Design and Potential of Non-Integrating Lentiviral Vectors
Abstract
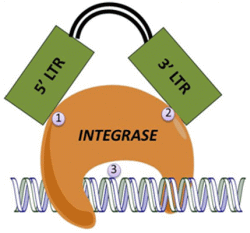
:1. Introduction
3. Prospects and Applications
| NILV Modification | Disease/Application | Envelope | Promoter | Transgene/Effector | Target | Ref. | |
|---|---|---|---|---|---|---|---|
| Vaccinations | D64V | West Nile Virus | VSV-G | CMV | West Nile Virus Envelope | Dendritic Cells | [93] |
| D64V | Malaria | VSV-G IND or NJ & Cocal Virus-G | CMV | Plasmodium yoelii Circumsporozoite Protein codon optimized | Dendritic Cells | [116] | |
| D64E | Hepatitis C Virus | HCV-E1E2-G | CMV | Hepatitis C Virus NS3 gene | Antigen Presenting Cells | [117] | |
| D116N | Human Papillomavirus | VSV-G | CMV | Human Papillomavirus 16 E7-Calreticulin fusion | Antigen Presenting Cells | [118] | |
| D64V | Thymoma | SVGmu | Ubiquitin-C | Ovalbumin, melanoma antigen hgp100 and HIV-1 subtype B gag | Dendritic Cells | [119] | |
| D64V, N120L, W235E & Δatt | Hepatitis B Virus | VSV-G | SFFV | Hepatitis B Virus surface antigen | Dendritic Cells | [120] | |
| D116N | Human Immunodeficiency Virus type 1 | VSV-G | CMV | HIV-1 JR-FL gp120 codon optimized | Antigen Presenting Cells | [114] | |
| Cell-Type Differentiation | D64V | Purification of hESC derived progenitors | VSV-G | APOA-II | Green Fluorescence Protein | Hepatic Progenitors | [121] |
| D64V | iPS Cell production | VSV-G | EF1α | OCT4, SOX2, NANOG, LIN28, n-Myc and SV40 Large T Antigen | Fibroblasts | [122] | |
| D64N & D116N | iPS Cell transgene excision | VSV-G | CMV | Cre recombinase | iPS Cells | [123] | |
| Site-Directed Integration | D64V | Retargetting HIV-1 | ampho MLV | SV40 | Integrase- E. coli LexA repressor fusion protein | E. coli LexA recognition sites | [124] |
| D64V | Directed Integration | VSV-G | SV40 | Integrase-Designed Polydactyl Zinc Finger Protein E2C fusion protein | E2C-recognition sequence | [125] | |
| D64V | Directed Integration | VSV-G | PGK | Yeast Flpx9 recombinase | Flp-recognition sites | [126] | |
| D64V | Transposon mediated random integration | VSV-G | CMV, SFFV & SV40 | Sleeping Beauty Transposase/Transposase Expression Cassette | Random Integration | [127] | |
| D64V | Homologous recombination mediated gene modification | VSV-G | N/A | Calmegin targeting cassette | Calmegin (clgn) gene | [128] | |
| K264E, F185A, D116A, D64A & H12A | Site-directed homologous recombination | VSV-G | CMV | I-SceI Nuclease/Homologous recombination repair matrix | I-SceI nuclease binding site | [84] | |
| D64V | Site-specific integration | VSV-G | PGK & SFFV | Zinc Finger Nuclease/ZFN donor template | ZFN-target site at IL2RG | [129] | |
| D64V | “Safe-site”-specific integration | VSV-G | SFFV, PGK & EF1α | Zinc Finger Nuclease/ZFN donor template-GFP expression cassette | CCR5 and AAVS1 loci | [130] | |
| D64V | Site-specific gene modification | VSV-G | EFS | Zinc Finger Nuclease/ZFN donor template | Adenosine Deaminase Locus | [131] | |
| D64V | Site-specific gene modification | VSV-G | CMV | Transcription Activator-Like Effector Nucleases/TALEN donor template | COL7A1 gene | [132] | |
| Persistent Episomal Expression | D64V, N120L, W235E, Q148A, K264R & Δatt | Stable gene transfer to muscle | VSV-G | SFFV | Green Fluorescence Protein | Muscle Tissue | [61] |
| D64E | Stable gene transfer to liver and brain | VSV-G | CMV & hAAT | Green Fluorescence Protein/Luciferase | Brain & Liver Tissue | [64] | |
| D64V | Stable gene transfer to liver | VSV-G | PGK & ET | Green Fluorescence Protein/Factor IX cDNA | Hepatocytes | [91] | |
| D64V | Stable gene transfer to liver | VSV-G | ET | Hyperfunctional coagulation factor IX | Hepatocytes | [133] | |
| D64V | Stable gene transfer to retina and brain | VSV-G | CMV & SFFV | Green Fluorescence Protein | Ocular & Brain Tissue | [89] | |
| D64E | Stable gene transfer to brain | VSV-G | CMV | Green Fluorescence Protein/Luciferase | Brain Tissue | [58] | |
| N region RRK motif to AAH | Stable gene transfer to brain | VSV-G | CMV | Green Fluorescence Protein | Neural cells | [62] | |
| D64V | Stable gene transfer to central nervous system | VSV-G, GP64 & Rabies-G | SFFV | Green Fluorescence Protein | Brain and Spinal Cord | [63] | |
| D64V | Stable gene transfer to spinal cord | VSV-G & Rabies-G | CMV | Green Fluorescence Protein | Spinal Cord | [134] |
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Cronin, J.; Zhang, X.Y.; Reiser, J. Altering the tropism of lentiviral vectors through pseudotyping. Curr. Gene Ther. 2005, 5, 387–398. [Google Scholar] [CrossRef]
- Clapham, P.R.; McKnight, A. Cell surface receptors, virus entry and tropism of primate lentiviruses. J. Gen. Virol. 2002, 83, 1809–1829. [Google Scholar]
