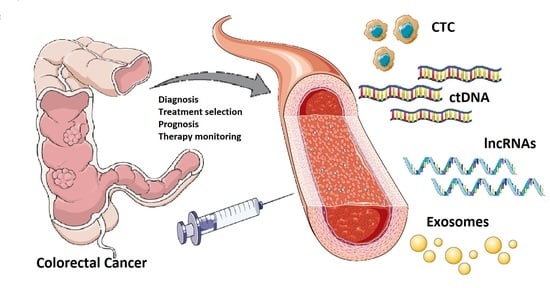
The Liquid Biopsy in the Management of Colorectal Cancer: An Overview
Abstract
:1. Introduction
2. Clinical Utility of Liquid Biopsies in Patients with Colorectal Cancer
2.1. Screening and Early Diagnosis
2.1.1. Circulating Tumor Cells (CTC) and Circulating Endothelial Cell Clusters (ECC)
2.1.2. Circulating Tumor DNA (ctDNA)
2.1.3. Serum, Fecal, and Salivary MicroRNAs (miRNAs)
2.2. Prognosis, Progression, and Response to Treatment
2.2.1. Circulating Tumor Cells (CTC)
2.2.2. Circulating Tumor DNA (ctDNA)
2.2.3. MicroRNAs (miRNAs)
2.2.4. Long Non-Coding RNAs (lncRNAs)
3. Current Issues and Limitations of Liquid Biopsy
4. Future Perspectives and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- GBD 2017 Colorectal Cancer Collaborators. The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2019, 4, 913–933. [Google Scholar] [CrossRef] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishehsari, F.; Mahdavinia, M.; Vacca, M.; Malekzadeh, R.; Mariani-Costantini, R. Epidemiological transition of colorectal cancer in developing countries: Environmental factors, molecular pathways, and opportunities for prevention. World J. Gastroenterol. 2014, 20, 6055–6072. [Google Scholar] [CrossRef] [PubMed]
- Hui, L. Quantifying the effects of aging and urbanization on major gastrointestinal diseases to guide preventative strategies. BMC Gastroenterol. 2018, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valli, A.; Harris, A.L.; Kessler, B.M. Hypoxia metabolism in ageing. Aging Albany N. Y. 2015, 7, 465–466. [Google Scholar] [CrossRef] [Green Version]
- Cancer Facts & Figures 2020|American Cancer Society. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2020.html (accessed on 14 July 2020).
- Stintzing, S. Management of colorectal cancer. F1000Prime Rep. 2014, 6, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayama, G.; Tanaka, C.; Kodera, Y. Current Options for the Diagnosis, Staging and Therapeutic Management of Colorectal Cancer. Gastrointest. Tumors 2013, 1, 25–32. [Google Scholar] [CrossRef] [Green Version]
- Bender, U.; Rho, Y.S.; Barrera, I.; Aghajanyan, S.; Acoba, J.; Kavan, P. Adjuvant therapy for stages II and III colon cancer: Risk stratification, treatment duration, and future directions. Curr. Oncol. 2019, 26, S43–S52. [Google Scholar] [CrossRef] [Green Version]
- Pfeiffer, P.; Köhne, C.-H.; Qvortrup, C. The changing face of treatment for metastatic colorectal cancer. Expert Rev. Anticancer Ther. 2019, 19, 61–70. [Google Scholar] [CrossRef]
- Moriarity, A.; O’Sullivan, J.; Kennedy, J.; Mehigan, B.; McCormick, P. Current targeted therapies in the treatment of advanced colorectal cancer: A review. Ther. Adv. Med. Oncol. 2016, 8, 276–293. [Google Scholar] [CrossRef] [Green Version]
- Norcic, G. Liquid biopsy in colorectal cancer-current status and potential clinical applications. Micromachines 2018, 9, 300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vacante, M.; Borzì, A.M.; Basile, F.; Biondi, A. Biomarkers in colorectal cancer: Current clinical utility and future perspectives. World J. Clin. Cases 2018, 6, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018, 10. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lázaro, D.; García Hernández, J.L.; García, A.C.; Córdova Martínez, A.; Mielgo-Ayuso, J.; Cruz-Hernández, J.J. Liquid biopsy as novel tool in precision medicine: Origins, properties, identification and clinical perspective of cancer’s biomarkers. Diagnostics 2020, 10, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carretero-González, A.; Otero, I.; Carril-Ajuria, L.; de Velasco, G.; Manso, L. Exosomes: Definition, role in tumor development and clinical implications. Cancer Microenviron. 2018, 11, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.-H.; Chen, Y.-C. Clinical significance of exosomes as potential biomarkers in cancer. World J. Clin. Cases 2019, 7, 171–190. [Google Scholar] [CrossRef] [PubMed]
- Lampis, A.; Ghidini, M.; Ratti, M.; Mirchev, M.B.; Okuducu, A.F.; Valeri, N.; Hahne, J.C. Circulating tumour DNAs and Non-Coding RNAs as liquid biopsies for the management of colorectal cancer patients. Gastrointest. Disord. 2020, 2, 22. [Google Scholar] [CrossRef]
- Osumi, H.; Shinozaki, E.; Yamaguchi, K.; Zembutsu, H. Clinical utility of circulating tumor DNA for colorectal cancer. Cancer Sci. 2019, 110, 1148–1155. [Google Scholar] [CrossRef] [Green Version]
- Mathai, R.A.; Vidya, R.V.S.; Reddy, B.S.; Thomas, L.; Udupa, K.; Kolesar, J.; Rao, M. Potential utility of liquid biopsy as a diagnostic and prognostic tool for the assessment of solid tumors: Implications in the precision oncology. J. Clin. Med. 2019, 8, 373. [Google Scholar] [CrossRef] [Green Version]
- Rubis, G.D.; Krishnan, S.R.; Bebawy, M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186. [Google Scholar] [CrossRef] [PubMed]
- Gold, B.; Cankovic, M.; Furtado, L.V.; Meier, F.; Gocke, C.D. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J. Mol. Diagn. 2015, 17, 209–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Li, W.; Wang, K.; Xu, C.; Hao, M.; Ding, L. Perspectives of the Application of Liquid Biopsy in Colorectal Cancer. Available online: https://www.hindawi.com/journals/bmri/2020/6843180/ (accessed on 16 July 2020).
