Untargeted Metabolomic Profiling of Colonic Mucosa in Individuals with Irritable Bowel Syndrome
Abstract
1. Introduction
2. Patients and Methods
2.1. Study Design
2.2. Sample Collection
2.3. Liquid Chromatography–Mass Spectrometry Analysis
2.4. Random Forest Classification and Feature Importance Analysis
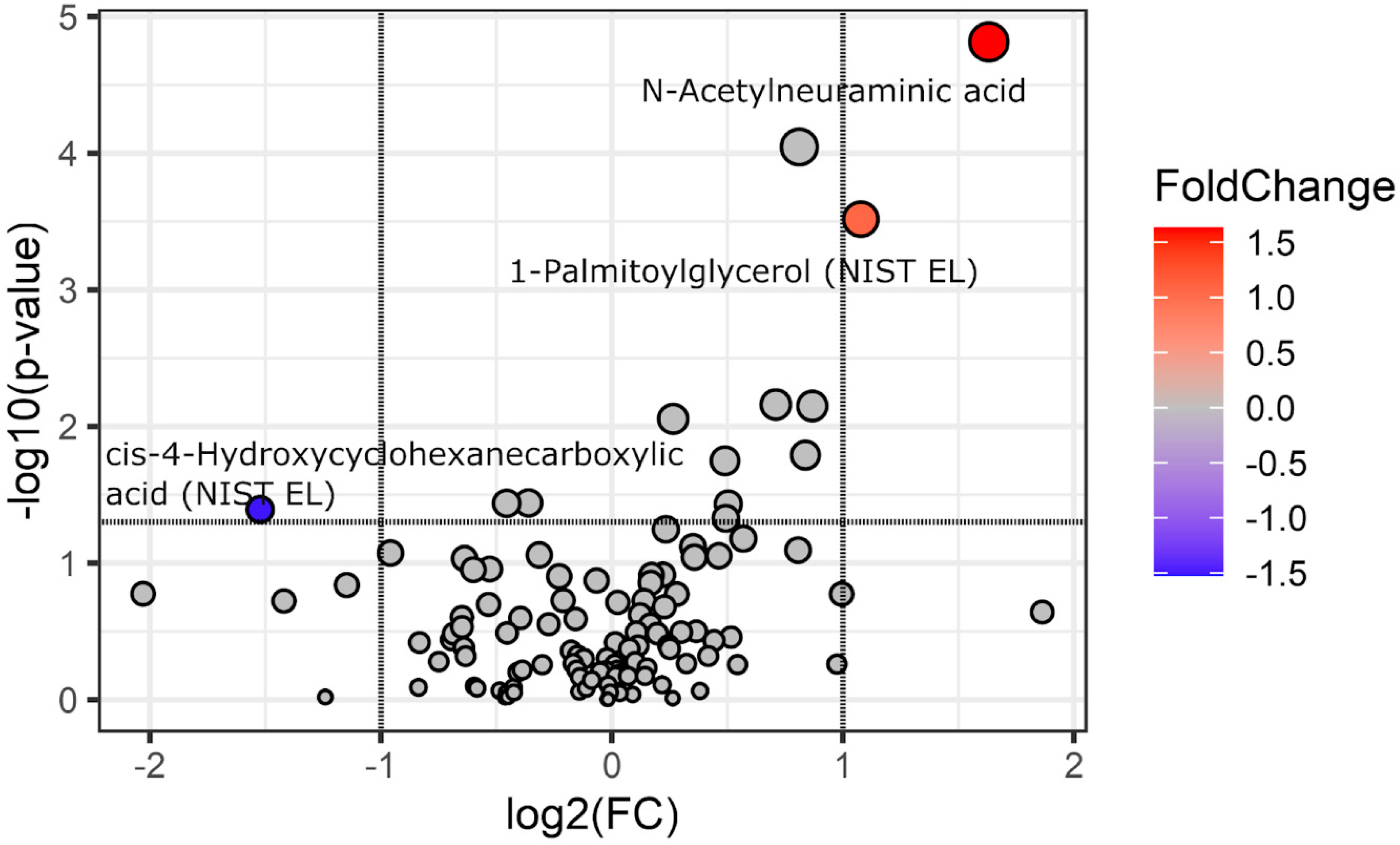
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Palsson, O.S.; Tack, J.; Drossman, D.A.; Le Nevé, B.; Quinquis, L.; Hassouna, R.; Ruddy, J.; Morris, C.B.; Sperber, A.D.; Bangdiwala, S.I.; et al. Worldwide Population Prevalence and Impact of Sub-Diagnostic Gastrointestinal Symptoms. Aliment. Pharmacol. Ther. 2024, 59, 852–864. [Google Scholar] [CrossRef]
- Jones, M.P.; Holtmann, G.J.; Tack, J.; Carbonne, F.; Chey, W.; Koloski, N.; Shah, A.; Bangdiwala, S.I.; Sperber, A.D.; Palsson, O.S.; et al. Diagnostic Classification Systems for Disorders of Gut-Brain Interaction Should Include Psychological Symptoms. Neurogastroenterol. Motil. 2024, 36, e14940. [Google Scholar] [CrossRef] [PubMed]
- Ballou, S.; Vasant, D.H.; Guadagnoli, L.; Reed, B.; Chiarioni, G.; Ten Cate, L.; Keefer, L.; Kinsinger, S.W. A Primer for the Gastroenterology Provider on Psychosocial Assessment of Patients with Disorders of Gut-Brain Interaction. Neurogastroenterol. Motil. 2024, 36, e14894. [Google Scholar] [CrossRef]
- Mulak, A.; Freud, T.; Waluga, M.; Bangdiwala, S.I.; Palsson, O.S.; Sperber, A.D. Sex- and Gender-Related Differences in the Prevalence and Burden of Disorders of Gut-Brain Interaction in Poland. Neurogastroenterol. Motil. 2023, 35, e14568. [Google Scholar] [CrossRef] [PubMed]
- Krynicka, P.; Kaczmarczyk, M.; Skonieczna-Żydecka, K.; Cembrowska-Lech, D.; Podsiadło, K.; Dąbkowski, K.; Gaweł, K.; Botke, N.; Zawada, I.; Ławniczak, M.; et al. The Burden of Irritable Bowel Syndrome and Functional Dyspepsia in Poland: A Cross-Sectional Study from West Pomeranian Voivodship. BMC Gastroenterol. 2025, 25, 8. [Google Scholar] [CrossRef]
- Palma, J.; Antoniewicz, J.; Borecki, K.; Tejchman, K.; Skonieczna-Żydecka, K.; Maciejewska-Markiewicz, D.; Ryterska, K.; Komorniak, N.; Czerwińska-Rogowska, M.; Wolska, A.; et al. Irritable Bowel Syndrome Prevalence among Participants of Woodstock Rock Festival in Poland Based on Rome IV Criteria Questionnaire. Int. J. Environ. Res. Public Health 2021, 18, 11464. [Google Scholar] [CrossRef] [PubMed]
