Aptamers and Their Significant Role in Cancer Therapy and Diagnosis
Abstract
:1. Introduction
2. Aptamers (Aptus + Meros)
- Stability over higher temperatures: The oligonucleotides are thermally stable when compared to protein antibodies, where as aptamers do not lose their tertiary structure over many cycles of amplification at higher temperatures, and, thus, establishing a benefit for us to use aptamers in the different screening process [28,29].
- Synthesis: Aptamers are synthesized chemically under controlled laboratory conditions, which will be useful for increased production and to avoid contamination by virus or bacteria. However, antibodies that are produced in biological conditions are mostly susceptible to viral or bacterial contamination affecting its quality [30].
- Modification: Compared to antibodies, aptamers can be easily altered chemically, especially with signaling molecules, such as probes, nanoparticles, and fluorophores, helping in the construction of signaling based biosensors [31].
3. SELEX

3.1. Target Molecules for SELEX
3.2. Oligonucleotide Library for SELEX
3.3. Selection, Amplification and Conditioning of Aptamers
3.4. Variations in SELEX Procedure
3.4.1. Atomic Force Microscopy (AFM)—SELEX
3.4.2. Automated SELEX
3.4.3. Cell SELEX
3.4.4. Capillary Electrophoresis (CE)—SELEX
3.4.5. Non-Equilibrium Capillary Electrophoresis of Equilibrium Mixtures (NECEEM)
3.4.6. FluMag—SELEX
3.4.7. Slow Off-Rate Modified Aptamer (SOMAmer)
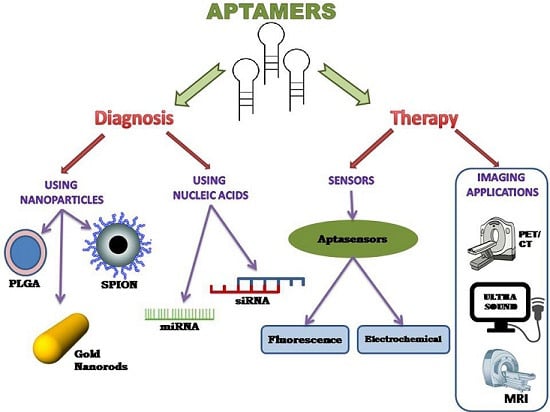
4. Aptamer-Based Therapy
4.1. Aptamer with Nanoparticles
4.2. Aptamer with Nucleic Acids
4.2.1. Aptamer si-RNA Therapy
4.2.2. Aptamer mi-RNA Therapy
5. Aptamer-Based Diagnosis
5.1. Sensors Using Aptamers
5.1.1. Electrochemical Aptasensors
5.1.2. Fluorescence Aptasensors
5.2. Imaging Using Aptamers
5.2.1. PET/CT Imaging
5.2.2. Ultrasound Imaging
5.2.3. Magnetic Resonance Imaging


6. Conclusions
7. Future Perspective
Acknowledgments
Conflicts of Interest
References
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2012. CA Cancer J. Clin. 2012, 62, 10–29. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-F.; Chang, H.-T.; Tan, W. Cancer cell targeting using multiple aptamers conjugated on nanorods. Anal. Chem. 2008, 80, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Murray, T.; Ward, E.; Samuels, A.; Tiwari, R.C.; Ghafoor, A.; Feuer, E.J.; Thun, M.J. Cancer statistics, 2005. CA Cancer J. Clin. 2005, 55, 10–30. [Google Scholar] [CrossRef] [PubMed]
- Basil, C.F.; Zhao, Y.; Zavaglia, K.; Jin, P.; Panelli, M.C.; Voiculescu, S.; Mandruzzato, S.; Lee, H.M.; Seliger, B.; Freedman, R.S.; et al. Common cancer biomarkers. Cancer Res. 2006, 66, 2953–2961. [Google Scholar] [CrossRef] [PubMed]
- Bast, R.C.; Lilja, H.; Urban, N.; Rimm, D.L.; Fritsche, H.; Gray, J.; Veltri, R.; Klee, G.; Allen, A.; Kim, N.; et al. Translational crossroads for biomarkers. Clin. Cancer Res. 2005, 11, 6103–6108. [Google Scholar] [CrossRef]
- Cho, K.; Wang, X.; Nie, S.; Chen, Z.; Shin, D.M. Therapeutic nanoparticles for drug delivery in cancer. Clin. Cancer Res. 2008, 14, 1310–1316. [Google Scholar] [CrossRef] [PubMed]
- Shapira, A.; Livney, Y.D.; Broxterman, H.J.; Assaraf, Y.G. Nanomedicine for targeted cancer therapy: Towards the overcoming of drug resistance. Drug Resist. Updat. 2011, 14, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Q.; Xu, X.; Bertrand, N.; Pridgen, E.; Swami, A.; Farokhzad, O.C. Interactions of nanomaterials and biological systems: Implications to personalized nanomedicine. Adv. Drug Deliv. Rev. 2012, 64, 1363–1384. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Huang, H.; Dong, S.; Ge, L.; Zhang, Y. Progress in aptamer-mediated drug delivery vehicles for cancer targeting and its implications in addressing chemotherapeutic challenges. Theranostics 2014, 4, 931–944. [Google Scholar] [CrossRef] [PubMed]