- Tsai, W.-S.; Nimgaonkar, A.; Segurado, O.; Chang, Y.; Hsieh, B.; Shao, H.-J.; Wu, J.; Lai, J.-M.; Javey, M.; Watson, D.; et al. Prospective clinical study of circulating tumor cells for colorectal cancer screening. J. Clin. Oncol. 2018, 36, 556. [Google Scholar] [CrossRef]
- Bork, U.; Rahbari, N.N.; Schölch, S.; Reissfelder, C.; Kahlert, C.; Büchler, M.W.; Weitz, J.; Koch, M. Circulating tumour cells and outcome in non-metastatic colorectal cancer: A prospective study. Br. J. Cancer 2015, 112, 1306–1313. [Google Scholar] [CrossRef]
- Gazzaniga, P.; Gianni, W.; Raimondi, C.; Gradilone, A.; Lo Russo, G.; Longo, F.; Gandini, O.; Tomao, S.; Frati, L. Circulating tumor cells in high-risk nonmetastatic colorectal cancer. Tumour Biol. 2013, 34, 2507–2509. [Google Scholar] [CrossRef]
- Tsai, W.-S.; Chen, J.-S.; Shao, H.-J.; Wu, J.-C.; Lai, J.-M.; Lu, S.-H.; Hung, T.-F.; Chiu, Y.-C.; You, J.-F.; Hsieh, P.-S.; et al. Circulating tumor cell count correlates with colorectal neoplasm progression and is a prognostic marker for distant metastasis in non-metastatic patients. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Musella, V.; Pietrantonio, F.; Di Buduo, E.; Iacovelli, R.; Martinetti, A.; Sottotetti, E.; Bossi, I.; Maggi, C.; Di Bartolomeo, M.; de Braud, F.; et al. Circulating tumor cells as a longitudinal biomarker in patients with advanced chemorefractory, RAS-BRAF wild-type colorectal cancer receiving cetuximab or panitumumab. Int. J. Cancer 2015, 137, 1467–1474. [Google Scholar] [CrossRef]
- Krebs, M.G.; Renehan, A.G.; Backen, A.; Gollins, S.; Chau, I.; Hasan, J.; Valle, J.W.; Morris, K.; Beech, J.; Ashcroft, L.; et al. Circulating Tumor Cell Enumeration in a Phase II Trial of a Four-Drug Regimen in Advanced Colorectal Cancer. Clin. Colorectal Cancer 2015, 14, 115.e2–122.e2. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.-L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra92. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Huang, T.; Cheng, F.; Huang, K.; Liu, M.; He, W.; Li, M.; Zhang, X.; Xu, M.; Chen, S.; et al. Monitoring colorectal cancer following surgery using plasma circulating tumor DNA. Oncol. Lett. 2018, 15, 4365–4375. [Google Scholar] [CrossRef]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.; Wang, Y.; Lee, M.; Wong, R.; Kosmider, S.; Ananda, S.; Cho, J.H.; Faragher, I.; McKendrick, J.J.; et al. Serial circulating tumor DNA (ctDNA) analysis as a prognostic marker and a real-time indicator of adjuvant chemotherapy (CT) efficacy in stage III colon cancer (CC). J. Clin. Oncol. 2018, 36, 3516. [Google Scholar] [CrossRef]
- Grasselli, J.; Elez, E.; Caratù, G.; Matito, J.; Santos, C.; Macarulla, T.; Vidal, J.; Garcia, M.; Viéitez, J.M.; Paéz, D.; et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann. Oncol. 2017, 28, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.; Rata, M.; Cunningham, D.; Koh, D.-M.; Tunariu, N.; Hahne, J.C.; Vlachogiannis, G.; Hedayat, S.; Marchetti, S.; Lampis, A.; et al. Functional imaging and circulating biomarkers of response to regorafenib in treatment-refractory metastatic colorectal cancer patients in a prospective phase II study. Gut 2018, 67, 1484–1492. [Google Scholar] [CrossRef]
- Flamini, E.; Mercatali, L.; Nanni, O.; Calistri, D.; Nunziatini, R.; Zoli, W.; Rosetti, P.; Gardini, N.; Lattuneddu, A.; Verdecchia, G.M.; et al. Free DNA and carcinoembryonic antigen serum levels: An important combination for diagnosis of colorectal cancer. Clin. Cancer Res. 2006, 12, 6985–6988. [Google Scholar] [CrossRef] [Green Version]
- Hao, T.B.; Shi, W.; Shen, X.J.; Qi, J.; Wu, X.H.; Wu, Y.; Tang, Y.Y.; Ju, S.Q. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br. J. Cancer 2014, 111, 1482–1489. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Fei, F.; Zhang, M.; Li, Y.; Zhang, X.; Zhu, S.; Zhang, S. The role of mSEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer 2019, 19, 450. [Google Scholar] [CrossRef] [Green Version]
- Link, A.; Balaguer, F.; Shen, Y.; Nagasaka, T.; Lozano, J.J.; Richard Boland, C.; Goel, A. Fecal microRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1766–1774. [Google Scholar] [CrossRef] [Green Version]
- Ya, G.; Wang, H.; Ma, Y.; Hu, A.; Ma, Y.; Hu, J.; Yu, Y. Serum miR-129 functions as a biomarker for colorectal cancer by targeting estrogen receptor (ER) β. Pharmazie 2017, 72, 107–112. [Google Scholar] [CrossRef]