- Kumar, M.; Keller, B.; Makalou, N.; Sutton, R.E. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 2001, 12, 1893–1905. [Google Scholar] [CrossRef]
- Sinn, P.L.; Sauter, S.L.; McCray, P.B., Jr. Gene therapy progress and prospects: Development of improved lentiviral and retroviral vectors—Design, biosafety, and production. Gene Ther. 2005, 12, 1089–1098. [Google Scholar] [CrossRef]
- Wanisch, K.; Yanez-Munoz, R.J. Integration-deficient lentiviral vectors: A slow coming of age. Mol. Ther. 2009, 17, 1316–1332. [Google Scholar] [CrossRef]
- Blomer, U.; Naldini, L.; Kafri, T.; Trono, D.; Verma, I.M.; Gage, F.H. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J. Virol. 1997, 71, 6641–6649. [Google Scholar]
- Abordo-Adesida, E.; Follenzi, A.; Barcia, C.; Sciascia, S.; Castro, M.G.; Naldini, L.; Lowenstein, P.R. Stability of lentiviral vector-mediated transgene expression in the brain in the presence of systemic antivector immune responses. Hum. Gene Ther. 2005, 16, 741–751. [Google Scholar] [CrossRef]
- Bischof, D.; Cornetta, K. Detection of replication competent retrovirus and lentivirus. In Methods in Molecular Biology: Genetic Modification of Hematopoietic Stem Cells: Methods and Protocols; Baum, C., Ed.; Humana Press, Inc.: Totowa, NJ, USA, 2008; pp. 243–263. [Google Scholar]
- Cavazzana-Calvo, M.; Payen, E.; Negre, O.; Wang, G.; Hehir, K.; Fusil, F.; Down, J.; Denaro, M.; Brady, T.; Westerman, K.; et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature 2010, 467, 318–322. [Google Scholar] [CrossRef]
- Aiuti, A.; Biasco, L.; Scaramuzza, S.; Ferrua, F.; Cicalese, M.P.; Baricordi, C.; Dionisio, F.; Calabria, A.; Giannelli, S.; Castiello, M.C.; et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science 2013, 341. [Google Scholar] [CrossRef]
- Biffi, A.; Montini, E.; Lorioli, L.; Cesani, M.; Fumagalli, F.; Plati, T.; Baldoli, C.; Martino, S.; Calabria, A.; Canale, S.; et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 2013, 341. [Google Scholar] [CrossRef]
- Cartier, N.; Hacein-Bey-Abina, S.; Bartholomae, C.C.; Veres, G.; Schmidt, M.; Kutschera, I.; Vidaud, M.; Abel, U.; Dal-Cortivo, L.; Caccavelli, L.; et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 2009, 326, 818–823. [Google Scholar] [CrossRef]
- Porter, D.L.; Levine, B.L.; Kalos, M.; Bagg, A.; June, C.H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 2011, 365, 725–733. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; von Kalle, C.; Schmidt, M.; Le Deist, F.; Wulffraat, N.; McIntyre, E.; Radford, I.; Villeval, J.L.; Fraser, C.C.; Cavazzana-Calvo, M.; et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003, 348, 255–256. [Google Scholar] [CrossRef]
- Stein, S.; Ott, M.G.; Schultze-Strasser, S.; Jauch, A.; Burwinkel, B.; Kinner, A.; Schmidt, M.; Kramer, A.; Schwable, J.; Glimm, H.; et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010, 16, 198–204. [Google Scholar] [CrossRef]
- Naldini, L.; Blomer, U.; Gallay, P.; Ory, D.; Mulligan, R.; Gage, F.H.; Verma, I.M.; Trono, D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 1996, 272, 263–267. [Google Scholar]
- Zufferey, R.; Dull, T.; Mandel, R.J.; Bukovsky, A.; Quiroz, D.; Naldini, L.; Trono, D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 1998, 72, 9873–9880. [Google Scholar]
- Zufferey, R.; Nagy, D.; Mandel, R.J.; Naldini, L.; Trono, D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997, 15, 871–875. [Google Scholar] [CrossRef]
- Dull, T.; Zufferey, R.; Kelly, M.; Mandel, R.J.; Nguyen, M.; Trono, D.; Naldini, L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998, 72, 8463–8471. [Google Scholar]
- Aiken, C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 1997, 71, 5871–5877. [Google Scholar]
- Braaten, D.; Luban, J. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 2001, 20, 1300–1309. [Google Scholar] [CrossRef]
- Franke, E.K.; Yuan, H.E.; Luban, J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 1994, 372, 359–362. [Google Scholar] [CrossRef]
- Luban, J. Human immunodeficiency virus type 1 gag protein binds to cyclophilin A and B. Cell 1993, 73, 1067–1078. [Google Scholar] [CrossRef]
- Luban, J. Absconding with the chaperone: Essential cyclophilin-Gag interaction in HIV-1 virions. Cell 1996, 87, 1157–1159. [Google Scholar] [CrossRef]
- Zhang, S.; Joseph, G.; Pollok, K.; Berthoux, L.; Sastry, L.; Luban, J.; Cornetta, K. The role of cyclophilin A and G2 cell cycle arrest in lentiviral gene transfer. Mol. Ther. 2006, 14, 546–554. [Google Scholar] [CrossRef]
- Kalpana, G.V.; Marmon, S.; Wang, W.; Crabtree, G.R.; Goff, S.P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 1994, 266, 2002–2006. [Google Scholar]
