The Peripheral and Intratumoral Immune Cell Landscape in Cancer Patients: A Proxy for Tumor Biology and a Tool for Outcome Prediction
Abstract
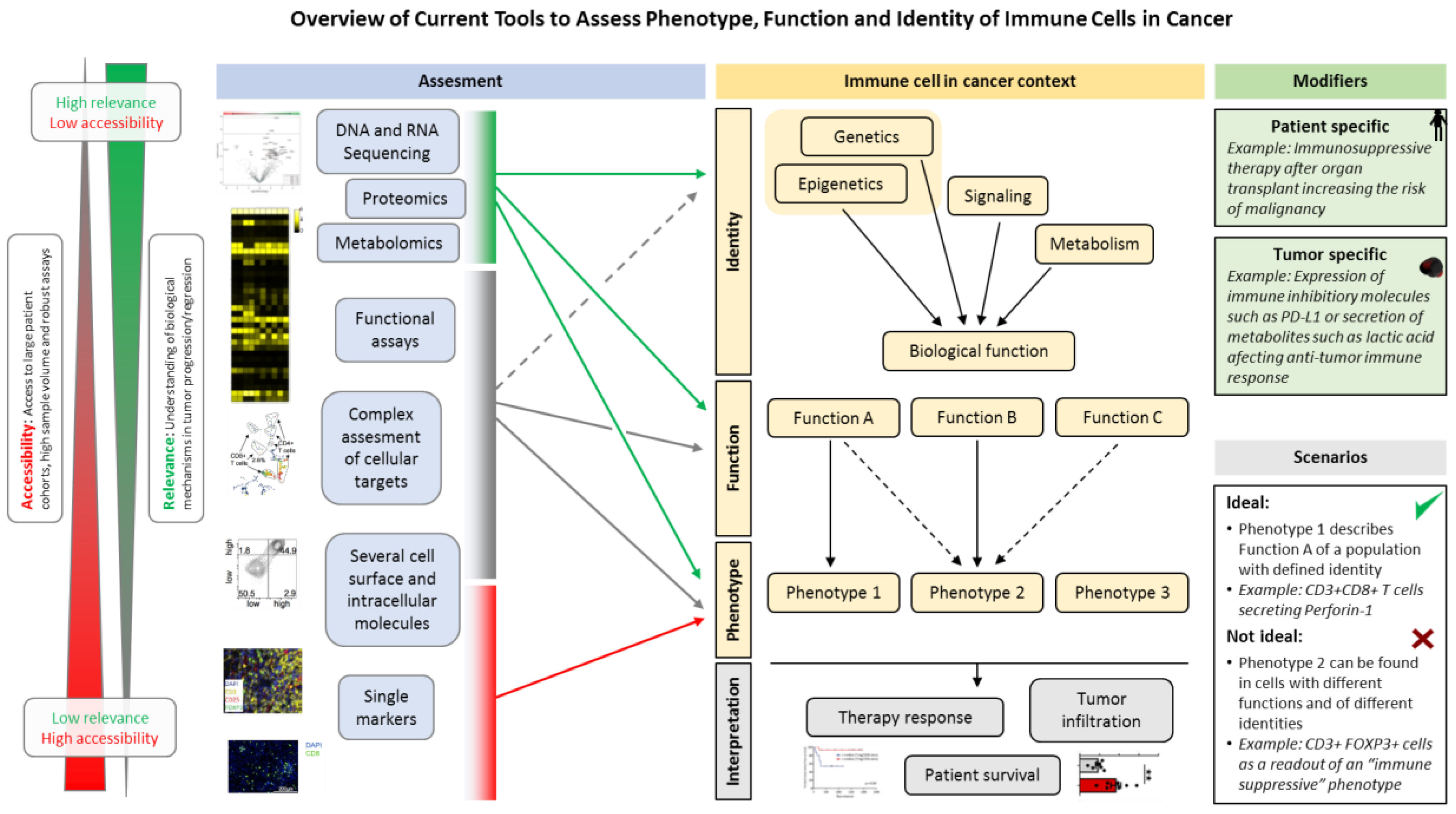
:1. Introduction
2. Phenotype and Function of Peripheral Blood Immune Cells as an Outcome Predictor
3. The Phenotype-Function Discrepancy and Its Implications for Cancer Immune Monitoring
4. Peripheral Immune Cell Subsets in Tumor Patients Receiving Immunotherapy
5. Immune Cells Provide Critical Information about the Tumor Microenvironment
6. Frequency and Phenotype of Tumor-Infiltrating Immune Cells as a Prognosis Predictor
7. Function and Metabolism of Immune Cells Dictate the Outcome of Tumor-Immune Cell Interactions
8. Tumoral Immune Composition and Therapy Response
9. Novel Tools to Improve Predictive Power of Immune Cell Assessment in Cancer Patients
9.1. Mass Cytometry
9.2. Assessment of TCR Clonality
9.3. Transcriptomics
9.4. Epigenomics
9.5. Multi-Layer Single Cell Data
9.6. Others
10. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dvorak, H.F. Tumors: Wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 1986, 315, 1650–1659. [Google Scholar] [PubMed]
- Siska, P.J.; Beckermann, K.E.; Mason, F.M.; Andrejeva, G.; Greenplate, A.R.; Sendor, A.B.; Chiang, Y.J.; Corona, A.L.; Gemta, L.F.; Vincent, B.G.; et al. Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Siska, P.J.; Johnpulle, R.A.N.; Zhou, A.; Bordeaux, J.; Kim, J.Y.; Dabbas, B.; Dakappagari, N.; Rathmell, J.C.; Rathmell, W.K.; Morgans, A.K.; et al. Deep exploration of the immune infiltrate and outcome prediction in testicular cancer by quantitative multiplexed immunohistochemistry and gene expression profiling. Oncoimmunology 2017, 6, e1305535. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, M.H.; Carmi, Y.; Reticker-Flynn, N.E.; Kwek, S.S.; Madhireddy, D.; Martins, M.M.; Gherardini, P.F.; Prestwood, T.R.; Chabon, J.; Bendall, S.C.; et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 2017, 168, 487–502. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Luo, G.; Lu, Y.; Jin, K.; Guo, M.; Xu, J.; Long, J.; Liu, L.; Yu, X.; Liu, C. The combination of systemic inflammation-based marker NLR and circulating regulatory T cells predicts the prognosis of resectable pancreatic cancer patients. Pancreatology 2016, 16, 1080–1084. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, J.; Cyman, M.; Slebioda, T.; Bemben, K.; Rutkowska, A.; Gruchala, M.; Kmiec, Z.; Pliszka, A.; Zaucha, R. Evaluation of peripheral blood T lymphocyte surface activation markers and transcription factors in patients with early stage non-small cell lung cancer. Cell. Immunol. 2017, 322, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Ihara, F.; Sakurai, D.; Horinaka, A.; Makita, Y.; Fujikawa, A.; Sakurai, T.; Yamasaki, K.; Kunii, N.; Motohashi, S.; Nakayama, T.; et al. CD45RA(-)Foxp3(high) regulatory T cells have a negative impact on the clinical outcome of head and neck squamous cell carcinoma. Cancer Immunol. Immunother. 2017, 66, 1275–1285. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Liang, H.; Xu, Y.; Liu, L.; Ren, X.; Zhang, S.; Wei, S.; Xu, P. The Role of Circulating T Follicular Helper Cells and Regulatory Cells in Non-Small Cell Lung Cancer Patients. Scand. J. Immunol. 2017, 86, 107–112. [Google Scholar] [PubMed]
- Xiao, Q.; Zhang, B.; Deng, X.; Wu, J.; Wang, H.; Wang, Y.; Wang, W. The Preoperative Neutrophil-To-Lymphocyte Ratio Is a Novel Immune Parameter for the Prognosis of Esophageal Basaloid Squamous Cell Carcinoma. PLoS ONE 2016, 11, e0168299. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Eo, W.; Jeon, H.; Park, S.; Chae, J. Prognostic Significance of Host-related Biomarkers for Survival in Patients with Advanced Non-Small Cell Lung Cancer. J. Cancer 2017, 8, 2974–2983. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Ma, J.; Yang, X.; Li, W. Peripheral CD4+ naive/memory ratio is an independent predictor of survival in non-small cell lung cancer. Oncotarget 2017, 8, 83650–83659. [Google Scholar] [PubMed]