- He, H.W.; Wang, N.N.; Yi, X.M.; Tang, C.P.; Wang, D. Low-level serum miR-24-2 is associated with the progression of colorectal cancer. Cancer Biomark. 2018, 21, 261–267. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Z.; Chen, W. Novel circulating microRNAs expression profile in colon cancer: A pilot study. Eur. J. Med. Res. 2017, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toiyama, Y.; Hur, K.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann. Surg. 2014, 259, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Zhao, Y.; Song, X.; Song, X.; Niu, L.; Xie, L. Tumor-derived exosomal miRNA-320d as a biomarker for metastatic colorectal cancer. J. Clin. Lab. Anal. 2019, 33. [Google Scholar] [CrossRef] [Green Version]
- Koga, Y.; Yamazaki, N.; Yamamoto, Y.; Yamamoto, S.; Saito, N.; Kakugawa, Y.; Otake, Y.; Matsumoto, M.; Matsumura, Y. Fecal miR-106a is a useful marker for colorectal cancer patients with false-negative results in immunochemical fecal occult blood test. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1844–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sazanov, A.A.; Kiselyova, E.V.; Zakharenko, A.A.; Romanov, M.N.; Zaraysky, M.I. Plasma and saliva miR-21 expression in colorectal cancer patients. J. Appl. Genet. 2017, 58, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Fu, F.; Jiang, W.; Zhou, L.; Chen, Z. Circulating exosomal miR-17-5p and miR-92a-3p predict pathologic stage and grade of colorectal cancer. Transl. Oncol. 2018, 11, 221–232. [Google Scholar] [CrossRef]
- Tsukamoto, M.; Iinuma, H.; Yagi, T.; Matsuda, K.; Hashiguchi, Y. Circulating exosomal MicroRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology 2017, 92, 360–370. [Google Scholar] [CrossRef]
- Liu, C.; Eng, C.; Shen, J.; Lu, Y.; Takata, Y.; Mehdizadeh, A.; Chang, G.J.; Rodriguez-Bigas, M.A.; Li, Y.; Chang, P.; et al. Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget 2016, 7, 76250–76260. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.; Jiang, Y.; Liang, C.; Cheng, M.; Jin, C.; Duan, Q.; Xu, D.; Yang, L.; Zhang, X.; Ren, B.; et al. Exosomal miR-6803-5p as potential diagnostic and prognostic marker in colorectal cancer. J. Cell. Biochem. 2018, 119, 4113–4119. [Google Scholar] [CrossRef]
- Liu, X.; Pan, B.; Sun, L.; Chen, X.; Zeng, K.; Hu, X.; Xu, T.; Xu, M.; Wang, S. Circulating exosomal miR-27a and miR-130a act as novel diagnostic and prognostic biomarkers of colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2018, 27, 746–754. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.-Y.; Gu, R.-H.; Yan, B. Downregulation of exosome-encapsulated miR-548c-5p is associated with poor prognosis in colorectal cancer. J. Cell. Biochem. 2018, 120, 1457–1463. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Liu, Y.; Zhang, J.; Bian, Z.; Yao, S.; Fei, B.; Zhou, L.; Yin, Y.; Huang, Z. A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother. Pharmacol. 2019, 84, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Yagi, T.; Iinuma, H.; Hayama, T.; Matsuda, K.; Nozawa, K.; Tsukamoto, M.; Shimada, R.; Akahane, T.; Tsuchiya, T.; Ozawa, T.; et al. Plasma exosomal microRNA-125b as a monitoring biomarker of resistance to mFOLFOX6-based chemotherapy in advanced and recurrent colorectal cancer patients. Mol. Clin. Oncol. 2019, 11, 416–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Han, D.; Yuan, Z.; Hu, H.; Zhao, Z.; Yang, R.; Jin, Y.; Zou, C.; Chen, Y.; Wang, G.; et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shang, J.; Zhang, Y.; Liu, S.; Peng, Y.; Zhou, Z.; Pan, H.; Wang, X.; Chen, L.; Zhao, Q. MEG3 is a prognostic factor for CRC and promotes chemosensitivity by enhancing oxaliplatin-induced cell apoptosis. Oncol. Rep. 2017, 38, 1383–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, F.; Liang, W.; Qian, J. The identification of CRNDE, H19, UCA1 and HOTAIR as the key lncRNAs involved in oxaliplatin or irinotecan resistance in the chemotherapy of colorectal cancer based on integrative bioinformatics analysis. Mol. Med. Rep. 2019, 20, 3583–3596. [Google Scholar] [CrossRef]
- Tang, J.; Yan, T.; Bao, Y.; Shen, C.; Yu, C.; Zhu, X.; Tian, X.; Guo, F.; Liang, Q.; Liu, Q.; et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat. Commun. 2019, 10, 3499. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, X.; Gao, S.; Jing, F.; Yang, Y.; Du, L.; Zheng, G.; Li, P.; Li, C.; Wang, C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget 2016, 7, 85551–85563. [Google Scholar] [CrossRef]
- Liang, Z.-X.; Liu, H.-S.; Wang, F.-W.; Xiong, L.; Zhou, C.; Hu, T.; He, X.-W.; Wu, X.-J.; Xie, D.; Wu, X.-R.; et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019, 10, 829. [Google Scholar] [CrossRef] [Green Version]