- Yung, E.; Sorin, M.; Pal, A.; Craig, E.; Morozov, A.; Delattre, O.; Kappes, J.; Ott, D.; Kalpana, G.V. Inhibition of HIV-1 virion production by a transdominant mutant of integrase interactor 1. Nat. Med. 2001, 7, 920–926. [Google Scholar] [CrossRef]
- Yung, E.; Sorin, M.; Wang, E.J.; Perumal, S.; Ott, D.; Kalpana, G.V. Specificity of interaction of INI1/hSNF5 with retroviral integrases and its functional significance. J. Virol. 2004, 78, 2222–2231. [Google Scholar] [CrossRef]
- Barry, S.C.; Harder, B.; Brzezinski, M.; Flint, L.Y.; Seppen, J.; Osborne, W.R. Lentivirus vectors encoding both central polypurine tract and posttranscriptional regulatory element provide enhanced transduction and transgene expression. Hum. Gene Ther. 2001, 12, 1103–1108. [Google Scholar] [CrossRef]
- Van Maele, B.; de Rijck, J.; de Clercq, E.; Debyser, Z. Impact of the central polypurine tract on the kinetics of human immunodeficiency virus type 1 vector transduction. J. Virol. 2003, 77, 4685–4694. [Google Scholar] [CrossRef]
- Zufferey, R.; Donello, J.E.; Trono, D.; Hope, T.J. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 1999, 73, 2886–2892. [Google Scholar]
- Dupuy, F.P.; Mouly, E.; Mesel-Lemoine, M.; Morel, C.; Abriol, J.; Cherai, M.; Baillou, C.; Negre, D.; Cosset, F.L.; Klatzmann, D.; et al. Lentiviral transduction of human hematopoietic cells by HIV-1- and SIV-based vectors containing a bicistronic cassette driven by various internal promoters. J. Gene Med. 2005, 7, 1158–1171. [Google Scholar] [CrossRef]
- Miyoshi, H.; Blomer, U.; Takahashi, M.; Gage, F.H.; Verma, I.M. Development of a self-inactivating lentivirus vector. J. Virol. 1998, 72, 8150–8157. [Google Scholar]
- Leath, A.; Cornetta, K. Developing novel lentiviral vectors into clinical products. Methods Enzymol. 2012, 507, 89–108. [Google Scholar] [CrossRef]
- Slepushkin, V.; Chang, N.; Cohen, R.; Gan, Y.; Jiang, B.; Deausen, E.; Berlinger, D.; Binder, G.; Andre, K.; Humeau, L.; et al. Large-scale purification of a lentiviral vector by size exclusion chromatography or mustang Q ion exchange capsule. Bioprocess. J. 2003, Sept./Oct., 89–95. [Google Scholar]
- Merten, O.; Charrier, S.; Laroudie, N.; Fauchille, S.; Dugue, C.; Jenny, C.; Audit, M.; Zanta-Boussif, M.; Chautard, H.; Radrizzani, M.; et al. Large scale manufacture and characterisation of a lentiviral vector produced for clinical ex vivo gene therapy application. Hum. Gene Ther. 2011, 22, 343–356. [Google Scholar] [CrossRef]
- Fassati, A.; Goff, S.P. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 2001, 75, 3626–3635. [Google Scholar] [CrossRef]
- Farnet, C.M.; Haseltine, W.A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 1991, 65, 1910–1915. [Google Scholar]
- Miller, M.D.; Farnet, C.M.; Bushman, F.D. Human immunodeficiency virus type 1 preintegration complexes: Studies of organization and composition. J. Virol. 1997, 71, 5382–5390. [Google Scholar]
- Bukrinsky, M.I.; Sharova, N.; McDonald, T.L.; Pushkarskaya, T.; Tarpley, W.G.; Stevenson, M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 1993, 90, 6125–6129. [Google Scholar] [CrossRef]
- Bukrinsky, M.I.; Sharova, N.; Dempsey, M.P.; Stanwick, T.L.; Bukrinskaya, A.G.; Haggerty, S.; Stevenson, M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. USA 1992, 89, 6580–6584. [Google Scholar] [CrossRef]
- De Rijck, J.; Vandekerckhove, L.; Christ, F.; Debyser, Z. Lentiviral nuclear import: A complex interplay between virus and host. BioEssays 2007, 29, 441–451. [Google Scholar] [CrossRef]
- Stevenson, M. Portals of entry: Uncovering HIV nuclear transport pathways. Trends Cell Biol. 1996, 6, 9–15. [Google Scholar] [CrossRef]
- Fassati, A. HIV infection of non-dividing cells: A divisive problem. Retrovirology 2006, 3, 74. [Google Scholar] [CrossRef]
- Chun, T.W.; Carruth, L.; Finzi, D.; Shen, X.; DiGiuseppe, J.A.; Taylor, H.; Hermankova, M.; Chadwick, K.; Margolick, J.; Quinn, T.C.; et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997, 387, 183–188. [Google Scholar] [CrossRef]
- Li, L.; Olvera, J.M.; Yoder, K.E.; Mitchell, R.S.; Butler, S.L.; Lieber, M.; Martin, S.L.; Bushman, F.D. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001, 20, 3272–3281. [Google Scholar] [CrossRef]
- Folger, K.R.; Wong, E.A.; Wahl, G.; Capecchi, M.R. Patterns of integration of DNA microinjected into cultured mammalian cells: Evidence for homologous recombination between injected plasmid DNA molecules. Mol. Cell. Biol. 1982, 2, 1372–1387. [Google Scholar]