- Shang, B.; Liu, Y.; Jiang, S.J.; Liu, Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: A systematic review and meta-analysis. Sci. Rep. 2015, 5, 15179. [Google Scholar] [CrossRef] [PubMed]
- Zhong, A.; Pan, X.; Shi, M. Expression of PD-1 by CD4(+)CD25(+)CD127(low) Treg cells in the peripheral blood of lung cancer patients. OncoTargets Ther. 2015, 8, 1831–1833. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.B.; Qian, Y.G.; Zheng, S.S. Prognostic significance of regulatory T lymphocytes in patients with hepatocellular carcinoma. J. Zhejiang Univ. Sci. B 2016, 17, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Vetsika, E.K.; Koinis, F.; Katsarou, A.; Gioulbasani, M.; Aggouraki, D.; Kentepozidis, N.; Georgoulias, V.; Kotsakis, A. Predictive/prognostic value of circulating regulatory T cell subset in untreated non-small lung cancer patients. Ann. Oncol. 2016, 27, 15–42. [Google Scholar] [CrossRef]
- Hamilton, G.; Rath, B. Circulating tumor cell interactions with macrophages: Implications for biology and treatment. Transl. Lung Cancer Res. 2017, 6, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Pan, G.; Tian, D.; Zhang, Y.; Li, T. Functional analysis of CD14(+)HLA-DR(-/low) myeloid-derived suppressor cells in patients with lung squamous cell carcinoma. Oncol. Lett. 2017, 14, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Pearce, H.; Hutton, P.; Chaudhri, S.; Porfiri, E.; Patel, P.; Viney, R.; Moss, P. Spontaneous CD4(+) and CD8(+) T-cell responses directed against cancer testis antigens are present in the peripheral blood of testicular cancer patients. Eur. J. Immunol. 2017, 47, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Retseck, J.; VanderWeele, R.; Lin, H.M.; Lin, Y.; Butterfield, L.H.; Tarhini, A.A. Phenotypic and functional testing of circulating regulatory T cells in advanced melanoma patients treated with neoadjuvant ipilimumab. J. Immunother. Cancer 2016, 4, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, R.S.; Shah, S.U.; Shrikhande, S.V.; Goel, M.; Dikshit, R.P.; Chiplunkar, S.V. IL17 producing gammadeltaT cells induce angiogenesis and are associated with poor survival in gallbladder cancer patients. Int. J. Cancer 2016, 139, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.Y.; Huang, D.Y.; Zhang, H.J.; Wang, S.; Chen, X.F. Function and regulation of LAG3 on CD4(+)CD25(-) T cells in non-small cell lung cancer. Exp. Cell Res. 2017, 360, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Santagata, S.; Napolitano, M.; D’Alterio, C.; Desicato, S.; Maro, S.D.; Marinelli, L.; Fragale, A.; Buoncervello, M.; Persico, F.; Gabriele, L.; et al. Targeting CXCR4 reverts the suppressive activity of T-regulatory cells in renal cancer. Oncotarget 2017, 8, 77110–77120. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, M.F.; Bohner, P.; Pieraerts, C.; Lhermitte, B.; Gourmaud, J.; Nobile, A.; Rotman, S.; Cesson, V.; Martin, V.; Legris, A.S.; et al. Immunoregulation of Dendritic Cell Subsets by Inhibitory Receptors in Urothelial Cancer. Eur. Urol. 2017, 71, 854–857. [Google Scholar] [CrossRef] [PubMed]
- Lang, C.; Wang, J.; Chen, L. CD25-expressing Th17 cells mediate CD8(+) T cell suppression in CTLA-4 dependent mechanisms in pancreatic ductal adenocarcinoma. Exp. Cell Res. 2017, 360, 384–389. [Google Scholar] [CrossRef] [PubMed]
- DeFalco, J.; Harbell, M.; Manning-Bog, A.; Baia, G.; Scholz, A.; Millare, B.; Sumi, M.; Zhang, D.; Chu, F.; Dowd, C.; et al. Non-progressing cancer patients have persistent B cell responses expressing shared antibody paratopes that target public tumor antigens. Clin. Immunol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, J.; Wang, J.; Vangundy, Z.; You, J.; Yildiz, V.; Yu, L.; Foote, I.P.; Branson, O.E.; Stiff, A.R.; Brooks, T.R.; et al. Nitric oxide mediated inhibition of antigen presentation from DCs to CD4(+) T cells in cancer and measurement of STAT1 nitration. Sci. Rep. 2017, 7, 15424. [Google Scholar] [CrossRef] [PubMed]
- Kamphorst, A.O.; Pillai, R.N.; Yang, S.; Nasti, T.H.; Akondy, R.S.; Wieland, A.; Sica, G.L.; Yu, K.; Koenig, L.; Patel, N.T.; et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc. Natl. Acad. Sci. USA 2017, 114, 4993–4998. [Google Scholar] [CrossRef] [PubMed]