- Hughes, B. Antibody-drug conjugates for cancer: Poised to deliver? Nat. Rev. Drug Discov. 2010, 9, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhu, X.; Lu, P.Y.; Rosato, R.R.; Tan, W.; Zu, Y. Oligonucleotide aptamers: New tools for targeted cancer therapy. Mol. Ther. Nucleic Acids 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Castanares, M.; Mukherjee, A.; Lupold, S.E. Nucleic acid aptamers: Clinical applications and promising new horizons. Curr. Med. Chem. 2011, 18, 4206–4214. [Google Scholar] [CrossRef]
- Tan, W.; Donovan, M.J.; Jiang, J. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev. 2013, 113, 2842–2862. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Tang, Z.; Mallikaratchy, P.; Xiao, Z.; Tan, W. Optimization and modifications of aptamers selected from live cancer cell lines. ChemBioChem 2007, 8, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Gomes, S.D.R.; Azéma, L.; Allard, M.; Toulmé, J.-J. Aptamers as imaging agents. Expert Opin. Med. Diagn. 2010, 4, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Rimmele, M. Nucleic acid aptamers as tools and drugs: Recent developments. ChemBioChem 2003, 4, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.S.; Szostak, J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999, 68, 611–647. [Google Scholar] [CrossRef]
- Reinemann, C.; Strehlitz, B. Aptamer-modified nanoparticles and their use in cancer diagnostics and treatment. Swiss Med. Wkly. 2014, 144. [Google Scholar] [CrossRef] [PubMed]
- Tombelli, S.; Minunni, M.; Mascini, M. Analytical applications of aptamers. Biosens. Bioelectron. 2005, 20, 2424–2434. [Google Scholar] [CrossRef] [PubMed]
- Berezovski, M.; Drabovich, A.; Krylova, S.M.; Musheev, M.; Okhonin, V.; Petrov, A.; Krylov, S.N. Nonequilibrium capillary electrophoresis of equilibrium mixtures: A universal tool for development of aptamers. J. Am. Chem. Soc. 2005, 127, 3165–3171. [Google Scholar] [CrossRef] [PubMed]
- Hermann, T.; Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef] [PubMed]
- Barros, A.L.B.; Ferreira, C.A. Aptamer functionalized nanoparticles for cancer targeting. J. Mol. Pharm. Org. Process Res. 2013, 1. [Google Scholar] [CrossRef]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-vegf aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Song, K.M.; Lee, S.; Ban, C. Aptamers and their biological applications. Sensors 2012, 12, 612–631. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, I.; Shafer, R.H. Effect of loop sequence and size on DNA aptamer stability. Biochemistry 2000, 39, 1462–1468. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Liang, Z.; Zhou, N. Design strategies for aptamer-based biosensors. Sensors 2010, 10, 4541–4557. [Google Scholar] [CrossRef] [PubMed]
- Que-Gewirth, N.S.; Sullenger, B.A. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007, 14, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Pieve, C.D.; Perkins, A.C.; Missailidis, S. Anti-MUC1 aptamers: Radiolabelling with 99mTc and biodistribution in MCF-7 tumour-bearing mice. Nucl. Med. Biol. 2009, 36, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Pendergrast, P.S.; Marsh, H.N.; Grate, D.; Healy, J.M.; Stanton, M. Nucleic acid aptamers for target validation and therapeutic applications. J. Biomol. Tech. 2005, 16, 224–234. [Google Scholar] [PubMed]
- Kim, Y.S.; Gu, M.B. Advances in aptamer screening and small molecule aptasensors. Adv. Biochem. Eng. Biotechnol. 2014, 140, 29–67. [Google Scholar] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. Selex—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Rudzinski, J.F.; Gong, Q.; Soh, H.T.; Atzberger, P.J. Influence of target concentration and background binding on in vitro selection of affinity reagents. PLoS ONE 2012, 7, e43940. [Google Scholar] [CrossRef] [PubMed]