- Jeanson, L.; Subra, F.; Vaganay, S.; Hervy, M.; Marangoni, E.; Bourhis, J.; Mouscadet, J.F. Effect of Ku80 depletion on the preintegrative steps of HIV-1 replication in human cells. Virology 2002, 300, 100–108. [Google Scholar] [CrossRef]
- Kilzer, J.M.; Stracker, T.; Beitzel, B.; Meek, K.; Weitzman, M.; Bushman, F.D. Roles of host cell factors in circularization of retroviral dna. Virology 2003, 314, 460–467. [Google Scholar] [CrossRef]
- Farnet, C.M.; Haseltine, W.A. Circularization of human immunodeficiency virus type 1 DNA in vitro. J. Virol. 1991, 65, 6942–6952. [Google Scholar]
- Gianni, A.M.; Smotkin, D.; Weinberg, R.A. Murine leukemia virus: Detection of unintegrated double-stranded DNA forms of the provirus. Proc. Natl. Acad. Sci. USA 1975, 72, 447–451. [Google Scholar] [CrossRef]
- Jacque, J.M.; Stevenson, M. The inner-nuclear-envelope protein emerin regulates HIV-1 infectivity. Nature 2006, 441, 641–645. [Google Scholar] [CrossRef]
- Shank, P.R.; Hughes, S.H.; Kung, H.J.; Majors, J.E.; Quintrell, N.; Guntaka, R.V.; Bishop, J.M.; Varmus, H.E. Mapping unintegrated avian sarcoma virus DNA: Termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell 1978, 15, 1383–1395. [Google Scholar] [CrossRef]
- Gilboa, E.; Mitra, S.W.; Goff, S.; Baltimore, D. A detailed model of reverse transcription and tests of crucial aspects. Cell 1979, 18, 93–100. [Google Scholar] [CrossRef]
- Klarmann, G.J.; Yu, H.; Chen, X.; Dougherty, J.P.; Preston, B.D. Discontinuous plus-strand DNA synthesis in human immunodeficiency virus type 1-infected cells and in a partially reconstituted cell-free system. J. Virol. 1997, 71, 9259–9269. [Google Scholar]
- Dina, D.; Benz, E.W., Jr. Structure of murine sarcoma virus DNA replicative intermediates synthesized in vitro. J. Virol. 1980, 33, 377–389. [Google Scholar]
- Junghans, R.P.; Boone, L.R.; Skalka, A.M. Products of reverse transcription in avian retrovirus analyzed by electron microscopy. J. Virol. 1982, 43, 544–554. [Google Scholar]
- Kantor, B.; Bayer, M.; Ma, H.; Samulski, J.; Li, C.; McCown, T.; Kafri, T. Notable reduction in illegitimate integration mediated by a PPT-deleted, nonintegrating lentiviral vector. Mol. Ther. 2011, 19, 547–556. [Google Scholar] [CrossRef]
- Ringold, G.M.; Yamamoto, K.R.; Shank, P.R.; Varmus, H.E. Mouse mammary tumor virus DNA in infected rat cells: Characterization of unintegrated forms. Cell 1977, 10, 19–26. [Google Scholar] [CrossRef]
- Shoemaker, C.; Goff, S.; Gilboa, E.; Paskind, M.; Mitra, S.W.; Baltimore, D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: Implications for retrovirus integration. Proc. Natl. Acad. Sci. USA 1980, 77, 3932–3936. [Google Scholar] [CrossRef]
- Apolonia, L.; Waddington, S.N.; Fernandes, C.; Ward, N.J.; Bouma, G.; Blundell, M.P.; Thrasher, A.J.; Collins, M.K.; Philpott, N.J. Stable gene transfer to muscle using non-integrating lentiviral vectors. Mol. Ther. 2007, 15, 1947–1954. [Google Scholar] [CrossRef]
- Philippe, S.; Sarkis, C.; Barkats, M.; Mammeri, H.; Ladroue, C.; Petit, C.; Mallet, J.; Serguera, C. Lentiviral vectors with a defective integrase allow efficient and sustained transgene expression in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 17684–17689. [Google Scholar] [CrossRef]
- Rahim, A.A.; Wong, A.M.; Howe, S.J.; Buckley, S.M.; Acosta-Saltos, A.D.; Elston, K.E.; Ward, N.J.; Philpott, N.J.; Cooper, J.D.; Anderson, P.N.; et al. Efficient gene delivery to the adult and fetal CNS using pseudotyped non-integrating lentiviral vectors. Gene Ther. 2009, 16, 509–520. [Google Scholar]
- Bayer, M.; Kantor, B.; Cockrell, A.; Ma, H.; Zeithaml, B.; Li, X.; McCown, T.; Kafri, T. A large U3 deletion causes increased in vivo expression from a nonintegrating lentiviral vector. Mol. Ther. 2008, 16, 1968–1976. [Google Scholar] [CrossRef]
- Zhou, H.; Rainey, G.J.; Wong, S.K.; Coffin, J.M. Substrate sequence selection by retroviral integrase. J. Virol. 2001, 75, 1359–1370. [Google Scholar] [CrossRef]
- Hindmarsh, P.; Leis, J. Retroviral DNA integration. Microbiol. Mol. Biol. Rev. 1999, 63, 836–843. [Google Scholar]
- Kulkosky, J.; Skalka, A.M. Molecular mechanism of retroviral DNA integration. Pharmacol. Ther. 1994, 61, 185–203. [Google Scholar] [CrossRef]
- Masuda, T.; Kuroda, M.J.; Harada, S. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J. Virol. 1998, 72, 8396–8402. [Google Scholar]
- Craigie, R.; Fujiwara, T.; Bushman, F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 1990, 62, 829–837. [Google Scholar] [CrossRef]
- Katz, R.A.; Merkel, G.; Kulkosky, J.; Leis, J.; Skalka, A.M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell 1990, 63, 87–95. [Google Scholar] [CrossRef]
- Mizuuchi, K. Polynucleotidyl transfer reactions in transpositional DNA recombination. J. Biol. Chem. 1992, 267, 21273–21276. [Google Scholar]