- Voong, K.R.; Feliciano, J.; Becker, D.; Levy, B. Beyond PD-L1 testing-emerging biomarkers for immunotherapy in non-small cell lung cancer. Ann. Transl. Med. 2017, 5, 376. [Google Scholar] [CrossRef] [PubMed]
- Wistuba-Hamprecht, K.; Martens, A.; Heubach, F.; Romano, E.; Geukes Foppen, M.; Yuan, J.; Postow, M.; Wong, P.; Mallardo, D.; Schilling, B.; et al. Peripheral CD8 effector-memory type 1 T-cells correlate with outcome in ipilimumab-treated stage IV melanoma patients. Eur. J. Cancer 2017, 73, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Tietze, J.K.; Angelova, D.; Heppt, M.V.; Reinholz, M.; Murphy, W.J.; Spannagl, M.; Ruzicka, T.; Berking, C. The proportion of circulating CD45RO(+)CD8(+) memory T cells is correlated with clinical response in melanoma patients treated with ipilimumab. Eur. J. Cancer 2017, 75, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Martens, A.; Wistuba-Hamprecht, K.; Geukes Foppen, M.; Yuan, J.; Postow, M.A.; Wong, P.; Romano, E.; Khammari, A.; Dreno, B.; Capone, M.; et al. Baseline Peripheral Blood Biomarkers Associated with Clinical Outcome of Advanced Melanoma Patients Treated with Ipilimumab. Clin. Cancer Res. 2016, 22, 2908–2918. [Google Scholar] [CrossRef] [PubMed]
- Tarhini, A.A.; Edington, H.; Butterfield, L.H.; Lin, Y.; Shuai, Y.; Tawbi, H.; Sander, C.; Yin, Y.; Holtzman, M.; Johnson, J.; et al. Immune monitoring of the circulation and the tumor microenvironment in patients with regionally advanced melanoma receiving neoadjuvant ipilimumab. PLoS ONE 2014, 9, e87705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kvistborg, P.; Philips, D.; Kelderman, S.; Hageman, L.; Ottensmeier, C.; Joseph-Pietras, D.; Welters, M.J.; van der Burg, S.; Kapiteijn, E.; Michielin, O.; et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci. Transl. Med. 2014, 6, 254ra128. [Google Scholar] [CrossRef] [PubMed]
- Singer, K.; Kastenberger, M.; Gottfried, E.; Hammerschmied, C.G.; Buttner, M.; Aigner, M.; Seliger, B.; Walter, B.; Schlosser, H.; Hartmann, A.; et al. Warburg phenotype in renal cell carcinoma: High expression of glucose-transporter 1 (GLUT-1) correlates with low CD8(+) T-cell infiltration in the tumor. Int. J. Cancer 2011, 128, 2085–2095. [Google Scholar] [CrossRef] [PubMed]
- Cham, C.M.; Driessens, G.; O’Keefe, J.P.; Gajewski, T.F. Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8+ T cells. Eur. J. Immunol. 2008, 38, 2438–2450. [Google Scholar] [CrossRef] [PubMed]
- Siska, P.J.; Rathmell, J.C. T cell metabolic fitness in antitumor immunity. Trends Immunol. 2015, 36, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Delitto, D.; Black, B.S.; Sorenson, H.L.; Knowlton, A.E.; Thomas, R.M.; Sarosi, G.A.; Moldawer, L.L.; Behrns, K.E.; Liu, C.; George, T.J.; et al. The inflammatory milieu within the pancreatic cancer microenvironment correlates with clinicopathologic parameters, chemoresistance and survival. BMC Cancer 2015, 15, 783. [Google Scholar] [CrossRef] [PubMed]
- Dawood, S.; Lei, X.; Dent, R.; Gupta, S.; Sirohi, B.; Cortes, J.; Cristofanilli, M.; Buchholz, T.; Gonzalez-Angulo, A.M. Survival of women with inflammatory breast cancer: A large population-based study. Ann. Oncol. 2014, 25, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, B.; Tucker, S.L.; Li, D.; Abbruzzese, J.L.; Kurzrock, R. Cytokines in pancreatic carcinoma: Correlation with phenotypic characteristics and prognosis. Cancer 2004, 101, 2727–2736. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Ogata, H.; Nishigaki, R.; Broide, D.H.; Karin, M. Tobacco smoke promotes lung tumorigenesis by triggering IKKbeta- and JNK1-dependent inflammation. Cancer Cell 2010, 17, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Dostert, C.; Petrilli, V.; Van Bruggen, R.; Steele, C.; Mossman, B.T.; Tschopp, J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 2008, 320, 674–677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhu, B.; Li, Y. Resolution of Cancer-Promoting Inflammation: A New Approach for Anticancer Therapy. Front. Immunol. 2017, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Balkwill, F.; Mantovani, A. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Angell, H.K.; Galon, J. The immune landscape of human tumors: Implications for cancer immunotherapy. Oncoimmunology 2014, 3, e27456. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Zitvogel, L.; Sautes-Fridman, C.; Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017, 14, 717–734. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Pages, F.; Sautes-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2017. [Google Scholar] [CrossRef]
