Roles of Interleukin-1 Receptor Antagonist in Prostate Cancer Progression
Abstract
1. Introduction
2. Experimental Section
2.1. Cell Lines and Analyses of the TME
2.2. RT-qPCR
2.3. Syngeneic Prostate Cancer Model
2.4. Colony-Formation Assay
2.5. DNA Synthesis Assay
2.6. Proliferation Assay
2.7. Western Blot Analysis
2.8. Statistical Analysis
2.9. Abbreviations
3. Results
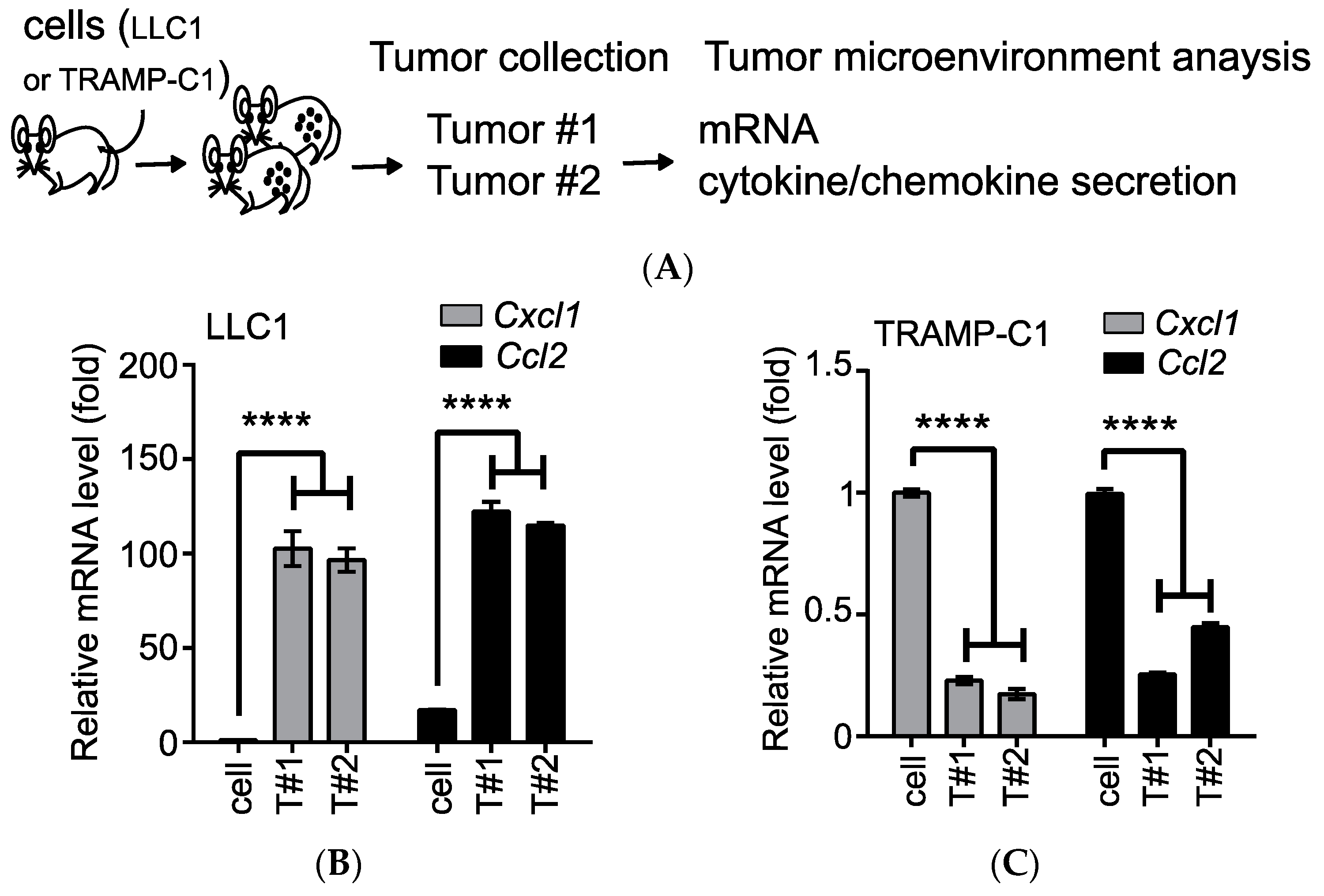
3.1. Confirmation of the IL1RN as a Tumor-Type-Specific Anti-Inflammatory Cytokine in the TRAMP-C1-Derived TME
3.2. Characterization of CD11b− TILs Responsible for IL1RN Secretion in the TRAMP-C1-Derived TME
3.3. Monitoring of Proliferation Effects of Conditioned Media from TILs/CD11b−
3.4. Confirmation of Proliferation Effects by Tumor-Associated Proinflammatory Chemokines and the IL1RN
3.5. The IL1RN Promotes Proliferation and Suppresses the Inhibitory Effects of IL1B in Human Prostate Cell Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Eisenberger, M.A.; Blumenstein, B.A.; Crawford, E.D.; Miller, G.; McLeod, D.G.; Loehrer, P.J.; Wilding, G.; Sears, K.; Culkin, D.J.; Thompson, I.M., Jr.; et al. Bilateral orchiectomy with or without flutamide for metastatic prostate cancer. N. Engl. J. Med. 1998, 339, 1036–1042. [Google Scholar] [CrossRef]
- Karantanos, T.; Corn, P.G.; Thompson, T.C. Prostate cancer progression after androgen deprivation therapy: Mechanisms of castrate resistance and novel therapeutic approaches. Oncogene 2013, 32, 5501–5511. [Google Scholar] [CrossRef] [PubMed]
- Mercader, M.; Bodner, B.K.; Moser, M.T.; Kwon, P.S.; Park, E.S.; Manecke, R.G.; Ellis, T.M.; Wojcik, E.M.; Yang, D.; Flanigan, R.C.; et al. T cell infiltration of the prostate induced by androgen withdrawal in patients with prostate cancer. Proc. Natl. Acad. Sci. USA 2001, 98, 14565–14570. [Google Scholar] [CrossRef] [PubMed]
- Ammirante, M.; Luo, J.L.; Grivennikov, S.; Nedospasov, S.; Karin, M. B-cell-derived lymphotoxin promotes castration-resistant prostate cancer. Nature 2010, 464, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Baek, S.H.; Bourk, E.M.; Ohgi, K.A.; Garcia-Bassets, I.; Sanjo, H.; Akira, S.; Kotol, P.F.; Glass, C.K.; Rosenfeld, M.G.; et al. Macrophage/cancer cell interactions mediate hormone resistance by a nuclear receptor derepression pathway. Cell 2006, 124, 615–629. [Google Scholar] [CrossRef]
