. Neurol. 2022, 18, 643–660. [Google Scholar] [CrossRef]
- Menon, N.K.; Dhopeshwarkar, G.A. Essential fatty acid deficiency and brain development. Prog. Lipid Res. 1982, 21, 309–326. [Google Scholar] [CrossRef]
- Barupal, D.K.; Baillie, R.; Fan, S.; Saykin, A.J.; Meikle, P.J.; Arnold, M.; Nho, K.; Fiehn, O.; Kaddurah-Daouk, R. Sets of coregulated serum lipids are associated with Alzheimer’s disease pathophysiology. Alzheimer’s Dement 2019, 11, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef]
- Kao, Y.C.; Ho, P.C.; Tu, Y.K.; Jou, I.M.; Tsai, K.J. Lipids and Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 1505. [Google Scholar] [CrossRef] [PubMed]
- Raulin, A.C.; Doss, S.V.; Trottier, Z.A.; Ikezu, T.C.; Bu, G.; Liu, C.C. ApoE in Alzheimer’s disease: Pathophysiology and therapeutic strategies. Mol. Neurodegener. 2022, 17, 72. [Google Scholar] [CrossRef]
- Marschallinger, J.; Iram, T.; Zardeneta, M.; Lee, S.E.; Lehallier, B.; Haney, M.S.; Pluvinage, J.V.; Mathur, V.; Hahn, O.; Morgens, D.W.; et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 2020, 23, 194–208. [Google Scholar] [CrossRef]
- Thorwald, M.A.; Silva, J.; Head, E.; Finch, C.E. Amyloid futures in the expanding pathology of brain aging and dementia. Alzheimer’s Dement 2023, 19, 2605–2617. [Google Scholar] [CrossRef]
- Laval, T.; Ouimet, M. A role for lipophagy in atherosclerosis. Nat. Rev. Cardiol. 2023, 20, 431–432. [Google Scholar] [CrossRef]
- Luo, R.; Su, L.Y.; Li, G.; Yang, J.; Liu, Q.; Yang, L.X.; Zhang, D.F.; Zhou, H.; Xu, M.; Fan, Y.; et al. Activation of PPARA-mediated autophagy reduces Alzheimer disease-like pathology and cognitive decline in a murine model. Autophagy 2020, 16, 52–69. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhao, Y.; Guo, H.; Li, Q.; Yan, C.; Li, Y.; He, S.; Wang, N.; Wang, Q. Impaired lipophagy induced-microglial lipid droplets accumulation contributes to the buildup of TREM1 in diabetes-associated cognitive impairment. Autophagy 2023, 19, 2639–2656. [Google Scholar] [CrossRef]
- Xu, Y.; Propson, N.E.; Du, S.; Xiong, W.; Zheng, H. Autophagy deficiency modulates microglial lipid homeostasis and aggravates tau pathology and spreading. Proc. Natl. Acad. Sci. USA 2021, 118, e2023418118. [Google Scholar] [CrossRef] [PubMed]
- Lian, P.; Cai, X.; Wang, C.; Liu, K.; Yang, X.; Wu, Y.; Zhang, Z.; Ma, Z.; Cao, X.; Xu, Y. Identification of metabolism-related subtypes and feature genes in Alzheimer’s disease. J. Transl. Med. 2023, 21, 628. [Google Scholar] [CrossRef]
- Zhang, Y.; Miao, Y.; Tan, J.; Chen, F.; Lei, P.; Zhang, Q. Identification of mitochondrial related signature associated with immune microenvironment in Alzheimer’s disease. J. Transl. Med. 2023, 21, 458. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Fu, C.; Wei, Y.; Xu, B.; Yang, R.; Li, C.; Qiu, M.; Yin, Y.; Qin, D. Ferroptosis-related biomarkers for Alzheimer’s disease: Identification by bioinformatic analysis in hippocampus. Front. Cell. Neurosci. 2022, 16, 1023947. [Google Scholar] [CrossRef]