- Sperber, A.D.; Bangdiwala, S.I.; Drossman, D.A.; Ghoshal, U.C.; Simren, M.; Tack, J.; Whitehead, W.E.; Dumitrascu, D.L.; Fang, X.; Fukudo, S.; et al. Worldwide Prevalence and Burden of Functional Gastrointestinal Disorders, Results of Rome Foundation Global Study. Gastroenterology 2021, 160, 99–114.e3. [Google Scholar] [CrossRef]
- Ionescu, V.A.; Gheorghe, G.; Georgescu, T.F.; Bacalbasa, N.; Gheorghe, F.; Diaconu, C.C. The Latest Data Concerning the Etiology and Pathogenesis of Irritable Bowel Syndrome. J. Clin. Med. 2024, 13, 5124. [Google Scholar] [CrossRef]
- Li, X.; Li, X.; Xiao, H.; Xu, J.; He, J.; Xiao, C.; Zhang, B.; Cao, M.; Hong, W. Meta-Analysis of Gut Microbiota Alterations in Patients with Irritable Bowel Syndrome. Front. Microbiol. 2024, 15, 1492349. [Google Scholar] [CrossRef]
- Barbaro, M.R.; Cremon, C.; Marasco, G.; Savarino, E.; Guglielmetti, S.; Bonomini, F.; Palombo, M.; Fuschi, D.; Rotondo, L.; Mantegazza, G.; et al. Molecular Mechanisms Underlying Loss of Vascular and Epithelial Integrity in Irritable Bowel Syndrome. Gastroenterology 2024, 167, 1152–1166. [Google Scholar] [CrossRef]
- Nazarewska, A.; Lewandowski, K.; Kaniewska, M.; Rosołowski, M.; Marlicz, W.; Rydzewska, G. Irritable Bowel Syndrome Following COVID-19: An Underestimated Consequence of SARS-CoV-2 Infection. Pol. Arch. Intern. Med. 2022, 132, 16323. [Google Scholar] [CrossRef]
- Nazarewska, A.; Lewandowski, K.; Kaniewska, M.; Tulewicz-Marti, E.; Więcek, M.; Szwarc, P.; Rosołowski, M.; Marlicz, W.; Rydzewska, G. Long-Lasting Dyspeptic Symptoms—Another Consequence of the COVID-19 Pandemic? Prz. Gastroenterol. 2023, 18, 175–182. [Google Scholar] [CrossRef]
- Fraser, K.; James, S.C.; Young, W.; Gearry, R.B.; Heenan, P.E.; Keenan, J.I.; Talley, N.J.; McNabb, W.C.; Roy, N.C. Characterisation of the Plasma and Faecal Metabolomes in Participants with Functional Gastrointestinal Disorders. Int. J. Mol. Sci. 2024, 25, 13465. [Google Scholar] [CrossRef] [PubMed]
- Kopera, K.; Gromowski, T.; Wydmański, W.; Skonieczna-Żydecka, K.; Muszyńska, A.; Zielińska, K.; Wierzbicka-Woś, A.; Kaczmarczyk, M.; Kadaj-Lipka, R.; Cembrowska-Lech, D.; et al. Gut Microbiome Dynamics and Predictive Value in Hospitalized COVID-19 Patients: A Comparative Analysis of Shallow and Deep Shotgun Sequencing. Front. Microbiol. 2024, 15, 1342749. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. 2024. Available online: https://www.R-project.org/ (accessed on 12 December 2024).