- Zhu, K.; Dobard, C.; Chow, S.A. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J. Virol. 2004, 78, 5045–5055. [Google Scholar] [CrossRef]
- Gallay, P.; Hope, T.; Chin, D.; Trono, D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 1997, 94, 9825–9830. [Google Scholar] [CrossRef]
- Engelman, A. In vivo analysis of retroviral integrase structure and function. Adv. Virus Res. 1999, 52, 411–426. [Google Scholar] [CrossRef]
- Saenz, D.T.; Loewen, N.; Peretz, M.; Whitwam, T.; Barraza, R.; Howell, K.G.; Holmes, J.M.; Good, M.; Poeschla, E.M. Unintegrated lentivirus DNA persistence and accessibility to expression in nondividing cells: Analysis with class I integrase mutants. J. Virol. 2004, 78, 2906–2920. [Google Scholar] [CrossRef]
- Wiskerchen, M.; Muesing, M.A. Human immunodeficiency virus type 1 integrase: Effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 1995, 69, 376–386. [Google Scholar]
- Banasik, M.B.; McCray, P.B., Jr. Integrase-defective lentiviral vectors: Progress and applications. Gene Ther. 2010, 17, 150–157. [Google Scholar] [CrossRef]
- Kulkosky, J.; Jones, K.S.; Katz, R.A.; Mack, J.P.; Skalka, A.M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 1992, 12, 2331–2338. [Google Scholar]
- Shibagaki, Y.; Chow, S.A. Central core domain of retroviral integrase is responsible for target site selection. J. Biol. Chem. 1997, 272, 8361–8369. [Google Scholar] [CrossRef]
- Nightingale, S.J.; Hollis, R.P.; Pepper, K.A.; Petersen, D.; Yu, X.J.; Yang, C.; Bahner, I.; Kohn, D.B. Transient gene expression by nonintegrating lentiviral vectors. Mol. Ther. 2006, 13, 1121–1132. [Google Scholar] [CrossRef]
- Leavitt, A.D.; Robles, G.; Alesandro, N.; Varmus, H.E. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J. Virol. 1996, 70, 721–728. [Google Scholar]
- Brown, H.E.; Chen, H.; Engelman, A. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: Effects on integration and cDNA synthesis. J. Virol. 1999, 73, 9011–9020. [Google Scholar]
- Masuda, T.; Planelles, V.; Krogstad, P.; Chen, I.S. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: Unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 1995, 69, 6687–6696. [Google Scholar]
- Cornu, T.I.; Cathomen, T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol. Ther. 2007, 15, 2107–2113. [Google Scholar] [CrossRef]
- Gaur, M.; Leavitt, A.D. Mutations in the human immunodeficiency virus type 1 integrase D,D(35)E motif do not eliminate provirus formation. J. Virol. 1998, 72, 4678–4685. [Google Scholar]
- Leavitt, A.D.; Shiue, L.; Varmus, H.E. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J. Biol. Chem. 1993, 268, 2113–2119. [Google Scholar]
- Nakajima, N.; Lu, R.; Engelman, A. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated dna recombination: Definition of permissive and nonpermissive T-cell lines. J. Virol. 2001, 75, 7944–7955. [Google Scholar] [CrossRef]
- Matrai, J.; Chuah, M.K.; VandenDriessche, T. Recent advances in lentiviral vector development and applications. Mol. Ther. 2010, 18, 477–490. [Google Scholar] [CrossRef]
- Yanez-Munoz, R.J.; Balaggan, K.S.; MacNeil, A.; Howe, S.J.; Schmidt, M.; Smith, A.J.; Buch, P.; MacLaren, R.E.; Anderson, P.N.; Barker, S.E.; et al. Effective gene therapy with nonintegrating lentiviral vectors. Nat. Med. 2006, 12, 348–353. [Google Scholar]
- Koyama, T.; Sun, B.; Tokunaga, K.; Tatsumi, M.; Ishizaka, Y. DNA damage enhances integration of HIV-1 into macrophages by overcoming integrase inhibition. Retrovirology 2013, 10, 21. [Google Scholar] [CrossRef]
- Matrai, J.; Cantore, A.; Bartholomae, C.C.; Annoni, A.; Wang, W.; Acosta-Sanchez, A.; Samara-Kuko, E.; De Waele, L.; Ma, L.; Genovese, P.; et al. Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology 2011, 53, 1696–1707. [Google Scholar] [CrossRef]
- Miller, D.G.; Petek, L.M.; Russell, D.W. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 2004, 36, 767–773. [Google Scholar] [CrossRef]
- Coutant, F.; Frenkiel, M.P.; Despres, P.; Charneau, P. Protective antiviral immunity conferred by a nonintegrative lentiviral vector-based vaccine. PLoS One 2008, 3, e3973. [Google Scholar] [CrossRef]
- Vargas, J., Jr.; Gusella, G.L.; Najfeld, V.; Klotman, M.E.; Cara, A. Novel integrase-defective lentiviral episomal vectors for gene transfer. Hum. Gene Ther. 2004, 15, 361–372. [Google Scholar] [CrossRef]