- Bénard, F.; Barkun, A.N.; Martel, M.; von Renteln, D. Systematic review of colorectal cancer screening guidelines for average-risk adults: Summarizing the current global recommendations. World J. Gastroenterol. 2018, 24, 124–138. [Google Scholar] [CrossRef]
- Marcuello, M.; Vymetalkova, V.; Neves, R.P.L.; Duran-Sanchon, S.; Vedeld, H.M.; Tham, E.; van Dalum, G.; Flügen, G.; Garcia-Barberan, V.; Fijneman, R.J.A.; et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. Asp. Med. 2019, 69, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Cima, I.; Kong, S.L.; Sengupta, D.; Tan, I.B.; Phyo, W.M.; Lee, D.; Hu, M.; Iliescu, C.; Alexander, I.; Goh, W.L.; et al. Tumor-derived circulating endothelial cell clusters in colorectal cancer. Sci. Transl. Med. 2016, 8, 345ra89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, F.; Wang, Q.; Dong, Q.; Wang, Y.; Zhang, L.; Zhang, J. Circulating tumor DNA in colorectal cancer: Opportunities and challenges. Am. J. Transl. Res. 2020, 12, 1044–1055. [Google Scholar]
- Wang, X.; Shi, X.-Q.; Zeng, P.-W.; Mo, F.-M.; Chen, Z.-H. Circulating cell free DNA as the diagnostic marker for colorectal cancer: A systematic review and meta-analysis. Oncotarget 2018, 9, 24514–24524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, H.; Zhao, Q.; Wei, W.; Zheng, L.; Yi, S.; Li, G.; Wang, W.; Sheng, H.; Pu, H.; Mo, H.; et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Nian, J.; Sun, X.; Ming, S.; Yan, C.; Ma, Y.; Feng, Y.; Yang, L.; Yu, M.; Zhang, G.; Wang, X. Diagnostic accuracy of methylated SEPT9 for blood-based colorectal cancer detection: A systematic review and meta-analysis. Clin. Transl. Gastroenterol. 2017, 8, e216. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. Lausanne 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Ragusa, M.; Statello, L.; Maugeri, M.; Majorana, A.; Barbagallo, D.; Salito, L.; Sammito, M.; Santonocito, M.; Angelica, R.; Cavallaro, A.; et al. Specific alterations of the microRNA transcriptome and global network structure in colorectal cancer after treatment with MAPK/ERK inhibitors. J. Mol. Med. 2012, 90, 1421–1438. [Google Scholar] [CrossRef]
- Chen, B.; Xia, Z.; Deng, Y.-N.; Yang, Y.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. Emerging microRNA biomarkers for colorectal cancer diagnosis and prognosis. Open Biol. 2019, 9, 180212. [Google Scholar] [CrossRef] [Green Version]
- Rapado-González, Ó.; Majem, B.; Muinelo-Romay, L.; Álvarez-Castro, A.; Santamaría, A.; Gil-Moreno, A.; López-López, R.; Suárez-Cunqueiro, M.M. Human salivary microRNAs in Cancer. J. Cancer 2018, 9, 638–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tellez-Gabriel, M.; Heymann, M.-F.; Heymann, D. Circulating tumor cells as a tool for assessing tumor heterogeneity. Theranostics 2019, 9, 4580–4594. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Gao, P.; Song, Y.; Sun, J.; Chen, X.; Zhao, J.; Liu, J.; Xu, H.; Wang, Z. Relationship between circulating tumor cells and tumor response in colorectal cancer patients treated with chemotherapy: A meta-analysis. BMC Cancer 2014, 14, 976. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasari, A.; Morris, V.K.; Allegra, C.J.; Atreya, C.; Benson, A.B.; Boland, P.; Chung, K.; Copur, M.S.; Corcoran, R.B.; Deming, D.A.; et al. ctDNA applications and integration in colorectal cancer: An NCI Colon and Rectal–Anal Task Forces whitepaper. Nat. Rev. Clin. Oncol. 2020, 1–14. [Google Scholar] [CrossRef]
- Osumi, H.; Shinozaki, E.; Yamaguchi, K. Circulating tumor DNA as a novel biomarker optimizing chemotherapy for colorectal cancer. Cancers 2020, 12, 1566. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef] [Green Version]
- Khan, K.H.; Cunningham, D.; Werner, B.; Vlachogiannis, G.; Spiteri, I.; Heide, T.; Mateos, J.F.; Vatsiou, A.; Lampis, A.; Damavandi, M.D.; et al. longitudinal liquid biopsy and mathematical modeling of clonal evolution forecast time to treatment failure in the PROSPECT-C Phase II colorectal cancer clinical trial. Cancer Discov. 2018, 8, 1270–1285. [Google Scholar] [CrossRef] [Green Version]
- Siravegna, G.; Sartore-Bianchi, A.; Nagy, R.J.; Raghav, K.; Odegaard, J.I.; Lanman, R.B.; Trusolino, L.; Marsoni, S.; Siena, S.; Bardelli, A. Plasma HER2 (ERBB2) Copy Number Predicts Response to HER2-targeted Therapy in Metastatic Colorectal Cancer. Clin. Cancer Res. 2019, 25, 3046–3053. [Google Scholar] [CrossRef] [Green Version]