- Moore, R.J.; Owens, D.M.; Stamp, G.; Arnott, C.; Burke, F.; East, N.; Holdsworth, H.; Turner, L.; Rollins, B.; Pasparakis, M.; et al. Mice deficient in tumor necrosis factor-alpha are resistant to skin carcinogenesis. Nat. Med. 1999, 5, 828–831. [Google Scholar] [CrossRef]
- Dinarello, C.A. Proinflammatory cytokines. Chest 2000, 118, 503–508. [Google Scholar] [CrossRef]
- Rosenzweig, J.M.; Lei, J.; Burd, I. Interleukin-1 receptor blockade in perinatal brain injury. Front. Pediatr. 2014, 2, 108. [Google Scholar] [CrossRef]
- Arend, W.P.; Malyak, M.; Guthridge, C.J.; Gabay, C. Interleukin-1 receptor antagonist: Role in biology. Annu. Rev. Immunol. 1998, 16, 27–55. [Google Scholar] [CrossRef]
- Lewis, A.M.; Varghese, S.; Xu, H.; Alexander, H.R. Interleukin-1 and cancer progression: The emerging role of interleukin-1 receptor antagonist as a novel therapeutic agent in cancer treatment. J. Transl. Med. 2006, 4, 48. [Google Scholar] [CrossRef]
- Elaraj, D.M.; Weinreich, D.M.; Varghese, S.; Puhlmann, M.; Hewitt, S.M.; Carroll, N.M.; Feldman, E.D.; Turner, E.M.; Alexander, H.R. The role of interleukin 1 in growth and metastasis of human cancer xenografts. Clin. Cancer Res. 2006, 12, 1088–1096. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.; Liou, G.Y. Macrophage Cytokines Enhance Cell Proliferation of Normal Prostate Epithelial Cells through Activation of ERK and Akt. Sci. Rep. 2018, 8, 7718. [Google Scholar] [CrossRef] [PubMed]
- Di Mitri, D.; Toso, A.; Chen, J.J.; Sarti, M.; Pinton, S.; Jost, T.R.; D’Antuono, R.; Montani, E.; Garcia-Escudero, R.; Guccini, I.; et al. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature 2014, 515, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Liou, G.Y.; Bastea, L.; Fleming, A.; Doppler, H.; Edenfield, B.H.; Dawson, D.W.; Zhang, L.; Bardeesy, N.; Storz, P. The Presence of Interleukin-13 at Pancreatic ADM/PanIN Lesions Alters Macrophage Populations and Mediates Pancreatic Tumorigenesis. Cell Rep. 2017, 19, 1322–1333. [Google Scholar] [CrossRef]
- Bertram, J.S.; Janik, P. Establishment of a cloned line of Lewis Lung Carcinoma cells adapted to cell culture. Cancer Lett. 1980, 11, 63–73. [Google Scholar] [CrossRef]
- Foster, B.A.; Gingrich, J.R.; Kwon, E.D.; Madias, C.; Greenberg, N.M. Characterization of prostatic epithelial cell lines derived from transgenic adenocarcinoma of the mouse prostate (TRAMP) model. Cancer Res. 1997, 57, 3325–3330. [Google Scholar]
- Guzman, C.; Bagga, M.; Kaur, A.; Westermarck, J.; Abankwa, D. ColonyArea: An ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS ONE 2014, 9, e92444. [Google Scholar] [CrossRef]