- Matkowski, R.; Gisterek, I.; Halon, A.; Lacko, A.; Szewczyk, K.; Staszek, U.; Pudelko, M.; Szynglarewicz, B.; Szelachowska, J.; Zolnierek, A.; et al. The prognostic role of tumor-infiltrating CD4 and CD8 T lymphocytes in breast cancer. Anticancer Res. 2009, 29, 2445–2451. [Google Scholar] [PubMed]
- Liu, S.; Lachapelle, J.; Leung, S.; Gao, D.; Foulkes, W.D.; Nielsen, T.O. CD8+ lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast Cancer Res. 2012, 14, R48. [Google Scholar] [CrossRef] [PubMed]
- Shah, W.; Yan, X.; Jing, L.; Zhou, Y.; Chen, H.; Wang, Y. A reversed CD4/CD8 ratio of tumor-infiltrating lymphocytes and a high percentage of CD4(+)FOXP3(+) regulatory T cells are significantly associated with clinical outcome in squamous cell carcinoma of the cervix. Cell. Mol. Immunol. 2011, 8, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, Z.; Wang, Z.; Wu, P.; Qiu, F.; Huang, J. Prognostic and predictive value of tumor-infiltrating lymphocytes in breast cancer: A systematic review and meta-analysis. Clin. Transl. Oncol. 2016, 18, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.D.; Zahurak, M.; Murphy, A.; Cornish, T.; Cuka, N.; Abdelfatah, E.; Yang, S.; Duncan, M.; Ahuja, N.; Taube, J.M.; et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut 2017, 66, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, N.A.; Becht, E.; Pages, F.; Skliris, G.; Verkarre, V.; Vano, Y.; Mejean, A.; Saint-Aubert, N.; Lacroix, L.; Natario, I.; et al. Orchestration and Prognostic Significance of Immune Checkpoints in the Microenvironment of Primary and Metastatic Renal Cell Cancer. Clin. Cancer Res. 2015, 21, 3031–3040. [Google Scholar] [CrossRef] [PubMed]
- Nakano, O.; Sato, M.; Naito, Y.; Suzuki, K.; Orikasa, S.; Aizawa, M.; Suzuki, Y.; Shintaku, I.; Nagura, H.; Ohtani, H. Proliferative activity of intratumoral CD8(+) T-lymphocytes as a prognostic factor in human renal cell carcinoma: Clinicopathologic demonstration of antitumor immunity. Cancer Res. 2001, 61, 5132–5136. [Google Scholar] [PubMed]
- Wolf, M.J.; Adili, A.; Piotrowitz, K.; Abdullah, Z.; Boege, Y.; Stemmer, K.; Ringelhan, M.; Simonavicius, N.; Egger, M.; Wohlleber, D.; et al. Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes. Cancer Cell. 2014, 26, 549–564. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Spaapen, R.M.; Zha, Y.; Williams, J.; Meng, Y.; Ha, T.T.; Gajewski, T.F. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci. Transl. Med. 2013, 5, 200ra116. [Google Scholar] [CrossRef] [PubMed]
- Hotta, K.; Sho, M.; Fujimoto, K.; Shimada, K.; Yamato, I.; Anai, S.; Konishi, N.; Hirao, Y.; Nonomura, K.; Nakajima, Y. Prognostic significance of CD45RO+ memory T cells in renal cell carcinoma. Br. J. Cancer 2011, 105, 1191–1196. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, N.A.; Becht, E.; Vano, Y.; Petitprez, F.; Lacroix, L.; Validire, P.; Sanchez-Salas, R.; Ingels, A.; Oudard, S.; Moatti, A.; et al. Tumor-Infiltrating and Peripheral Blood T-cell Immunophenotypes Predict Early Relapse in Localized Clear Cell Renal Cell Carcinoma. Clin. Cancer Res. 2017, 23, 4416–4428. [Google Scholar] [CrossRef] [PubMed]
- Mei, Z.; Liu, Y.; Liu, C.; Cui, A.; Liang, Z.; Wang, G.; Peng, H.; Cui, L.; Li, C. Tumour-infiltrating inflammation and prognosis in colorectal cancer: Systematic review and meta-analysis. Br. J. Cancer 2014, 110, 1595–1605. [Google Scholar] [CrossRef] [PubMed]
- Coca, S.; Perez-Piqueras, J.; Martinez, D.; Colmenarejo, A.; Saez, M.A.; Vallejo, C.; Martos, J.A.; Moreno, M. The prognostic significance of intratumoral natural killer cells in patients with colorectal carcinoma. Cancer 1997, 79, 2320–2328. [Google Scholar] [CrossRef]