- Tabarzad, M.; Kazemi, B.; Vahidi, H.; Aboofazeli, R.; Shahhosseini, S.; Nafissi-Varcheh, N. Challenges to design and develop of DNA aptamers for protein targets. I. Optimization of asymmetric PCR for generation of a single stranded DNA library. Iran. J. Pharm. Res. 2014, 13, 133–141. [Google Scholar] [PubMed]
- Gopinath, S. Methods developed for selex. Anal. Bioanal. Chem. 2007, 387, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Parekh, P.; Turner, P.; Moyer, R.W.; Tan, W. Generating aptamers for recognition of virus-infected cells. Clin. Chem. 2009, 55, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Nikolaus, N.; Strehlitz, B. Capture-selex: Selection of DNA aptamers for aminoglycoside antibiotics. J. Anal. Methods Chem. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Goringer, H.U.; Homann, M.; Lorger, M. In vitro selection of high-affinity nucleic acid ligands to parasite target molecules. Int. J. Parasitol. 2003, 33, 1309–1317. [Google Scholar] [CrossRef]
- Heilkenbrinker, A.; Reinemann, C.; Stoltenburg, R.; Walter, J.-G.; Jochums, A.; Stahl, F.; Zimmermann, S.; Strehlitz, B.; Scheper, T. Identification of the target binding site of ethanolamine-binding aptamers and its exploitation for ethanolamine detection. Anal. Chem. 2014, 87, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Avino, A.; Fabrega, C.; Tintore, M.; Eritja, R. Thrombin binding aptamer, more than a simple aptamer: Chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012, 18, 2036–2047. [Google Scholar] [CrossRef] [PubMed]
- Hicke, B.J.; Marion, C.; Chang, Y.-F.; Gould, T.; Lynott, C.K.; Parma, D.; Schmidt, P.G.; Warren, S. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 2001, 276, 48644–48654. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.C.; Hayhurst, A.; Hesselberth, J.; Bayer, T.S.; Georgiou, G.; Ellington, A.D. Automated selection of aptamers against protein targets translated in vitro: From gene to aptamer. Nucleic Acids Res. 2002, 30. [Google Scholar] [CrossRef]
- Wang, J.; Li, G. Aptamers against cell surface receptors: Selection, modification and application. Curr. Med. Chem. 2011, 18, 4107–4116. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.K.; Griffiths, C.; Lea, S.M.; James, W. Structural characterization of an anti-gp120RNA aptamer that neutralizes R5 strains of HIV-1. RNA 2005, 11, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Binning, J.M.; Leung, D.W.; Amarasinghe, G.K. Aptamers in virology: Recent advances and challenges. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Biroccio, A.; Hamm, J.; Incitti, I.; de Francesco, R.; Tomei, L. Selection of RNA aptamers that are specific and high-affinity ligands of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 2002, 76, 3688–3696. [Google Scholar] [CrossRef] [PubMed]
- Bellecave, P.; Cazenave, C.; Rumi, J.; Staedel, C.; Cosnefroy, O.; Andreola, M.L.; Ventura, M.; Tarrago-Litvak, L.; Astier-Gin, T. Inhibition of hepatitis C virus (HCV) RNA polymerase by DNA aptamers: Mechanism of inhibition of in vitro RNA synthesis and effect on HCV-infected cells. Antimicrob. Agents Chemother. 2008, 52, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.C.; DeFeo-Fraulini, T.; Hutabarat, R.M.; Horvath, C.J.; Merlino, P.G.; Marsh, H.N.; Healy, J.M.; Boufakhreddine, S.; Holohan, T.V.; Schaub, R.G. First-in-human evaluation of anti von willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation 2007, 116, 2678–2686. [Google Scholar] [CrossRef]
- Huang, R.H.; Fremont, D.H.; Diener, J.L.; Schaub, R.G.; Sadler, J.E. A structural explanation for the antithrombotic activity of ARC1172, a DNA aptamer that binds von willebrand factor domain A1. Structure 2009, 17, 1476–1484. [Google Scholar] [CrossRef] [PubMed]