- Zhao, D.-Y.; Qi, Q.-Q.; Long, X.; Li, X.; Chen, F.-X.; Yu, Y.-B.; Zuo, X.-L. Ultrastructure of Intestinal Mucosa in Diarrhea-Predominant Irritable Bowel Syndrome. Physiol. Int. 2019, 106, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Vich Vila, A.; Zhang, J.; Liu, M.; Faber, K.N.; Weersma, R.K. Untargeted Faecal Metabolomics for the Discovery of Biomarkers and Treatment Targets for Inflammatory Bowel Diseases. Gut 2024, 73, 1909–1920. [Google Scholar] [CrossRef]
- Gao, R.; Wu, C.; Zhu, Y.; Kong, C.; Zhu, Y.; Gao, Y.; Zhang, X.; Yang, R.; Zhong, H.; Xiong, X.; et al. Integrated Analysis of Colorectal Cancer Reveals Cross-Cohort Gut Microbial Signatures and Associated Serum Metabolites. Gastroenterology 2022, 163, 1024–1037.e9. [Google Scholar] [CrossRef]
- Han, L.; Zhao, L.; Zhou, Y.; Yang, C.; Xiong, T.; Lu, L.; Deng, Y.; Luo, W.; Chen, Y.; Qiu, Q.; et al. Altered Metabolome and Microbiome Features Provide Clues in Understanding Irritable Bowel Syndrome and Depression Comorbidity. ISME J. 2022, 16, 983–996. [Google Scholar] [CrossRef]
- Lin, A.-Z.; Fu, X.; Jiang, Q.; Zhou, X.; Hwang, S.H.; Yin, H.-H.; Ni, K.-D.; Pan, Q.-J.; He, X.; Zhang, L.-T.; et al. Metabolomics Reveals Soluble Epoxide Hydrolase as a Therapeutic Target for High-Sucrose Diet-Mediated Gut Barrier Dysfunction. Proc. Natl. Acad. Sci. USA 2024, 121, e2409841121. [Google Scholar] [CrossRef]
- Calzadilla, N.; Qazi, A.; Sharma, A.; Mongan, K.; Comiskey, S.; Manne, J.; Youkhana, A.G.; Khanna, S.; Saksena, S.; Dudeja, P.K.; et al. Mucosal Metabolomic Signatures in Chronic Colitis: Novel Insights into the Pathophysiology of Inflammatory Bowel Disease. Metabolites 2023, 13, 873. [Google Scholar] [CrossRef]
- Caceres Lessa, A.Y.; Edwinson, A.; Sato, H.; Yang, L.; Berumen, A.; Breen-Lyles, M.; Byale, A.; Ryks, M.; Keehn, A.; Camilleri, M.; et al. Transcriptomic and Metabolomic Correlates of Increased Colonic Permeability in Postinfection Irritable Bowel Syndrome. Clin. Gastroenterol. Hepatol. 2024, 23, 632–643.e13. [Google Scholar] [CrossRef] [PubMed]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The Neuroactive Potential of the Human Gut Microbiota in Quality of Life and Depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, B.M.R.; Gralnek, I.M.; Bolus, R.; Chang, L.; Dulai, G.S.; Naliboff, B.; Mayer, E.A. Is a Negative Colonoscopy Associated with Reassurance or Improved Health-Related Quality of Life in Irritable Bowel Syndrome? Gastrointest. Endosc. 2005, 62, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Lin, Q.; Luo, F.; Wang, H. Insights into the Structure, Metabolism, Biological Functions and Molecular Mechanisms of Sialic Acid: A Review. Foods 2024, 13, 145. [Google Scholar] [CrossRef]