- To, K.K.; Tong, C.W.; Wu, M.; Cho, W.C. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J. Gastroenterol. 2018, 24, 2949–2973. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, L.; Zhang, M.; Zhang, Y.; Fan, D.; Jiang, J.; Ye, L.; Fang, X.; Chen, X.; Fan, S.; et al. Prognostic value of high-expression of miR-17-92 cluster in various tumors: Evidence from a meta-analysis. Sci. Rep. 2017, 7, 8375. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Wang, Y.; Zhang, H.; Wang, F. miR-1290 Contributes to Colorectal Cancer Cell Proliferation by Targeting INPP4B. Oncol. Res. 2018, 26, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Scola, L.; Zanghì, A.; Biondi, A.; Di Cataldo, A.; Libra, M.; Candido, S. Integrated analysis of colorectal cancer microRNA datasets: Identification of microRNAs associated with tumor development. Aging Albany N. Y 2018, 10, 1000–1014. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, M.; Majorana, A.; Statello, L.; Maugeri, M.; Salito, L.; Barbagallo, D.; Guglielmino, M.R.; Duro, L.R.; Angelica, R.; Caltabiano, R.; et al. Specific alterations of microRNA transcriptome and global network structure in colorectal carcinoma after cetuximab treatment. Mol. Cancer Ther. 2010, 9, 3396–3409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagano, T.; Fraser, P. No-nonsense functions for long noncoding RNAs. Cell 2011, 145, 178–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Tang, B.; Xiao, Y.-F.; Xie, R.; Li, B.-S.; Dong, H.; Zhou, J.-Y.; Yang, S.-M. Long non-coding RNAs in colorectal cancer. Oncotarget 2015, 7, 5226–5239. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, H.; Al-Ghafari, A.; Choudhry, H.; Al Doghaither, H. Roles of long non-coding RNAs in colorectal cancer tumorigenesis: A Review. Mol. Clin. Oncol. 2019, 11, 167–172. [Google Scholar] [CrossRef]
- He, Q.; Long, J.; Yin, Y.; Li, Y.; Lei, X.; Li, Z.; Zhu, W. Emerging Roles of lncRNAs in the Formation and Progression of Colorectal Cancer. Front. Oncol. 2020, 9. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Wang, M.; Ma, N.; Xu, Y.; Jiang, Y.; Gao, X. Long noncoding RNAs: Novel players in colorectal cancer. Cancer Lett. 2015, 361, 13–21. [Google Scholar] [CrossRef]
- Luo, J.; Qu, J.; Wu, D.-K.; Lu, Z.-L.; Sun, Y.-S.; Qu, Q. Long non-coding RNAs: A rising biotarget in colorectal cancer. Oncotarget 2017, 8, 22187–22202. [Google Scholar] [CrossRef] [Green Version]
- Valli, A.; Morotti, M.; Zois, C.E.; Albers, P.K.; Soga, T.; Feldinger, K.; Fischer, R.; Frejno, M.; McIntyre, A.; Bridges, E.; et al. Adaptation to HIF1α Deletion in Hypoxic Cancer Cells by Upregulation of GLUT14 and Creatine Metabolism. Mol. Cancer Res. 2019, 17, 1531–1544. [Google Scholar] [CrossRef] [Green Version]
- Barbagallo, C.; Brex, D.; Caponnetto, A.; Cirnigliaro, M.; Scalia, M.; Magnano, A.; Caltabiano, R.; Barbagallo, D.; Biondi, A.; Cappellani, A.; et al. LncRNA UCA1, Upregulated in CRC Biopsies and Downregulated in Serum Exosomes, Controls mRNA Expression by RNA-RNA Interactions. Mol. Ther. Nucleic Acids 2018, 12, 229–241. [Google Scholar] [CrossRef] [Green Version]
- Beije, N.; Martens, J.W.M.; Sleijfer, S. Incorporating liquid biopsies into treatment decision-making: Obstacles and possibilities. Drug Discov. Today 2019, 24, 1715–1719. [Google Scholar] [CrossRef]
- Kolenčík, D.; Shishido, S.N.; Pitule, P.; Mason, J.; Hicks, J.; Kuhn, P. Liquid Biopsy in Colorectal Carcinoma: Clinical Applications and Challenges. Cancers 2020, 12, 1376. [Google Scholar] [CrossRef]
- Leers, M.P.G. Circulating tumor DNA and their added value in molecular oncology. Clin. Chem. Lab. Med. CCLM 2020, 58, 152–161. [Google Scholar] [CrossRef] [Green Version]
- Hong, B.; Zu, Y. Detecting circulating tumor cells: Current challenges and new trends. Theranostics 2013, 3, 377–394. [Google Scholar] [CrossRef] [Green Version]
- Mamdouhi, T.; Twomey, J.D.; McSweeney, K.M.; Zhang, B. Fugitives on the run: Circulating tumor cells (CTCs) in metastatic diseases. Cancer Metastasis Rev. 2019, 38, 297–305. [Google Scholar] [CrossRef] [Green Version]
- Millner, L.M.; Linder, M.W.; Valdes, R. Circulating tumor cells: A review of present methods and the need to identify heterogeneous phenotypes. Ann. Clin. Lab. Sci. 2013, 43, 295–304. [Google Scholar]
- Barrière, G.; Tartary, M.; Rigaud, M. Epithelial mesenchymal transition: A new insight into the detection of circulating tumor cells. ISRN Oncol. 2012, 2012. [Google Scholar] [CrossRef] [Green Version]