- Dyke, C.K.; Steinhubl, S.R.; Kleiman, N.S.; Cannon, R.O.; Aberle, L.G.; Lin, M.; Myles, S.K.; Melloni, C.; Harrington, R.A.; Alexander, J.H.; et al. First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: A phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation 2006, 114, 2490–2497. [Google Scholar] [CrossRef] [PubMed]
- Marshall, K.A.; Ellington, A.D. In vitro selection of RNA aptamers. Methods Enzymol. 2000, 318, 193–214. [Google Scholar] [PubMed]
- Burmeister, P.E.; Lewis, S.D.; Silva, R.F.; Preiss, J.R.; Horwitz, L.R.; Pendergrast, P.S.; McCauley, T.G.; Kurz, J.C.; Epstein, D.M.; Wilson, C.; et al. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 2005, 12, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Bassett, S.E.; Fennewald, S.M.; King, D.J.; Li, X.; Herzog, N.K.; Shope, R.; Aronson, J.F.; Luxon, B.A.; Gorenstein, D.G. Combinatorial selection and edited combinatorial selection of phosphorothioate aptamers targeting human nuclear factor-κb RelA/p50 and RelA/RelA. Biochemistry 2004, 43, 9105–9115. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, K.K.; Wengel, J. Locked nucleic acid and aptamers. Nucleic Acid Ther. 2012, 22, 366–370. [Google Scholar] [PubMed]
- Hagiwara, K.; Fujita, H.; Kasahara, Y.; Irisawa, Y.; Obika, S.; Kuwahara, M. In vitro selection of DNA-based aptamers that exhibit RNA-like conformations using a chimeric oligonucleotide library that contains two different xeno-nucleic acids. Mol. BioSyst. 2015, 11, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Stormo, G.D. Combining selex with quantitative assays to rapidly obtain accurate models of protein–DNA interactions. Nucleic Acids Res. 2005, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Battig, M.; Wang, Y. Aptamer-based molecular recognition for biosensor development. Anal. Bioanal. Chem. 2010, 398, 2471–2480. [Google Scholar] [CrossRef] [PubMed]
- Naimuddin, M.; Kitamura, K.; Kinoshita, Y.; Honda-Takahashi, Y.; Murakami, M.; Ito, M.; Yamamoto, K.; Hanada, K.; Husimi, Y.; Nishigaki, K. Selection-by-function: Efficient enrichment of cathepsin e inhibitors from a DNA library. J. Mol. Recognit. 2007, 20, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Marimuthu, C.; Tang, T.H.; Tominaga, J.; Tan, S.C.; Gopinath, S.C. Single-stranded DNA (ssDNA) production in DNA aptamer generation. Analyst 2012, 137, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; DeRosa, M.C. Challenges and opportunities for small molecule aptamer development. J. Nucleic Acids 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Basnar, B.; Elnathan, R.; Willner, I. Following aptamer−thrombin binding by force measurements. Anal. Chem. 2006, 78, 3638–3642. [Google Scholar] [CrossRef] [PubMed]
- Miyachi, Y.; Shimizu, N.; Ogino, C.; Kondo, A. Selection of DNA aptamers using atomic force microscopy. Nucleic Acids Res. 2009, 38. [Google Scholar] [CrossRef] [PubMed]
- Eulberg, D.; Buchner, K.; Maasch, C.; Klussmann, S. Development of an automated in vitro selection protocol to obtain RNA-based aptamers: Identification of a biostable substance p antagonist. Nucleic Acids Res. 2005, 33. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Liu, C.; Tan, W. Aptamers generated by cell selex for biomarker discovery. Biomark. Med. 2009, 3, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Han, D.; Ocsoy, I.; Tan, W. Aptamers selected by cell-selex for application in cancer studies. Bioanalysis 2010, 2, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Mendonsa, S.D.; Bowser, M.T. In vitro selection of high-affinity DNA ligands for human ige using capillary electrophoresis. Anal. Chem. 2004, 76, 5387–5392. [Google Scholar] [CrossRef] [PubMed]