- Cozar, J.M.; Canton, J.; Tallada, M.; Concha, A.; Cabrera, T.; Garrido, F.; Ruiz-Cabello Osuna, F. Analysis of NK cells and chemokine receptors in tumor infiltrating CD4 T lymphocytes in human renal carcinomas. Cancer Immunol. Immunother. 2005, 54, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Maehara, Y.; Tokunaga, E.; Koga, T.; Kakeji, Y.; Sugimachi, K. Prognostic significance of natural killer cell activity in patients with gastric carcinoma: A multivariate analysis. Am. J. Gastroenterol. 2001, 96, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Villegas, F.R.; Coca, S.; Villarrubia, V.G.; Jimenez, R.; Chillon, M.J.; Jareno, J.; Zuil, M.; Callol, L. Prognostic significance of tumor infiltrating natural killer cells subset CD57 in patients with squamous cell lung cancer. Lung Cancer 2002, 35, 23–28. [Google Scholar] [CrossRef]
- Xu, B.; Chen, L.; Li, J.; Zheng, X.; Shi, L.; Wu, C.; Jiang, J. Prognostic value of tumor infiltrating NK cells and macrophages in stage II+III esophageal cancer patients. Oncotarget 2016, 7, 74904–74916. [Google Scholar] [CrossRef] [PubMed]
- Boddupalli, C.S.; Bar, N.; Kadaveru, K.; Krauthammer, M.; Pornputtapong, N.; Mai, Z.; Ariyan, S.; Narayan, D.; Kluger, H.; Deng, Y.; et al. Interlesional diversity of T cell receptors in melanoma with immune checkpoints enriched in tissue-resident memory T cells. JCI Insight 2016, 1, e88955. [Google Scholar] [CrossRef] [PubMed]
- Reissfelder, C.; Stamova, S.; Gossmann, C.; Braun, M.; Bonertz, A.; Walliczek, U.; Grimm, M.; Rahbari, N.N.; Koch, M.; Saadati, M.; et al. Tumor-specific cytotoxic T lymphocyte activity determines colorectal cancer patient prognosis. J. Clin. Investig. 2015, 125, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Duan, Y.; Cheng, X.; Chen, X.; Xie, W.; Long, H.; Lin, Z.; Zhu, B. IL-17 is associated with poor prognosis and promotes angiogenesis via stimulating VEGF production of cancer cells in colorectal carcinoma. Biochem. Biophys. Res. Commun. 2011, 407, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.M.; Gao, Q.; Shi, G.M.; Zhang, X.; Wang, J.; Jiang, J.H.; Wang, X.Y.; Shi, Y.H.; Ding, Z.B.; Fan, J.; et al. Intratumoral IL-17(+) cells and neutrophils show strong prognostic significance in intrahepatic cholangiocarcinoma. Ann. Surg. Oncol. 2012, 19, 2506–2514. [Google Scholar] [CrossRef] [PubMed]
- Barlow, J.; Yandell, D.; Weaver, D.; Casey, T.; Plaut, K. Higher stromal expression of transforming growth factor-beta type II receptors is associated with poorer prognosis breast tumors. Breast Cancer Res. Treat. 2003, 79, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.O.; Siska, P.J.; Contreras, D.C.; Rathmell, J.C. Nutrients and the microenvironment to feed a T cell army. Semin. Immunol. 2016, 28, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Siska, P.J.; Rathmell, J.C. Metabolic Signaling Drives IFN-gamma. Cell Metab. 2016, 24, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Siska, P.J.; van der Windt, G.J.; Kishton, R.J.; Cohen, S.; Eisner, W.; MacIver, N.J.; Kater, A.P.; Weinberg, J.B.; Rathmell, J.C. Suppression of Glut1 and Glucose Metabolism by Decreased Akt/mTORC1 Signaling Drives T Cell Impairment in B Cell Leukemia. J. Immunol. 2016, 197, 2532–2540. [Google Scholar] [CrossRef] [PubMed]
- Scharping, N.E.; Menk, A.V.; Moreci, R.S.; Whetstone, R.D.; Dadey, R.E.; Watkins, S.C.; Ferris, R.L.; Delgoffe, G.M. The Tumor Microenvironment Represses T Cell Mitochondrial Biogenesis to Drive Intratumoral T Cell Metabolic Insufficiency and Dysfunction. Immunity 2016, 45, 701–703. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.D.; O’Sullivan, D.; Klein Geltink, R.I.; Curtis, J.D.; Chang, C.H.; Sanin, D.E.; Qiu, J.; Kretz, O.; Braas, D.; van der Windt, G.J.; et al. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell 2016, 166, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.C.; Liu, P.S. Metabolic communication in tumors: A new layer of immunoregulation for immune evasion. J. Immunother. Cancer 2016, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Renner, K.; Singer, K.; Koehl, G.E.; Geissler, E.K.; Peter, K.; Siska, P.J.; Kreutz, M. Metabolic Hallmarks of Tumor and Immune Cells in the Tumor Microenvironment. Front. Immunol. 2017, 8, 248. [Google Scholar] [CrossRef] [PubMed]