- Castro-Giner, F.; Gkountela, S.; Donato, C.; Alborelli, I.; Quagliata, L.; Ng, C.K.Y.; Piscuoglio, S.; Aceto, N. Cancer diagnosis using a liquid biopsy: Challenges and expectations. Diagnostics 2018, 8, 31. [Google Scholar] [CrossRef] [Green Version]
- Elazezy, M.; Joosse, S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 2018, 16, 370–378. [Google Scholar] [CrossRef]
- D’Haene, N.; Fontanges, Q.; De Nève, N.; Blanchard, O.; Melendez, B.; Delos, M.; Dehou, M.-F.; Maris, C.; Nagy, N.; Rousseau, E.; et al. Clinical application of targeted next-generation sequencing for colorectal cancer patients: A multicentric Belgian experience. Oncotarget 2018, 9, 20761–20768. [Google Scholar] [CrossRef]
- Qin, D. Next-generation sequencing and its clinical application. Cancer Biol. Med. 2019, 16, 4–10. [Google Scholar] [CrossRef]
- Narayan, A.; Carriero, N.J.; Gettinger, S.N.; Kluytenaar, J.; Kozak, K.R.; Yock, T.I.; Muscato, N.E.; Ugarelli, P.; Decker, R.H.; Patel, A.A. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012, 72, 3492–3498. [Google Scholar] [CrossRef] [Green Version]
- Couraud, S.; Vaca-Paniagua, F.; Villar, S.; Oliver, J.; Schuster, T.; Blanché, H.; Girard, N.; Trédaniel, J.; Guilleminault, L.; Gervais, R.; et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: A proof-of-concept study from BioCAST/IFCT-1002. Clin. Cancer Res. 2014, 20, 4613–4624. [Google Scholar] [CrossRef] [Green Version]
- Fontanges, Q.; De Mendonca, R.; Salmon, I.; Le Mercier, M.; D’Haene, N. Clinical application of targeted next generation sequencing for colorectal cancers. Int. J. Mol. Sci. 2016, 17, 2117. [Google Scholar] [CrossRef]
- Grada, A.; Weinbrecht, K. Next-generation sequencing: Methodology and application. J. Investig. Dermatol. 2013, 133, e11. [Google Scholar] [CrossRef] [Green Version]
- Ng, C.; Li, H.; Wu, W.K.K.; Wong, S.H.; Yu, J. Genomics and metagenomics of colorectal cancer. J. Gastrointest. Oncol. 2019, 10, 1164–1170. [Google Scholar] [CrossRef]
- Kim, R.Y.; Xu, H.; Myllykangas, S.; Ji, H. Genetic-based biomarkers and next-generation sequencing: The future of personalized care in colorectal cancer. Pers. Med. 2011, 8, 331–345. [Google Scholar] [CrossRef] [Green Version]
- Wadapurkar, R.M.; Vyas, R. Computational analysis of next generation sequencing data and its applications in clinical oncology. Inform. Med. Unlocked 2018, 11, 75–82. [Google Scholar] [CrossRef]
- Rachiglio, A.M.; Abate, R.E.; Sacco, A.; Pasquale, R.; Fenizia, F.; Lambiase, M.; Morabito, A.; Montanino, A.; Rocco, G.; Romano, C.; et al. Limits and potential of targeted sequencing analysis of liquid biopsy in patients with lung and colon carcinoma. Oncotarget 2016, 7, 66595–66605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Vecchio, F.; Mastroiaco, V.; Di Marco, A.; Compagnoni, C.; Capece, D.; Zazzeroni, F.; Capalbo, C.; Alesse, E.; Tessitore, A. Next-generation sequencing: Recent applications to the analysis of colorectal cancer. J. Transl. Med. 2017, 15, 246. [Google Scholar] [CrossRef] [Green Version]
- Soda, N.; Rehm, B.H.A.; Sonar, P.; Nguyen, N.-T.; Shiddiky, M.J.A. Advanced liquid biopsy technologies for circulating biomarker detection. J. Mater. Chem. B 2019, 7, 6670–6704. [Google Scholar] [CrossRef]
- Risberg, B.; Tsui, D.W.Y.; Biggs, H.; Ruiz-Valdepenas Martin de Almagro, A.; Dawson, S.-J.; Hodgkin, C.; Jones, L.; Parkinson, C.; Piskorz, A.; Marass, F.; et al. Effects of Collection and Processing Procedures on Plasma Circulating Cell-Free DNA from Cancer Patients. J. Mol. Diagn. 2018, 20, 883–892. [Google Scholar] [CrossRef] [Green Version]
- Grölz, D.; Hauch, S.; Schlumpberger, M.; Guenther, K.; Voss, T.; Sprenger-Haussels, M.; Oelmüller, U. Liquid Biopsy Preservation Solutions for Standardized Pre-Analytical Workflows—Venous Whole Blood and Plasma. Curr. Pathobiol. Rep. 2018, 6, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Harouaka, R.; Kang, Z.; Zheng, S.; Cao, L. Circulating tumor cells: Advances in isolation and analysis, and challenges for clinical applications. Pharmacol. Ther. 2014, 141, 209–221. [Google Scholar] [CrossRef] [Green Version]
- Tamkovich, S.N.; Cherepanova, A.V.; Kolesnikova, E.V.; Rykova, E.Y.; Pyshnyi, D.V.; Vlassov, V.V.; Laktionov, P.P. Circulating DNA and DNase activity in human blood. Ann. N. Y. Acad. Sci. 2006, 1075, 191–196. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.B.; Siu, L.L.; et al. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019, 30, 1580–1590. [Google Scholar] [CrossRef] [Green Version]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef]