- Wang, Y.; Wan, X.; Du, S. Integrated analysis revealing a novel stemness-metabolism-related gene signature for predicting prognosis and immunotherapy response in hepatocellular carcinoma. Front. Immunol. 2023, 14, 1100100. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Xie, K.; Zhang, J.; Yang, L.; Zhou, H.; Zhang, L.; Peng, R. Comprehensive analysis of mitochondrial dysfunction and necroptosis in intracranial aneurysms from the perspective of predictive, preventative, and personalized medicine. Apoptosis 2023, 28, 1452–1468. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Qi, L.; Sun, Z.; Liu, J.; Lv, Z.; Chen, L.; Huang, B.; Zhu, S.; Liu, Y.; Li, Y. Evaluation and analysis of incidence and risk factors of lower extremity venous thrombosis after urologic surgeries: A prospective two-center cohort study using LASSO-logistic regression. Int. J. Surg. 2021, 89, 105948. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Huang, Y.; Liu, F.; Wang, H. Identification of immune-related genes in diagnosing atherosclerosis with rheumatoid arthritis through bioinformatics analysis and machine learning. Front. Immunol. 2023, 14, 1126647. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Hu, X.; Lin, R.; Tang, Z.; Ye, Z.; Mao, R.; Chen, W.; Zhou, Y. Identification of shared gene signatures and molecular mechanisms between chronic kidney disease and ulcerative colitis. Front. Immunol. 2023, 14, 1078310. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhou, Y.; Zhou, Y.; Tang, X.; Tang, L.; Wang, J. Identification of crucial genes for predicting the risk of atherosclerosis with system lupus erythematosus based on comprehensive bioinformatics analysis and machine learning. Comput. Biol. Med. 2023, 152, 106388. [Google Scholar] [CrossRef]
- Yu, E.; Zhang, M.; Xu, G.; Liu, X.; Yan, J. Consensus cluster analysis of apoptosis-related genes in patients with osteoarthritis and their correlation with immune cell infiltration. Front. Immunol. 2023, 14, 1202758. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhou, H.; Hu, P.; Pan, Y.; Wang, S.; Liu, L.; Liu, X. Identification and validation of immune and oxidative stress-related diagnostic markers for diabetic nephropathy by WGCNA and machine learning. Front. Immunol. 2023, 14, 1084531. [Google Scholar] [CrossRef]
- Merlo, S.; Spampinato, S.; Canonico, P.L.; Copani, A.; Sortino, M.A. Alzheimer’s disease: Brain expression of a metabolic disorder? Trends Endocrinol. Metab. 2010, 21, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Horton, P. Next-generation bioinformatics: Connecting bases to genes, networks and disease. Brief. Bioinform. 2014, 15, 137. [Google Scholar] [CrossRef]
- Punzo, F.; Mientjes, E.J.; Rohe, C.F.; Scianguetta, S.; Amendola, G.; Oostra, B.A.; Bertoli-Avella, A.M.; Perrotta, S. A mutation in the acyl-coenzyme A binding domain-containing protein 5 gene (ACBD5) identified in autosomal dominant thrombocytopenia. J. Thromb. Haemost. 2010, 8, 2085–2087. [Google Scholar] [CrossRef]
- Granadeiro, L.; Zarralanga, V.E.; Rosa, R.; Franquinho, F.; Lamas, S.; Brites, P. Ataxia with giant axonopathy in Acbd5-deficient mice halted by adeno-associated virus gene therapy. Brain 2023, 147, 1457–1473. [Google Scholar] [CrossRef]
- Costello, J.L.; Castro, I.G.; Hacker, C.; Schrader, T.A.; Metz, J.; Zeuschner, D.; Azadi, A.S.; Godinho, L.F.; Costina, V.; Findeisen, P.; et al. ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J. Cell Biol. 2017, 216, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Hua, R.; Cheng, D.; Coyaud, É.; Freeman, S.; Di Pietro, E.; Wang, Y.; Vissa, A.; Yip, C.M.; Fairn, G.D.; Braverman, N.; et al. VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J. Cell Biol. 2017, 216, 367–377. [Google Scholar] [CrossRef]