- Wang, S.; Peng, R.; Qin, S.; Liu, Y.; Yang, H.; Ma, J. Effects of Oligosaccharide-Sialic Acid (OS) Compound on Maternal-Newborn Gut Microbiome, Glucose Metabolism and Systematic Immunity in Pregnancy: Protocol for a Randomised Controlled Study. BMJ Open 2019, 9, e026583. [Google Scholar] [CrossRef]
- Liu, Q.; Yu, Z.; Tian, F.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Surface Components and Metabolites of Probiotics for Regulation of Intestinal Epithelial Barrier. Microb. Cell Factories 2020, 19, 23. [Google Scholar] [CrossRef]
- Li, W.; Tang, X.; Liu, H.; Liu, K.; Tian, Z.; Zhao, Y. Protective Effect of 1,3-Dioleoyl-2-Palmitoylglycerol against DSS-Induced Colitis via Modulating Gut Microbiota and Maintaining Intestinal Epithelial Barrier Integrity. Food Funct. 2024, 15, 8700–8711. [Google Scholar] [CrossRef]
- McGuckin, M.A.; Eri, R.; Simms, L.A.; Florin, T.H.J.; Radford-Smith, G. Intestinal Barrier Dysfunction in Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 2009, 15, 100–113. [Google Scholar] [CrossRef]
- Adams, L.; Li, X.; Burchmore, R.; Goodwin, R.J.A.; Wall, D.M. Microbiome-Derived Metabolite Effects on Intestinal Barrier Integrity and Immune Cell Response to Infection. Microbiology 2024, 170, 001504. [Google Scholar] [CrossRef]
- Choo, C.; Mahurkar-Joshi, S.; Dong, T.S.; Lenhart, A.; Lagishetty, V.; Jacobs, J.P.; Labus, J.S.; Jaffe, N.; Mayer, E.A.; Chang, L. Colonic Mucosal Microbiota Is Associated with Bowel Habit Subtype and Abdominal Pain in Patients with Irritable Bowel Syndrome. Am. J. Physiol. Gastrointest. Liver Physiol. 2022, 323, G134–G143. [Google Scholar] [CrossRef]
- Iribarren, C.; Savolainen, O.; Sapnara, M.; Törnblom, H.; Simrén, M.; Magnusson, M.K.; Öhman, L. Temporal Stability of Fecal Metabolomic Profiles in Irritable Bowel Syndrome. Neurogastroenterol. Motil. 2024, 36, e14741. [Google Scholar] [CrossRef] [PubMed]
- Kirk, D.; Louca, P.; Attaye, I.; Zhang, X.; Wong, K.E.; Michelotti, G.A.; Falchi, M.; Valdes, A.M.; Williams, F.M.K.; Menni, C. Multifluid Metabolomics Identifies Novel Biomarkers for Irritable Bowel Syndrome. Metabolites 2025, 15, 121. [Google Scholar] [CrossRef] [PubMed]
- Vakili, O.; Adibi Sedeh, P.; Pourfarzam, M. Metabolic Biomarkers in Irritable Bowel Syndrome Diagnosis. Clin. Chim. Acta 2024, 560, 119753. [Google Scholar] [CrossRef]