- Haselmann, V.; Ahmad-Nejad, P.; Geilenkeuser, W.J.; Duda, A.; Gabor, M.; Eichner, R.; Patton, S.; Neumaier, M. Results of the first external quality assessment scheme (EQA) for isolation and analysis of circulating tumour DNA (ctDNA). Clin. Chem. Lab. Med. 2018, 56, 220–228. [Google Scholar] [CrossRef]
- Vivancos, A.; Aranda, E.; Benavides, M.; Élez, E.; Gómez-España, M.A.; Toledano, M.; Alvarez, M.; Parrado, M.R.C.; García-Barberán, V.; Diaz-Rubio, E. Comparison of the Clinical Sensitivity of the Idylla Platform and the OncoBEAM RAS CRC Assay for KRAS Mutation Detection in Liquid Biopsy Samples. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
| Study (Year) | Biomarkers | Sample Size | Methods | Statistical Significance (p Value), Sensitivity/Specificity (%) and/or Hazard Ratio | Potential Clinical Applications |
|---|---|---|---|---|---|
| Tsai et al. (2018) [25] | CTC | n = 620 (n = 438 adenoma, polyps, or stage I–IV CRC, n = 182 healthy controls). | CellMax biomimetic platform (CMx) | All subjects: Sn 84.0/Sp 97.3 Precancerous lesions: Sn 76.6/Sp 97.3 CRC: Sn 86.9/Sp 97.3 | Screening |
| Bork et al. (2015) [26] | CTC | Total n = 287 (n = 239 stage I–III CRC) | CellSearch | OS: HR 5.5 (95% CI 2.3–13.6, p < 0.001) PFS: HR 12.7 (95% CI 5.2–31.1, p < 0.001) | Prognostic in non-mCRC |
| Gazzaniga et al. (2013) [27] | CTC | n = 37 high-risk stage II or III CRC | CellSearch | The presence of CTC was detected in 8 of 37 patients (22%) 87.5% of CTC-positive patients had N1–2 disease and stage III CRC | Selection of high-risk stage II CRC patient candidates for adjuvant chemotherapy |
| Tsai et al. (2016) [28] | CTC | n = 158 (n = 27 healthy, n = 21 benign, n = 95 non-mCRC, n = 15 m-CRC) | CellMax biomimetic platform (CMx) | CRC: Sn 63.0/Sp 82.0 All colorectal neoplasms, including adenomatous polyps, dysplastic polyps, and CRC: Sn 61.0/Sp 94.0 | Prognostic in non-mCRC at high risk of early recurrence |
| Musella et al. (2015) [29] | CTC | n = 38 advanced RAS-BRAF-wild-type CRC receiving third-line therapy with cetuximab-irinotecan or panitumumab. | AdnaTest ColonCancerSelect | OS: HR 8.06 (95% CI, 2.54–25.59, p < 0.001) PFS: HR 6.10 (95% CI, 2.49–14.96, p < 0.001) | Prognostic and predictive in CRC patients treated with anti-EGFR monoclonal antibodies |
| Krebs et al. (2014) [30] | CTC | n = 48 (CTC enumeration performed only in 42 patients) | CellSearch | ORR: 71% Median OS for high and low CTC count: 18.7 and 22.3 months (log-rank test, p < 0.038) | Prognostic in CRC patients treated with irinotecan, oxaliplatin, and tegafur-uracil with leucovorin and cetuximab |
| Tie et al. (2016) [31] | ctDNA | n = 230 resected stage II colon cancer | Safe-SeqS | Postoperative recurrence at 36 months: Sn 48.0/Sp 100.0 | Monitoring of MRD and identification of CRC patients at very high risk of recurrence |
| Sun et al. (2018) [32] | ctDNA | n = 11 CRC treated surgically | NGS | n = 7: decreased mutation rates in postoperative vs. preoperative period n = 4: no mutations n = 1 patient with metastatic rectal cancer: the rate of TP53 mutation increased from 8.95 (preoperative) to 71.4% (postoperative) | Prognostic and Predictive |
| Tie et al. (2015) [33] | ctDNA | n = 53 mCRC patients receiving standard first-line chemotherapy | Safe-SeqS | 10-fold change ctDNA threshold: Sn 75.0/Sp 64.0 | Predictive during first-line chemotherapy |
| Tie et al. (2018) [34] | ctDNA | n = 95 stage III colon cancer receiving adjuvant chemotherapy | Safe-SeqS | Inferior RFS: in case of positive ctDNA post-surgery (HR 3.52, p = 0.004). Superior RFS: when ctDNA became undetectable after chemotherapy (HR 5.11, p = 0.02). Inferior RFS: when ctDNA status changed from negative to positive after chemotherapy (HR 5.30, p = 0.006). Inferior RFS: positive ctDNA after adjuvant chemotherapy completion (HR 7.14, p < 0.001) | Prognostic and therapy monitoring in stage III colon cancer |
| Grasselli et al. (2017) [35] | ctDNA | n = 146 mCRC patients | SoC PCR and Digital PCR (BEAMing) | ctDNA BEAMing RAS testing showed 89.7% agreement with SoC (Kappa index 0.80, 95% CI 0.71–0.90) BEAMing in tissue showed 90.9% agreement with SoC (Kappa index 0.83, 95% CI 0.74–0.92) | Predictive and anti-EGFR treatment selection |
| Khan et al. (2018) [36] | ctDNA | n = 27 RAS mutant mCRC | Digital-droplet PCR | PFS: HR 0.21 (95% CI 0.06–0.71, p = 0.01) | Predictive of duration of anti-angiogenic response to regorafenib |