- West, N.R.; Milne, K.; Truong, P.T.; Macpherson, N.; Nelson, B.H.; Watson, P.H. Tumor-infiltrating lymphocytes predict response to anthracycline-based chemotherapy in estrogen receptor-negative breast cancer. Breast Cancer Res. 2011, 13, R126. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, K.; Nirei, T.; Sunami, E.; Nagawa, H.; Kitayama, J. Density of CD4(+) and CD8(+) T lymphocytes in biopsy samples can be a predictor of pathological response to chemoradiotherapy (CRT) for rectal cancer. Radiat. Oncol. 2011, 6, 49. [Google Scholar] [CrossRef] [PubMed]
- Beuselinck, B.; Job, S.; Becht, E.; Karadimou, A.; Verkarre, V.; Couchy, G.; Giraldo, N.; Rioux-Leclercq, N.; Molinie, V.; Sibony, M.; et al. Molecular subtypes of clear cell renal cell carcinoma are associated with sunitinib response in the metastatic setting. Clin. Cancer Res. 2015, 21, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Chevrier, S.; Levine, J.H.; Zanotelli, V.R.T.; Silina, K.; Schulz, D.; Bacac, M.; Ries, C.H.; Ailles, L.; Jewett, M.A.S.; Moch, H.; et al. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell 2017, 169, 736–749. [Google Scholar] [CrossRef] [PubMed]
- Leelatian, N.; Doxie, D.B.; Greenplate, A.R.; Mobley, B.C.; Lehman, J.M.; Sinnaeve, J.; Kauffmann, R.M.; Werkhaven, J.A.; Mistry, A.M.; Weaver, K.D.; et al. Single cell analysis of human tissues and solid tumors with mass cytometry. Cytom. B Clin. Cytom. 2017, 92, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Lavin, Y.; Kobayashi, S.; Leader, A.; Amir, E.D.; Elefant, N.; Bigenwald, C.; Remark, R.; Sweeney, R.; Becker, C.D.; Levine, J.H.; et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell 2017, 169, 750–765. [Google Scholar] [CrossRef] [PubMed]
- Lowther, D.E.; Goods, B.A.; Lucca, L.E.; Lerner, B.A.; Raddassi, K.; van Dijk, D.; Hernandez, A.L.; Duan, X.; Gunel, M.; Coric, V.; et al. PD-1 marks dysfunctional regulatory T cells in malignant gliomas. JCI Insight 2016, 1, e85935. [Google Scholar] [CrossRef] [PubMed]
- Krieg, C.; Nowicka, M.; Guglietta, S.; Schindler, S.; Hartmann, F.J.; Weber, L.M.; Dummer, R.; Robinson, M.D.; Levesque, M.P.; Becher, B. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zhang, Q.; Wu, S.; Zhang, X.; Wang, M.; He, F.; Wei, T.; Yang, J.; Lou, Y.; Cai, Z.; et al. Characteristics of Tumor Infiltrating Lymphocyte and Circulating Lymphocyte Repertoires in Pancreatic Cancer by the Sequencing of T Cell Receptors. Sci. Rep. 2015, 5, 13664. [Google Scholar] [CrossRef] [PubMed]
- Munson, D.J.; Egelston, C.A.; Chiotti, K.E.; Parra, Z.E.; Bruno, T.C.; Moore, B.L.; Nakano, T.A.; Simons, D.L.; Jimenez, G.; Yim, J.H.; et al. Identification of shared TCR sequences from T cells in human breast cancer using emulsion RT-PCR. Proc. Natl. Acad. Sci. USA 2016, 113, 8272–8277. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, A.M.; Emerson, R.O.; Scherer, D.; Habermann, N.; Buck, K.; Staffa, J.; Desmarais, C.; Halama, N.; Jaeger, D.; Schirmacher, P.; et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol. Immunother. 2013, 62, 1453–1461. [Google Scholar] [CrossRef] [PubMed]
- Emerson, R.O.; Sherwood, A.M.; Rieder, M.J.; Guenthoer, J.; Williamson, D.W.; Carlson, C.S.; Drescher, C.W.; Tewari, M.; Bielas, J.H.; Robins, H.S. High-throughput sequencing of T-cell receptors reveals a homogeneous repertoire of tumour-infiltrating lymphocytes in ovarian cancer. J. Pathol. 2013, 231, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Sittig, S.P.; Kollgaard, T.; Gronbaek, K.; Idorn, M.; Hennenlotter, J.; Stenzl, A.; Gouttefangeas, C.; Thor Straten, P. Clonal expansion of renal cell carcinoma-infiltrating T lymphocytes. Oncoimmunology 2013, 2, e26014. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pages, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Mandal, R.; Senbabaoglu, Y.; Desrichard, A.; Havel, J.J.; Dalin, M.G.; Riaz, N.; Lee, K.W.; Ganly, I.; Hakimi, A.A.; Chan, T.A.; et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight 2016, 1, e89829. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Zheng, L.; Yoo, J.K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q.; et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017, 169, 1342–1356. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., 2nd; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.; Eum, H.H.; Lee, H.O.; Lee, K.M.; Lee, H.B.; Kim, K.T.; Ryu, H.S.; Kim, S.; Lee, J.E.; Park, Y.H.; et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017, 8, 15081. [Google Scholar] [CrossRef] [PubMed]
- Allan, S.E.; Crome, S.Q.; Crellin, N.K.; Passerini, L.; Steiner, T.S.; Bacchetta, R.; Roncarolo, M.G.; Levings, M.K. Activation-induced FOXP3 in human T effector cells does not suppress proliferation or cytokine production. Int. Immunol. 2007, 19, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ioan-Facsinay, A.; van der Voort, E.I.; Huizinga, T.W.; Toes, R.E. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur. J. Immunol. 2007, 37, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Floess, S.; Freyer, J.; Siewert, C.; Baron, U.; Olek, S.; Polansky, J.; Schlawe, K.; Chang, H.D.; Bopp, T.; Schmitt, E.; et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007, 5, e38. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Schmidl, C.; Rendeiro, A.F.; Sheffield, N.C.; Bock, C. ChIPmentation: Fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 2015, 12, 963–965. [Google Scholar] [CrossRef] [PubMed]
- Mognol, G.P.; Spreafico, R.; Wong, V.; Scott-Browne, J.P.; Togher, S.; Hoffmann, A.; Hogan, P.G.; Rao, A.; Trifari, S. Exhaustion-associated regulatory regions in CD8(+) tumor-infiltrating T cells. Proc. Natl. Acad. Sci. USA 2017, 114, E2776–E2785. [Google Scholar] [CrossRef] [PubMed]
- Philip, M.; Fairchild, L.; Sun, L.; Horste, E.L.; Camara, S.; Shakiba, M.; Scott, A.C.; Viale, A.; Lauer, P.; Merghoub, T.; et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 2017, 545, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Farlik, M.; Sheffield, N.C.; Nuzzo, A.; Datlinger, P.; Schonegger, A.; Klughammer, J.; Bock, C. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015, 10, 1386–1397. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, S.A.; Lee, H.J.; Angermueller, C.; Krueger, F.; Saadeh, H.; Peat, J.; Andrews, S.R.; Stegle, O.; Reik, W.; Kelsey, G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 2014, 11, 817–820. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Wu, B.; Litzenburger, U.M.; Ruff, D.; Gonzales, M.L.; Snyder, M.P.; Chang, H.Y.; Greenleaf, W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Cusanovich, D.A.; Daza, R.; Adey, A.; Pliner, H.A.; Christiansen, L.; Gunderson, K.L.; Steemers, F.J.; Trapnell, C.; Shendure, J. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015, 348, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.; Ding, Y.; Song, J.; Zhao, K. Genome-wide mapping of DNase I hypersensitive sites in rare cell populations using single-cell DNase sequencing. Nat. Protoc. 2017, 12, 2342–2354. [Google Scholar] [CrossRef] [PubMed]
- Rotem, A.; Ram, O.; Shoresh, N.; Sperling, R.A.; Goren, A.; Weitz, D.A.; Bernstein, B.E. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 2015, 33, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Medaglia, C.; Giladi, A.; Stoler-Barak, L.; De Giovanni, M.; Salame, T.M.; Biram, A.; David, E.; Li, H.; Iannacone, M.; Shulman, Z.; et al. Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science 2017, 358, 1622–1626. [Google Scholar] [CrossRef] [PubMed]
- Peterson, V.M.; Zhang, K.X.; Kumar, N.; Wong, J.; Li, L.; Wilson, D.C.; Moore, R.; McClanahan, T.K.; Sadekova, S.; Klappenbach, J.A. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 2017, 35, 936–939. [Google Scholar] [CrossRef] [PubMed]