- Darmostuk, M.; Rimpelová, S.; Gbelcová, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. Flumag-selex as an advantageous method for DNA aptamer selection. Anal. Bioanal. Chem. 2005, 383, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed]
- Monsuez, J.-J.; Charniot, J.-C.; Vignat, N.; Artigou, J.-Y. Cardiac side-effects of cancer chemotherapy. Int. J. Cardiol. 2010, 144, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Brannon-Peppas, L.; Blanchette, J.O. Nanoparticle and targeted systems for cancer therapy. Adv. Drug Del. Rev. 2012, 64, 206–212. [Google Scholar] [CrossRef]
- Farokhzad, O.C.; Jon, S.; Khademhosseini, A.; Tran, T.-N.T.; LaVan, D.A.; Langer, R. Nanoparticle-aptamer bioconjugates: A new approach for targeting prostate cancer cells. Cancer Res. 2004, 64, 7668–7672. [Google Scholar] [CrossRef] [PubMed]
- Farokhzad, O.C.; Cheng, J.; Teply, B.A.; Sherifi, I.; Jon, S.; Kantoff, P.W.; Richie, J.P.; Langer, R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 6315–6320. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.; Gu, F.X.; Langer, R.; Farokhzad, O.C.; Lippard, S.J. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEGnanoparticles. Proc. Natl. Acad. Sci. USA 2008, 105, 17356–17361. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Hu, Y.; Duan, J.; Yuan, W.; Wang, C.; Xu, H.; Yang, X.D. Novel aptamer-nanoparticle bioconjugates enhances delivery of anticancer drug to MUC1-positive cancer cells in vitro. PLoS ONE 2011, 6, e24077. [Google Scholar] [CrossRef] [PubMed]
- Aravind, A.; Jeyamohan, P.; Nair, R.; Veeranarayanan, S.; Nagaoka, Y.; Yoshida, Y.; Maekawa, T.; Kumar, D.S. As1411 aptamer tagged PLGA-lecithin-PEG nanoparticles for tumor cell targeting and drug delivery. Biotechnol. Bioeng. 2012, 109, 2920–2931. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; You, J.; Zeng, Z.; Li, C.; Zu, Y. An ultra pH-sensitive and aptamer-equipped nanoscale drug-delivery system for selective killing of tumor cells. Small 2013, 9, 3477–3484. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sefah, K.; Altman, M.B.; Chen, T.; You, M.; Zhao, Z.; Huang, C.Z.; Tan, W. Aptamer-conjugated nanorods for targeted photothermal therapy of prostate cancer stem cells. Chem. Asian J. 2013, 8, 2417–2422. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Shigdar, S.; Qiao, G.; Wang, T.; Kouzani, A.Z.; Zhou, S.-F.; Kong, L.; Li, Y.; Pu, C.; Duan, W. Nucleic acid aptamer-guided cancer therapeutics and diagnostics: The next generation of cancer medicine. Theranostics 2015, 5, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Z.; Bagalkot, V.; Vasilliou, C.C.; Gu, F.; Alexis, F.; Zhang, L.; Shaikh, M.; Yuet, K.; Cima, M.J.; Langer, R.; et al. Superparamagnetic iron oxide nanoparticle-aptamer bioconjugates for combined prostate cancer imaging and therapy. ChemMedChem 2008, 3, 1311–1315. [Google Scholar] [CrossRef] [PubMed]
- Delfaut, E.M.; Beltran, J.; Johnson, G.; Rousseau, J.; Marchandise, X.; Cotten, A. Fat suppression in mr imaging: Techniques and pitfalls. Radiographics 1999, 19, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Jalalian, S.H.; Taghdisi, S.M.; Shahidi Hamedani, N.; Kalat, S.A.; Lavaee, P.; Zandkarimi, M.; Ghows, N.; Jaafari, M.R.; Naghibi, S.; Danesh, N.M.; et al. Epirubicin loaded super paramagnetic iron oxide nanoparticle-aptamer bioconjugate for combined colon cancer therapy and imaging in vivo. Eur. J. Pharm. Sci. 2013, 50, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Shu, D.; Haque, F.; Guo, P. Fabrication of pRNA nanoparticles to deliver therapeutic RNAs and bioactive compounds into tumor cells. Nat. Protoc. 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Haque, F.; Hallahan, B.; Reif, R.; Li, H. Uniqueness, advantages, challenges, solutions, and perspectives in therapeutics applying RNA nanotechnology. Nucleic Acid Ther. 2012, 22, 226–245. [Google Scholar] [PubMed]