- Kou, J.; Kovacs, G.G.; Höftberger, R.; Kulik, W.; Brodde, A.; Forss-Petter, S.; Hönigschnabl, S.; Gleiss, A.; Brügger, B.; Wanders, R.; et al. Peroxisomal alterations in Alzheimer’s disease. Acta Neuropathol. 2011, 122, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Keulers, T.G.; Libregts, S.F.; Beaumont, J.E.J.; Savelkouls, K.G.; Bussink, J.; Duimel, H.; Dubois, L.; Zonneveld, M.I.; López-Iglesias, C.; Bezstarosti, K.; et al. Secretion of pro-angiogenic extracellular vesicles during hypoxia is dependent on the autophagy-related protein GABARAPL1. J Extracell. Vesicles 2021, 10, e12166. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Wang, L.; Wang, P.; Xue, Y.; Li, X.; Qiao, X.; Zhang, X.; Xu, T.; Liu, G.; et al. Autophagy impairment mediated by S-nitrosation of ATG4B leads to neurotoxicity in response to hyperglycemia. Autophagy 2017, 13, 1145–1160. [Google Scholar] [CrossRef] [PubMed]
- Mei, T.; Li, Y.; Orduña Dolado, A.; Li, Z.; Andersson, R.; Berliocchi, L.; Rasmussen, L.J. Pooled analysis of frontal lobe transcriptomic data identifies key mitophagy gene changes in Alzheimer’s disease brain. Front. Aging Neurosci. 2023, 15, 1101216. [Google Scholar] [CrossRef]
- Vianello, C.; Salluzzo, M.; Anni, D.; Boriero, D.; Buffelli, M.; Carboni, L. Increased Expression of Autophagy-Related Genes in Alzheimer’s Disease-Type 2 Diabetes Mellitus Comorbidity Models in Cells. Int. J. Environ. Res. Public Health 2023, 20, 4540. [Google Scholar] [CrossRef] [PubMed]
- Chakrama, F.Z.; Seguin-Py, S.; Le Grand, J.N.; Fraichard, A.; Delage-Mourroux, R.; Despouy, G.; Perez, V.; Jouvenot, M.; Boyer-Guittaut, M. GABARAPL1 (GEC1) associates with autophagic vesicles. Autophagy 2010, 6, 495–505. [Google Scholar] [CrossRef]
- Nixon, R.A.; Rubinsztein, D.C. Mechanisms of autophagy-lysosome dysfunction in neurodegenerative diseases. Nat. Rev. Mol. Cell Biol. 2024, 25, 926–946. [Google Scholar] [CrossRef] [PubMed]
- Stricher, F.; Macri, C.; Ruff, M.; Muller, S. HSPA8/HSC70 chaperone protein: Structure, function, and chemical targeting. Autophagy 2013, 9, 1937–1954. [Google Scholar] [CrossRef]
- Ho, P.W.; Leung, C.T.; Liu, H.; Pang, S.Y.; Lam, C.S.; Xian, J.; Li, L.; Kung, M.H.; Ramsden, D.B.; Ho, S.L. Age-dependent accumulation of oligomeric SNCA/α-synuclein from impaired degradation in mutant LRRK2 knockin mouse model of Parkinson disease: Role for therapeutic activation of chaperone-mediated autophagy (CMA). Autophagy 2020, 16, 347–370. [Google Scholar] [CrossRef]
- Xu, X.; Sun, Y.; Cen, X.; Shan, B.; Zhao, Q.; Xie, T.; Wang, Z.; Hou, T.; Xue, Y.; Zhang, M.; et al. Metformin activates chaperone-mediated autophagy and improves disease pathologies in an Alzheimer disease mouse model. Protein Cell 2021, 12, 769–787. [Google Scholar] [CrossRef]
- Fontaine, S.N.; Zheng, D.; Sabbagh, J.J.; Martin, M.D.; Chaput, D.; Darling, A.; Trotter, J.H.; Stothert, A.R.; Nordhues, B.A.; Lussier, A.; et al. DnaJ/Hsc70 chaperone complexes control the extracellular release of neurodegenerative-associated proteins. EMBO J. 2016, 35, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Cuervo, A.M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 2015, 17, 759–770. [Google Scholar] [CrossRef]
- Lan, Z.Q.; Ge, Z.Y.; Lv, S.K.; Zhao, B.; Li, C.X. The regulatory role of lipophagy in central nervous system diseases. Cell Death Discov. 2023, 9, 229. [Google Scholar] [CrossRef] [PubMed]
- Galasko, D. Active immunization in Alzheimer’s disease. Nat. Aging 2021, 1, 493–495. [Google Scholar] [CrossRef]
- Dai, L.; Shen, Y. Insights into T-cell dysfunction in Alzheimer’s disease. Aging Cell 2021, 20, e13511. [Google Scholar] [CrossRef]