- Li, L.; Lu, Y. Optimizing a 3D Culture System to Study the Interaction between Epithelial Breast Cancer and Its Surrounding Fibroblasts. J. Cancer 2011, 2, 458–466. [Google Scholar] [CrossRef]
- Fan, Y.C.; Chen, W.Y.; Lee, K.D.; Tsai, Y.C. Tumor-infiltrating Leukocytes Suppress Local Inflammation Via Interleukin-1 Receptor Antagonist in a Syngeneic Prostate Cancer Model. Biology 2020, 9, 67. [Google Scholar] [CrossRef]
- Yoshimura, T.; Liu, M.; Chen, X.; Li, L.; Wang, J.M. Crosstalk between Tumor Cells and Macrophages in Stroma Renders Tumor Cells as the Primary Source of MCP-1/CCL2 in Lewis Lung Carcinoma. Front. Immunol. 2015, 6, 332. [Google Scholar] [CrossRef]
- Tsai, Y.C.; Chen, W.Y.; Abou-Kheir, W.; Zeng, T.; Yin, J.J.; Bahmad, H.; Lee, Y.C.; Liu, Y.N. Androgen deprivation therapy-induced epithelial-mesenchymal transition of prostate cancer through downregulating SPDEF and activating CCL2. Biochim. Biophys. Acta 2018, 1864, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The three Es of cancer immunoediting. Annu. Rev. Immunol. 2004, 22, 329–360. [Google Scholar] [CrossRef] [PubMed]
- Rajan, P.; Sudbery, I.M.; Villasevil, M.E.; Mui, E.; Fleming, J.; Davis, M.; Ahmad, I.; Edwards, J.; Sansom, O.J.; Sims, D.; et al. Next-generation sequencing of advanced prostate cancer treated with androgen-deprivation therapy. Eur. Urol. 2014, 66, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Boot, R.G.; Blommaart, E.F.; Swart, E.; Ghauharali-van der Vlugt, K.; Bijl, N.; Moe, C.; Place, A.; Aerts, J.M. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J. Biol. Chem. 2001, 276, 6770–6778. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Y.-C.; Lee, K.-D.; Tsai, Y.-C. Roles of Interleukin-1 Receptor Antagonist in Prostate Cancer Progression. Biomedicines 2020, 8, 602. https://doi.org/10.3390/biomedicines8120602
Fan Y-C, Lee K-D, Tsai Y-C. Roles of Interleukin-1 Receptor Antagonist in Prostate Cancer Progression. Biomedicines. 2020; 8(12):602. https://doi.org/10.3390/biomedicines8120602
Chicago/Turabian StyleFan, Yu-Ching, Kuan-Der Lee, and Yuan-Chin Tsai. 2020. "Roles of Interleukin-1 Receptor Antagonist in Prostate Cancer Progression" Biomedicines 8, no. 12: 602. https://doi.org/10.3390/biomedicines8120602
APA StyleFan, Y.-C., Lee, K.-D., & Tsai, Y.-C. (2020). Roles of Interleukin-1 Receptor Antagonist in Prostate Cancer Progression. Biomedicines, 8(12), 602. https://doi.org/10.3390/biomedicines8120602