- Liu, J.; Guo, S.; Cinier, M.; Shlyakhtenko, L.S.; Shu, Y.; Chen, C.; Shen, G.; Guo, P. Fabrication of stable and rnase-resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging. ACS Nano 2011, 5, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Guo, P. The emerging field of RNA nanotechnology. Nat. Nano 2010, 5, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Thiel, K.W.; Giangrande, P.H. Intracellular delivery of RNA-based therapeutics using aptamers. Ther. Deliv. 2010, 1, 849–861. [Google Scholar] [CrossRef] [PubMed]
- McNamara, J.O., 2nd; Andrechek, E.R.; Wang, Y.; Viles, K.D.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006, 24, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Thiel, K.W.; Hernandez, L.I.; Dassie, J.P.; Thiel, W.H.; Liu, X.; Stockdale, K.R.; Rothman, A.M.; Hernandez, F.J.; McNamara, J.O., 2nd; Giangrande, P.H. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012, 40, 6319–6337. [Google Scholar] [CrossRef] [PubMed]
- Wullner, U.; Neef, I.; Eller, A.; Kleines, M.; Tur, M.K.; Barth, S. Cell-specific induction of apoptosis by rationally designed bivalent aptamer-siRNA transcripts silencing eukaryotic elongation factor 2. Curr. Cancer Drug Targets 2008, 8, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Cinier, M.; Shu, D.; Guo, P. Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods 2011, 54, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Shu, Y.; Guo, P.; Smith, D.D.; Rossi, J.J. Dual functional RNA nanoparticles containing phi29 motor pRNA and anti-gp120 aptamer for cell-type specific delivery and HIV-1 inhibition. Methods 2011, 54, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.; Zhang, Y.; Zhu, X.; Shan, N.; Chen, Y. Anticancer role of MUC1 aptamer-miR-29b chimera in epithelial ovarian carcinoma cells through regulation of pten methylation. Target. Oncol. 2012, 7, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Esposito, C.L.; Cerchia, L.; Catuogno, S.; De Vita, G.; Dassie, J.P.; Santamaria, G.; Swiderski, P.; Condorelli, G.; Giangrande, P.H.; de Franciscis, V. Multifunctional aptamer-mirna conjugates for targeted cancer therapy. Mol. Ther. 2014, 22, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, C.K. Aptasensors—the future of biosensing? Anal. Bioanal. Chem. 2002, 372, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Zayats, M.; Huang, Y.; Gill, R.; Ma, C.A.; Willner, I. Label-free and reagentless aptamer-based sensors for small molecules. J. Am. Chem. Soc. 2006, 128, 13666–13667. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, S.; Obubuafo, A.; Soper, S.A.; Spivak, D.A. Surface immobilization methods for aptamer diagnostic applications. Anal. Bioanal. Chem. 2008, 390, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Willner, I.; Zayats, M. Electronic aptamer-based sensors. Angew. Chem. Int. Ed. Engl. 2007, 46, 6408–6418. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.O.; So, H.M.; Jeon, E.K.; Chang, H.; Won, K.; Kim, Y.H. Aptamers as molecular recognition elements for electrical nanobiosensors. Anal. Bioanal. Chem. 2008, 390, 1023–1032. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Chen, Y.; Ren, J.; Qu, X. A graphene functionalized electrochemical aptasensor for selective label-free detection of cancer cells. Biomaterials 2011, 32, 2930–2937. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Tian, D.; Gu, J.; Cui, H. A novel electrochemiluminescence aptasensor for protein based on a sensitive N-(aminobutyl)-N-ethylisoluminol-functionalized gold nanoprobe. Analyst 2011, 136, 3244–3251. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Lin, Y.; Tang, H.; Pang, D. A graphene oxide-based fluorescent aptasensor for the turn-on detection of epithelial tumor marker mucin 1. Nanoscale 2012, 4, 2054–2059. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Tan, Y.; Guo, Q.; Wang, K.; Yuan, B.; Wan, J.; Zhao, X. A fluorescent aptasensor for sensitive detection of human hepatocellular carcinoma smmc-7721 cells based on graphene oxide. Anal. Methods 2014, 6, 6809–6814. [Google Scholar] [CrossRef]