- Xu, H.; Jia, J. Single-Cell RNA Sequencing of Peripheral Blood Reveals Immune Cell Signatures in Alzheimer’s Disease. Front. Immunol. 2021, 12, 645666. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, F.C.; Cozachenco, D.; Heimfarth, L.; Fortuna, J.T.S.; de Freitas, G.B.; de Sousa, J.M.; Alves-Leon, S.V.; Leite, R.E.P.; Suemoto, C.K.; Grinberg, L.T.; et al. Synaptic proteasome is inhibited in Alzheimer’s disease models and associates with memory impairment in mice. Commun. Biol. 2023, 6, 1127. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Lipton, S.A. ‘SNO’-Storms Compromise Protein Activity and Mitochondrial Metabolism in Neurodegenerative Disorders. Trends Endocrinol. Metab. 2017, 28, 879–892. [Google Scholar] [CrossRef]
- Lu, L.; Kotowska, A.M.; Kern, S.; Fang, M.; Rudd, T.R.; Alexander, M.R.; Scurr, D.J.; Zhu, Z. Metabolomic and Proteomic Analysis of ApoE4-Carrying H4 Neuroglioma Cells in Alzheimer’s Disease Using OrbiSIMS and LC-MS/MS. Anal. Chem. 2024, 96, 11760–11770. [Google Scholar] [CrossRef]
- Rousseaux, M.W.; Revelli, J.P.; Vazquez-Velez, G.E.; Kim, J.Y.; Craigen, E.; Gonzales, K.; Beckinghausen, J.; Zoghbi, H.Y. Depleting Trim28 in adult mice is well tolerated and reduces levels of α-synuclein and tau. eLife 2018, 7, e36768. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.D.; Kim, W.K.; Huong Vu, G. Molecular mechanisms implicated in protein changes in the Alzheimer’s disease human hippocampus. Mech. Ageing Dev. 2024, 219, 111930. [Google Scholar] [CrossRef]
- Sun, J.; Li, L.; Xiong, L.; Chen, F.; She, L.; Tang, H.; Zeng, Y.; Duan, Y.; Li, L.; Wang, W.; et al. Parthenolide alleviates cognitive dysfunction and neurotoxicity via regulation of AMPK/GSK3β(Ser9)/Nrf2 signaling pathway. Biomed. Pharmacother. 2023, 169, 115909. [Google Scholar] [CrossRef]






| Term | Adjusted p-Value | Odds Ratio | Combined Score | Genes |
|---|---|---|---|---|
| 1,4-chrysenequinone | 0.004290494 | 886.7555556 | 9781.874312 | HSPA8; GABARAPL1 |
| thiostrepton | 0.014931296 | 305.6461538 | 2736.968135 | HSPA8; GABARAPL1 |
| parthenolide | 0.014931296 | 266.4161074 | 2313.934025 | HSPA8; GABARAPL1 |
| 15-delta prostaglandin J2 | 0.016810729 | 216.5464481 | 1792.825598 | HSPA8; GABARAPL1 |
| puromycin | 0.029464858 | 145.0367647 | 1087.028637 | HSPA8; GABARAPL1 |
| menadione | 0.03643546 | 118.4638554 | 841.1150706 | HSPA8; GABARAPL1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, H.; Zheng, S.; Sun, S.; Shi, X.; Wang, X.; Yang, Y.; Ma, R.; Li, G. Identification of Lipophagy-Related Gene Signature for Diagnosis and Risk Prediction of Alzheimer’s Disease. Biomedicines 2025, 13, 362. https://doi.org/10.3390/biomedicines13020362
Guo H, Zheng S, Sun S, Shi X, Wang X, Yang Y, Ma R, Li G. Identification of Lipophagy-Related Gene Signature for Diagnosis and Risk Prediction of Alzheimer’s Disease. Biomedicines. 2025; 13(2):362. https://doi.org/10.3390/biomedicines13020362
Chicago/Turabian StyleGuo, Hongxiu, Siyi Zheng, Shangqi Sun, Xueying Shi, Xiufeng Wang, Yang Yang, Rong Ma, and Gang Li. 2025. "Identification of Lipophagy-Related Gene Signature for Diagnosis and Risk Prediction of Alzheimer’s Disease" Biomedicines 13, no. 2: 362. https://doi.org/10.3390/biomedicines13020362
APA StyleGuo, H., Zheng, S., Sun, S., Shi, X., Wang, X., Yang, Y., Ma, R., & Li, G. (2025). Identification of Lipophagy-Related Gene Signature for Diagnosis and Risk Prediction of Alzheimer’s Disease. Biomedicines, 13(2), 362. https://doi.org/10.3390/biomedicines13020362