- Vargas, J., Jr.; Klotman, M.E.; Cara, A. Conditionally replicating lentiviral-hybrid episomal vectors for suicide gene therapy. Antivir. Res. 2008, 80, 288–294. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; Garrigue, A.; Wang, G.P.; Soulier, J.; Lim, A.; Morillon, E.; Clappier, E.; Caccavelli, L.; Delabesse, E.; Beldjord, K.; et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Investig. 2008, 118, 3132–3142. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; Von Kalle, C.; Schmidt, M.; McCormack, M.P.; Wulffraat, N.; Leboulch, P.; Lim, A.; Osborne, C.S.; Pawliuk, R.; Morillon, E.; et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003, 302, 415–419. [Google Scholar] [CrossRef]
- Howe, S.J.; Mansour, M.R.; Schwarzwaelder, K.; Bartholomae, C.; Hubank, M.; Kempski, H.; Brugman, M.H.; Pike-Overzet, K.; Chatters, S.J.; de Ridder, D.; et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Investig. 2008, 118, 3143–3150. [Google Scholar] [CrossRef]
- Boztug, K.; Schmidt, M.; Schwarzer, A.; Banerjee, P.P.; Diez, I.A.; Dewey, R.A.; Bohm, M.; Nowrouzi, A.; Ball, C.R.; Glimm, H.; et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N. Engl. J. Med. 2010, 363, 1918–1927. [Google Scholar] [CrossRef]
- Persons, D.A.; Baum, C. Solving the problem of gamma-retroviral vectors containing long terminal repeats. Mol. Ther. 2011, 19, 229–231. [Google Scholar] [CrossRef]
- Montini, E.; Cesana, D.; Schmidt, M.; Sanvito, F.; Ponzoni, M.; Bartholomae, C.; Sergi Sergi, L.; Benedicenti, F.; Ambrosi, A.; Di Serio, C.; et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 2006, 24, 687–696. [Google Scholar] [CrossRef]
- Ramezani, A.; Hawley, T.S.; Hawley, R.G. Combinatorial incorporation of enhancer-blocking components of the chicken beta-globin 5'HS4 and human T-cell receptor alpha/delta BEAD-1 insulators in self-inactivating retroviral vectors reduces their genotoxic potential. Stem Cells 2008, 26, 3257–3266. [Google Scholar] [CrossRef]
- Ryu, B.Y.; Evans-Galea, M.V.; Gray, J.T.; Bodine, D.M.; Persons, D.A.; Nienhuis, A.W. An experimental system for the evaluation of retroviral vector design to diminish the risk for proto-oncogene activation. Blood 2008, 111, 1866–1875. [Google Scholar] [CrossRef]
- Zychlinski, D.; Schambach, A.; Modlich, U.; Maetzig, T.; Meyer, J.; Grassman, E.; Mishra, A.; Baum, C. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol. Ther. 2008, 16, 718–725. [Google Scholar] [CrossRef]
- Zhou, S.; Mody, D.; DeRavin, S.S.; Hauer, J.; Lu, T.; Ma, Z.; Hacein-Bey Abina, S.; Gray, J.T.; Greene, M.R.; Cavazzana-Calvo, M.; et al. A self-inactivating lentiviral vector for SCID-X1 gene therapy that does not activate LMO2 expression in human T cells. Blood 2010, 116, 900–908. [Google Scholar] [CrossRef]
- Schambach, A.; Zychlinski, D.; Ehrnstroem, B.; Baum, C. Biosafety features of lentiviral vectors. Hum. Gene Ther. 2013, 24, 132–142. [Google Scholar] [CrossRef]
- Berger, G.; Turpin, J.; Cordeil, S.; Tartour, K.; Nguyen, X.N.; Mahieux, R.; Cimarelli, A. Functional analysis of the relationship between Vpx and the restriction factor SAMHD1. J. Biol. Chem. 2012, 287, 41210–41217. [Google Scholar] [CrossRef]
- Goldstone, D.C.; Ennis-Adeniran, V.; Hedden, J.J.; Groom, H.C.; Rice, G.I.; Christodoulou, E.; Walker, P.A.; Kelly, G.; Haire, L.F.; Yap, M.W.; et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 2011, 480, 379–382. [Google Scholar] [CrossRef]
- Hrecka, K.; Hao, C.; Gierszewska, M.; Swanson, S.K.; Kesik-Brodacka, M.; Srivastava, S.; Florens, L.; Washburn, M.P.; Skowronski, J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 2011, 474, 658–661. [Google Scholar] [CrossRef]
- Laguette, N.; Sobhian, B.; Casartelli, N.; Ringeard, M.; Chable-Bessia, C.; Segeral, E.; Yatim, A.; Emiliani, S.; Schwartz, O.; Benkirane, M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 2011, 474, 654–657. [Google Scholar] [CrossRef]
- Berger, G.; Goujon, C.; Darlix, J.L.; Cimarelli, A. SIVMAC Vpx improves the transduction of dendritic cells with nonintegrative HIV-1-derived vectors. Gene Ther. 2009, 16, 159–163. [Google Scholar] [CrossRef]
- Negri, D.R.; Rossi, A.; Blasi, M.; Michelini, Z.; Leone, P.; Chiantore, M.V.; Baroncelli, S.; Perretta, G.; Cimarelli, A.; Klotman, M.E.; et al. Simian immunodeficiency virus-Vpx for improving integrase defective lentiviral vector-based vaccines. Retrovirology 2012, 9, 69. [Google Scholar] [CrossRef]
- Suwanmanee, T.; Hu, G.; Gui, T.; Bartholomae, C.C.; Kutschera, I.; von Kalle, C.; Schmidt, M.; Monahan, P.E.; Kafri, T. Integration-deficient lentiviral vectors expressing codon-optimized R338L human FIX restore normal hemostasis in hemophilia B mice. Mol. Ther. 2013. [Google Scholar] [CrossRef]
- Negri, D.R.; Michelini, Z.; Baroncelli, S.; Spada, M.; Vendetti, S.; Buffa, V.; Bona, R.; Leone, P.; Klotman, M.E.; Cara, A. Successful immunization with a single injection of non-integrating lentiviral vector. Mol. Ther. 2007, 15, 1716–1723. [Google Scholar] [CrossRef]