- Hong, H.; Goel, S.; Zhang, Y.; Cai, W. Molecular imaging with nucleic acid aptamers. Curr. Med. Chem. 2011, 18, 4195–4205. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; He, X.; Wang, K.; Wu, X.; Ye, X.; Guo, Q.; Tan, W.; Qing, Z.; Yang, X.; Zhou, B. Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration. Proc. Natl. Acad. Sci. USA 2011, 108, 3900–3905. [Google Scholar] [CrossRef] [PubMed]
- Wadas, T.J.; Wong, E.H.; Weisman, G.R.; Anderson, C.J. Copper chelation chemistry and its role in copper radiopharmaceuticals. Curr. Pharm. Des. 2007, 13, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, H.; Bates, P.J.; Malik, T.; Li, X.F.; Trent, J.O.; Ng, C.K. Aptamer imaging with cu-64 labeled as1411: Preliminary assessment in lung cancer. Nucl. Med. Biol. 2014, 41, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, O.; Yan, X.; Niu, G.; Weiss, I.D.; Ma, Y.; Szajek, L.P.; Shen, B.; Kiesewetter, D.O.; Chen, X. PETimaging of tenascin-c with a radiolabeled single-stranded DNA aptamer. J. Nucl. Med. 2015, 56, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Jeong, Y.Y.; Jon, S. A drug-loaded aptamer-gold nanoparticle bioconjugate for combined CT imaging and therapy of prostate cancer. ACS Nano 2010, 4, 3689–3696. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Huang, Y.F.; Yeh, C.K. Aptamer-conjugated nanobubbles for targeted ultrasound molecular imaging. Langmuir 2011, 27, 6971–6976. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuka, M.A.; Mattrey, R.F.; Esener, S.C.; Cha, J.N.; Goodwin, A.P. Aptamer-crosslinked microbubbles: Smart contrast agents for thrombin-activated ultrasound imaging. Adv. Mater. 2012, 24, 6010–6016. [Google Scholar] [CrossRef] [PubMed]
- You, X.G.; Tu, R.; Peng, M.L.; Bai, Y.J.; Tan, M.; Li, H.J.; Guan, J.; Wen, L.J. Molecular magnetic resonance probe targeting vegf165: Preparation and in vitro and in vivo evaluation. Contrast Med. Mol. Imaging 2014, 9, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.K.; Kim, D.; Lee, I.H.; So, J.S.; Jeong, Y.Y.; Jon, S. Image-guided prostate cancer therapy using aptamer-functionalized thermally cross-linked superparamagnetic iron oxide nanoparticles. Small 2011, 7, 2241–2249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Liu, M.; Tong, X.; Sun, N.; Zhou, L.; Cao, Y.; Wang, J.; Zhang, H.; Pei, R. Aptamer-modified temperature-sensitive liposomal contrast agent for MRimaging. Biomacromolecules 2015. [Google Scholar] [CrossRef]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prakash, J.S.; Rajamanickam, K. Aptamers and Their Significant Role in Cancer Therapy and Diagnosis. Biomedicines 2015, 3, 248-269. https://doi.org/10.3390/biomedicines3030248
Prakash JS, Rajamanickam K. Aptamers and Their Significant Role in Cancer Therapy and Diagnosis. Biomedicines. 2015; 3(3):248-269. https://doi.org/10.3390/biomedicines3030248
Chicago/Turabian StylePrakash, Joy Sebastian, and Karunanithi Rajamanickam. 2015. "Aptamers and Their Significant Role in Cancer Therapy and Diagnosis" Biomedicines 3, no. 3: 248-269. https://doi.org/10.3390/biomedicines3030248
APA StylePrakash, J. S., & Rajamanickam, K. (2015). Aptamers and Their Significant Role in Cancer Therapy and Diagnosis. Biomedicines, 3(3), 248-269. https://doi.org/10.3390/biomedicines3030248