- Baumgartner, M.; Lang, M.; Holley, H.; Crepaz, D.; Hausmann, B.; Pjevac, P.; Moser, D.; Haller, F.; Hof, F.; Beer, A.; et al. Mucosal Biofilms Are an Endoscopic Feature of Irritable Bowel Syndrome and Ulcerative Colitis. Gastroenterology 2021, 161, 1245–1256.e20. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | IBS N = 44 1 | Other N = 69 1 | p 2 |
|---|---|---|---|
| Sex | 0.175 | ||
| Females | 30 (68%) | 38 (55%) | |
| Males | 14 (32%) | 31 (45%) | |
| Age (years) | 0.029 | ||
| Mean (SD) | 52 (16) | 59 (13) | |
| Body mass (kg) | 0.741 | ||
| Mean (SD) | 77 (18) | 77 (15) | |
| Height (cm) | 0.538 | ||
| Mean (SD) | 168 (10) | 168 (13) | |
| BMI (kg/m2) | 0.932 | ||
| Mean (SD) | 27.0 (5.4) | 27.7 (8.5) | |
| DM (Yes/No) | 7 (16%) | 9 (13%) | 0.783 |
| Hypertension (Yes/No) | 14 (32%) | 32 (46%) | 0.169 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krynicka, P.; Kaczmarczyk, M.; Skonieczna-Żydecka, K.; Styburski, D.; Podsiadło, K.; Cembrowska-Lech, D.; Dąbkowski, K.; Deskur, A.; Rogoza-Mateja, W.; Ławniczak, M.; et al. Untargeted Metabolomic Profiling of Colonic Mucosa in Individuals with Irritable Bowel Syndrome. Biomedicines 2025, 13, 629. https://doi.org/10.3390/biomedicines13030629
Krynicka P, Kaczmarczyk M, Skonieczna-Żydecka K, Styburski D, Podsiadło K, Cembrowska-Lech D, Dąbkowski K, Deskur A, Rogoza-Mateja W, Ławniczak M, et al. Untargeted Metabolomic Profiling of Colonic Mucosa in Individuals with Irritable Bowel Syndrome. Biomedicines. 2025; 13(3):629. https://doi.org/10.3390/biomedicines13030629
Chicago/Turabian StyleKrynicka, Patrycja, Mariusz Kaczmarczyk, Karolina Skonieczna-Żydecka, Daniel Styburski, Konrad Podsiadło, Danuta Cembrowska-Lech, Krzysztof Dąbkowski, Anna Deskur, Wiesława Rogoza-Mateja, Małgorzata Ławniczak, and et al. 2025. "Untargeted Metabolomic Profiling of Colonic Mucosa in Individuals with Irritable Bowel Syndrome" Biomedicines 13, no. 3: 629. https://doi.org/10.3390/biomedicines13030629
APA StyleKrynicka, P., Kaczmarczyk, M., Skonieczna-Żydecka, K., Styburski, D., Podsiadło, K., Cembrowska-Lech, D., Dąbkowski, K., Deskur, A., Rogoza-Mateja, W., Ławniczak, M., Białek, A., Koulaouzidis, A., & Marlicz, W. (2025). Untargeted Metabolomic Profiling of Colonic Mucosa in Individuals with Irritable Bowel Syndrome. Biomedicines, 13(3), 629. https://doi.org/10.3390/biomedicines13030629