- Pelascini, L.P.; Janssen, J.M.; Goncalves, M.A. Histone deacetylase inhibition activates transgene expression from integration-defective lentiviral vectors in dividing and non-dividing cells. Hum. Gene Ther. 2013, 24, 78–96. [Google Scholar] [CrossRef]
- Coutant, F.; Sanchez David, R.Y.; Felix, T.; Boulay, A.; Caleechurn, L.; Souque, P.; Thouvenot, C.; Bourgouin, C.; Beignon, A.S.; Charneau, P. A nonintegrative lentiviral vector-based vaccine provides long-term sterile protection against malaria. PLoS One 2012, 7, e48644. [Google Scholar] [CrossRef]
- Deng, Y.; Guan, J.; Wen, B.; Zhu, N.; Chen, H.; Song, J.; Yang, Y.; Wang, Y.; Tan, W. Induction of broadly neutralising HCV antibodies in mice by integration-deficient lentiviral vector-based pseudotyped particles. PLoS One 2013, 8, e62684. [Google Scholar]
- Grasso, F.; Negri, D.R.; Mochi, S.; Rossi, A.; Cesolini, A.; Giovannelli, A.; Chiantore, M.V.; Leone, P.; Giorgi, C.; Cara, A. Successful therapeutic vaccination with integrase defective lentiviral vector expressing nononcogenic human papillomavirus E7 protein. Int. J. Cancer 2012, 132, 335–344. [Google Scholar]
- Hu, B.; Dai, B.; Wang, P. Vaccines delivered by integration-deficient lentiviral vectors targeting dendritic cells induces strong antigen-specific immunity. Vaccine 2010, 28, 6675–6683. [Google Scholar] [CrossRef]
- Karwacz, K.; Mukherjee, S.; Apolonia, L.; Blundell, M.P.; Bouma, G.; Escors, D.; Collins, M.K.; Thrasher, A.J. Nonintegrating lentivector vaccines stimulate prolonged T-cell and antibody responses and are effective in tumor therapy. J. Virol. 2009, 83, 3094–3103. [Google Scholar] [CrossRef]
- Yang, G.; Si-Tayeb, K.; Corbineau, S.; Vernet, R.; Gayon, R.; Dianat, N.; Martinet, C.; Clay, D.; Goulinet-Mainot, S.; Tachdjian, G.; et al. Integration-deficient lentivectors: An effective strategy to purify and differentiate human embryonic stem cell-derived hepatic progenitors. BMC Biol. 2013, 11, 86. [Google Scholar] [CrossRef]
- Mali, P.; Ye, Z.; Hommond, H.H.; Yu, X.; Lin, J.; Chen, G.; Zou, J.; Cheng, L. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells 2008, 26, 1998–2005. [Google Scholar] [CrossRef]
- Papapetrou, E.P.; Sadelain, M. Generation of transgene-free human induced pluripotent stem cells with an excisable single polycistronic vector. Nat. Protocols 2011, 6, 1251–1273. [Google Scholar] [CrossRef]
- Holmes-Son, M.L.; Chow, S.A. Correct integration mediated by integrase-LexA fusion proteins incorporated into HIV-1. Mol. Ther. 2002, 5, 360–370. [Google Scholar] [CrossRef]
- Tan, W.; Dong, Z.; Wilkinson, T.A.; Barbas, C.F., 3rd; Chow, S.A. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J. Virol. 2006, 80, 1939–1948. [Google Scholar] [CrossRef]
- Moldt, B.; Staunstrup, N.H.; Jakobsen, M.; Yanez-Munoz, R.J.; Mikkelsen, J.G. Genomic insertion of lentiviral DNA circles directed by the yeast Flp recombinase. BMC Biotechnol. 2008, 8, 60. [Google Scholar] [CrossRef]
- Vink, C.A.; Gaspar, H.B.; Gabriel, R.; Schmidt, M.; McIvor, R.S.; Thrasher, A.J.; Qasim, W. Sleeping beauty transposition from nonintegrating lentivirus. Mol. Ther. 2009, 17, 1197–1204. [Google Scholar] [CrossRef]
- Okada, Y.; Ueshin, Y.; Hasuwa, H.; Takumi, K.; Okabe, M.; Ikawa, M. Targeted gene modification in mouse ES cells using integrase-defective lentiviral vectors. Genesis 2009, 47, 217–223. [Google Scholar] [CrossRef]
- Lombardo, A.; Genovese, P.; Beausejour, C.M.; Colleoni, S.; Lee, Y.L.; Kim, K.A.; Ando, D.; Urnov, F.D.; Galli, C.; Gregory, P.D.; et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007, 25, 1298–1306. [Google Scholar] [CrossRef]
- Lombardo, A.; Cesana, D.; Genovese, P.; di Stefano, B.; Provasi, E.; Colombo, D.F.; Neri, M.; Magnani, Z.; Cantore, A.; Lo Riso, P.; et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods 2011, 8, 861–869. [Google Scholar] [CrossRef]
- Joglekar, A.V.; Hollis, R.P.; Kuftinec, G.; Senadheera, S.; Chan, R.; Kohn, D.B. Integrase-defective lentiviral vectors as a delivery platform for targeted modification of adenosine deaminase locus. Mol. Ther. 2013, 21, 1705–1717. [Google Scholar] [CrossRef]
- Osborn, M.J.; Starker, C.G.; McElroy, A.N.; Webber, B.R.; Riddle, M.J.; Xia, L.; DeFeo, A.P.; Gabriel, R.; Schmidt, M.; von Kalle, C.; et al. TALEN-based gene correction for epidermolysis bullosa. Mol. Ther. 2013, 21, 1151–1159. [Google Scholar] [CrossRef]
- Cantore, A.; Nair, N.; Della Valle, P.; di Matteo, M.; Matrai, J.; Sanvito, F.; Brombin, C.; di Serio, C.; D’Angelo, A.; Chuah, M.; et al. Hyperfunctional coagulation factor IX improves the efficacy of gene therapy in hemophilic mice. Blood 2012, 120, 4517–4520. [Google Scholar] [CrossRef]