- Stoeckius, M.; Hafemeister, C.; Stephenson, W.; Houck-Loomis, B.; Chattopadhyay, P.K.; Swerdlow, H.; Satija, R.; Smibert, P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 2017, 14, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Angermueller, C.; Clark, S.J.; Lee, H.J.; Macaulay, I.C.; Teng, M.J.; Hu, T.X.; Krueger, F.; Smallwood, S.; Ponting, C.P.; Voet, T.; et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 2016, 13, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Pott, S. Simultaneous measurement of chromatin accessibility, DNA methylation, and nucleosome phasing in single cells. eLife 2017, 6, e23203. [Google Scholar] [CrossRef] [PubMed]
- Darmanis, S.; Gallant, C.J.; Marinescu, V.D.; Niklasson, M.; Segerman, A.; Flamourakis, G.; Fredriksson, S.; Assarsson, E.; Lundberg, M.; Nelander, S.; et al. Simultaneous Multiplexed Measurement of RNA and Proteins in Single Cells. Cell Rep. 2016, 14, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Guo, H.; Cao, C.; Li, X.; Hu, B.; Zhu, P.; Wu, X.; Wen, L.; Tang, F.; Huang, Y.; et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016, 26, 304–319. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Zheng, J.; Yu, Q.; Chen, W.; Xing, J.; Chen, C.; Tian, R. High throughput and accurate serum proteome profiling by integrated sample preparation technology and single-run data independent mass spectrometry analysis. J. Proteom. 2017, 174, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hou, H.; Chen, H.; Liu, Y.; Wang, A.; Hu, Q. Serum metabolomic of laryngeal cancer based on liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. Biomed. Chromatogr. 2017. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhu, J.; Li, W.; Zhang, Z.; Xiong, P.; Wang, H.; Zhang, J. Serum microRNA panel excavated by machine learning as a potential biomarker for the detection of gastric cancer. Oncol. Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wang, T.; Liu, Y.; Su, Z.; Lu, P.; Chen, X.; Hu, D. LncRNA PVT1 as an effective biomarker for cancer diagnosis and detection based on transcriptome data and meta-analysis. Oncotarget 2017, 8, 75455–75466. [Google Scholar] [CrossRef] [PubMed]
- Hocker, J.R.; Deb, S.J.; Li, M.; Lerner, M.R.; Lightfoot, S.A.; Quillet, A.A.; Hanas, R.J.; Reinersman, M.; Thompson, J.L.; Vu, N.T.; et al. Serum Monitoring and Phenotype Identification of Stage I Non-Small Cell Lung Cancer Patients. Cancer Investig. 2017, 35, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Del Boccio, P.; Perrotti, F.; Rossi, C.; Cicalini, I.; Di Santo, S.; Zucchelli, M.; Sacchetta, P.; Genovesi, D.; Pieragostino, D. Serum lipidomic study reveals potential early biomarkers for predicting response to chemoradiation therapy in advanced rectal cancer: A pilot study. Adv. Radiat. Oncol. 2017, 2, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Intasqui, P.; Bertolla, R.P.; Sadi, M.V. Prostate cancer proteomics: Clinically useful protein biomarkers and future perspectives. Expert Rev. Proteom. 2018, 15, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Vetizou, M.; Pitt, J.M.; Daillere, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillere, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2017. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schnell, A.; Schmidl, C.; Herr, W.; Siska, P.J. The Peripheral and Intratumoral Immune Cell Landscape in Cancer Patients: A Proxy for Tumor Biology and a Tool for Outcome Prediction. Biomedicines 2018, 6, 25. https://doi.org/10.3390/biomedicines6010025
Schnell A, Schmidl C, Herr W, Siska PJ. The Peripheral and Intratumoral Immune Cell Landscape in Cancer Patients: A Proxy for Tumor Biology and a Tool for Outcome Prediction. Biomedicines. 2018; 6(1):25. https://doi.org/10.3390/biomedicines6010025
Chicago/Turabian StyleSchnell, Annette, Christian Schmidl, Wolfgang Herr, and Peter J. Siska. 2018. "The Peripheral and Intratumoral Immune Cell Landscape in Cancer Patients: A Proxy for Tumor Biology and a Tool for Outcome Prediction" Biomedicines 6, no. 1: 25. https://doi.org/10.3390/biomedicines6010025
APA StyleSchnell, A., Schmidl, C., Herr, W., & Siska, P. J. (2018). The Peripheral and Intratumoral Immune Cell Landscape in Cancer Patients: A Proxy for Tumor Biology and a Tool for Outcome Prediction. Biomedicines, 6(1), 25. https://doi.org/10.3390/biomedicines6010025