- Peluffo, H.; Foster, E.; Ahmed, S.G.; Lago, N.; Hutson, T.H.; Moon, L.; Yip, P.; Wanisch, K.; Caraballo-Miralles, V.; Olmos, G.; et al. Efficient gene expression from integration-deficient lentiviral vectors in the spinal cord. Gene Ther. 2013, 20, 645–657. [Google Scholar] [CrossRef]
- Michelini, Z.; Negri, D.R.; Baroncelli, S.; Spada, M.; Leone, P.; Bona, R.; Klotman, M.E.; Cara, A. Development and use of SIV-based Integrase defective lentiviral vector for immunization. Vaccine 2009, 27, 4622–4629. [Google Scholar] [CrossRef]
- Daenthanasanmak, A.; Salguero, G.; Borchers, S.; Figueiredo, C.; Jacobs, R.; Sundarasetty, B.S.; Schneider, A.; Schambach, A.; Eiz-Vesper, B.; Blasczyk, R.; et al. Integrase-defective lentiviral vectors encoding cytokines induce differentiation of human dendritic cells and stimulate multivalent immune responses in vitro and in vivo. Vaccine 2012, 30, 5118–5131. [Google Scholar] [CrossRef]
- Papapetrou, E.P.; Tomishima, M.J.; Chambers, S.M.; Mica, Y.; Reed, E.; Menon, J.; Tabar, V.; Mo, Q.; Studer, L.; Sadelain, M. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 12759–12764. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [Green Version]
- Wernig, M.; Meissner, A.; Foreman, R.; Brambrink, T.; Ku, M.; Hochedlinger, K.; Bernstein, B.E.; Jaenisch, R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007, 448, 318–324. [Google Scholar] [CrossRef]
- Okita, K.; Ichisaka, T.; Yamanaka, S. Generation of germline-competent induced pluripotent stem cells. Nature 2007, 448, 313–317. [Google Scholar] [CrossRef]
- Soldner, F.; Hockemeyer, D.; Beard, C.; Gao, Q.; Bell, G.W.; Cook, E.G.; Hargus, G.; Blak, A.; Cooper, O.; Mitalipova, M.; et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 2009, 136, 964–977. [Google Scholar] [CrossRef]
- Stadtfeld, M.; Nagaya, M.; Utikal, J.; Weir, G.; Hochedlinger, K. Induced pluripotent stem cells generated without viral integration. Science 2008, 322, 945–949. [Google Scholar] [CrossRef]
- Yu, J.; Hu, K.; Smuga-Otto, K.; Tian, S.; Stewart, R.; Sukvin, II; Thomson, J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science 2009, 324, 797–801. [Google Scholar] [CrossRef]
- Goulaouic, H.; Chow, S.A. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J. Virol. 1996, 70, 37–46. [Google Scholar]
- Holmes-Son, M.L.; Chow, S.A. Integrase-lexA fusion proteins incorporated into human immunodeficiency virus type 1 that contains a catalytically inactive integrase gene are functional to mediate integration. J. Virol. 2000, 74, 11548–11556. [Google Scholar] [CrossRef]
- Moldt, B.; Miskey, C.; Staunstrup, N.H.; Gogol-Doring, A.; Bak, R.O.; Sharma, N.; Mates, L.; Izsvak, Z.; Chen, W.; Ivics, Z.; et al. Comparative genomic integration profiling of Sleeping Beauty transposons mobilized with high efficacy from integrase-defective lentiviral vectors in primary human cells. Mol. Ther. 2011, 19, 1499–1510. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Ventura, A.; Jacks, T. MicroRNAs and cancer: Short RNAs go a long way. Cell 2009, 136, 586–591. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Trang, P.; Wiggins, J.F.; Patrawala, L.; Cheng, A.; Ford, L.; Weidhaas, J.B.; Brown, D.; Bader, A.G.; Slack, F.J. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 2008, 7, 759–764. [Google Scholar] [CrossRef]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar]
- Hsu, S.H.; Wang, B.; Kota, J.; Yu, J.; Costinean, S.; Kutay, H.; Yu, L.; Bai, S.; La Perle, K.; Chivukula, R.R.; et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Investig. 2012, 122, 2871–2883. [Google Scholar] [CrossRef]
- Kota, J.; Chivukula, R.R.; O’Donnell, K.A.; Wentzel, E.A.; Montgomery, C.L.; Hwang, H.W.; Chang, T.C.; Vivekanandan, P.; Torbenson, M.; Clark, K.R.; et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009, 137, 1005–1017. [Google Scholar] [CrossRef]
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Shaw, A.; Cornetta, K. Design and Potential of Non-Integrating Lentiviral Vectors. Biomedicines 2014, 2, 14-35. https://doi.org/10.3390/biomedicines2010014
Shaw A, Cornetta K. Design and Potential of Non-Integrating Lentiviral Vectors. Biomedicines. 2014; 2(1):14-35. https://doi.org/10.3390/biomedicines2010014
Chicago/Turabian StyleShaw, Aaron, and Kenneth Cornetta. 2014. "Design and Potential of Non-Integrating Lentiviral Vectors" Biomedicines 2, no. 1: 14-35. https://doi.org/10.3390/biomedicines2010014
APA StyleShaw, A., & Cornetta, K. (2014). Design and Potential of Non-Integrating Lentiviral Vectors. Biomedicines, 2(1), 14-35. https://doi.org/10.3390/biomedicines2010014