| Flamini et al. (2006) [37] | ctDNA | n = 75 healthy subjects n = 75 CRC | qPCR | ctDNA alone: Sn 81.3/Sp 73.3 ctDNA + CEA: Sn 84.0/Sp 88.0 | Diagnosis of early-stage CRC |
| Hao et al. (2014) [38] | ctDNA | n = 104 primary CRC, n = 85 operated CRC, n = 16 recurrent/mCRC, n = 63 intestinal polyps, n = 110 normal controls | ALU-qPCR | ALU115: Sn 69.23/Sp 99.09 ALU247/115: Sn 73.08/Sp 97.27 | Early complementary diagnosis, monitoring of progression and prognosis of CRC |
| Sun et al. (2019) [39] | mSEPT9 DNA | n = 650 | Epigenomics AG for Epi proColon 2.0 | CRC: Sn 73.0/Sp 94.5 Polyps and adenoma: Sn 17.1/Sp 94.5 | Screening and recurrence monitoring |
| Link et al. (2010) [40] | Fecal miRNAs | n = 8 healthy controls, n = 29 normal colonoscopies, colon adenomas, and CRCs | TaqMan qRT-PCR | Increased expression of miR-21 and miR-106a in CRC and adenomas vs. normal controls (p < 0.05) | Screening |
| Ya et al. (2017) [41] | Serum miR-129 | n = 18 female patients with CRC | Real-time PCR | Contribution to carcinogenesis by targeting ERβ (p < 0.01) | Development of therapeutic agents |
| He et al. (2018) [42] | Serum miR-24-2 | n = 68 healthy subjects, n = 228 CRC | Real-time qRT-PCR | Higher levels in CRC than healthy subjects (p < 0.05) | Negative biomarker in the diagnosis of the progression of CRC |
| Wang et al. (2017) [43] | Serum miR-31, miR-141, miR-224-3p, miR-576-5p, and miR-4669 | n = 44 healthy subjects, n = 50 CRC. Double-blind validation using sera from 30 CRC, 30 colonic polyps, 30 healthy controls | Real-time PCR | AUC = 0.995 (microarrays) AUC = 0.964 (double-blind validation test) | Panel for diagnosis of CRC |
| Toiyama et al. (2014) [44] | Serum miR-200c | Total n = 446 colorectal specimens. First phase: n = 12 stage I and IV CRC. Second phase: n = 182 CRC, n = 24 controls. Third phase: n = 156 tumor tissues from 182 CRC and an independent set of 20 matched primary CRC and corresponding liver mts | Real-time qRT-PCR | Correlation with lymph node mts (p = 0.0026), distant mts (p = 0.0023), and prognosis (p = 0.0064) Predictor for lymph node mts (OR 4.81, 95% CI 1.98–11.7, p = 0.0005) and tumor recurrence (HR 4.51, 95% CI 1.56–13.01, p = 0.005) Prognostic (HR 2.67, 95% CI 1.28–5.67, p = 0.01) | Prognostic and predictive of metastasis |
| Tang et al. (2019) [45] | Exosomal miR-320d | n = 34 mCRC, n = 108 non-mCRC | qPCR | miR-320d: AUC = 0.633, p = 0.019 miR-320d + CEA: AUC = 0.804 | Predictive of metastasis |
| Koga et al. (2013) [46] | Fecal miR-106a | n = 117 CRC, n = 107 healthy subjects | Real-time RT-PCR | FmiRT: Sn 34.2/Sp 97.2. iFOBT + FmiRT: Sn 70.9/Sp 96.3 | Screening |
| Sazanov et al. (2017) [47] | Plasma and saliva miR-21 | Plasma: total n = 65 CRC (n = 34 controls, n = 6 stage II, n = 16 stage III, n = 9 stage IV) Saliva: total n = 68 CRC (n = 34 controls, n = 6 stage II, n = 18 stage III, n = 10 stage IV) | Real-time qRT-PCR | Plasma: Sn 65/Sp 85 Saliva: Sn 97/Sp 91 | Screening |
| Fu et al. (2018) [48] | Exosomal miR-17-5p and miR-92a-3p | n = 10 normal controls, n = 18 CRC, n = 11 mCRC | Real-time qPCR | miR-17-5p: AUC = 0.897 (95% CI 0.800–0.994) for CRC, and 0.841 (95% CI 0.720–0.962) for mts miR-92a-3p: AUC = 0.845 (95% CI 0.724–0.966) for CRC and 0.854 (95% CI 0.735–0.973) for mts miR-17-5p + miR-92a-3p: AUC = 0.910 (95% CI 0.820–1) for CRC and 0.841 (95% CI 0.718–0.964) for mts | Prognostic |
| Tsukamoto et al. (2017) [49] | Exosomal miR-21 | Total n = 326 CRC (n = 51 stage I, n = 110 stage II, n = 98 stage III, n = 67 stage IV) | TaqMan miRNA assays | OS: HR 2.28 (95% CI 1.81–5.74, p < 0.01) DFS: HR 2.34 (95% CI 1.87–4.60, p < 0.01) | Prediction of recurrence and poor prognosis in CRC patients with TNM stage II, III, or IV |
| Liu et al. (2016) [50] | Exosomal miR-4772-3p | n = 84 stage II–III colon cancer | Real-time qRT-PCR | AUC = 0.72 (95% CI 0.59–0.85, p = 0.001) | Prognostic for tumor recurrence in stage II and III colon cancer patients |
| Yan et al. (2018) [51] | Exosomal miR-6803-5p | n = 168 CRC | qRT-PCR | OS: HR 2.93 (95% CI 1.35–6.37, p < 0.007) DFS: HR 3.26 (95% CI 1.56–6.81, p < 0.002) AUC = 0.7399 | Diagnostic and prognostic |
| Liu et al. (2018) [52] | Exosomal miR-27a and miR-130a | Training phase: n = 40 healthy subjects n = 40 stage I CRC. Validation phase: n = 40 stage I, n = 20 stage II, n = 14 stage III, n = 6 stage IV CRC, n = 40 healthy subjects. External validation phase: 50 stage I CRC, 50 adenomas, 50 healthy subjects | qRT-PCR | miR-27a: AUC = 0.773 Sn 75/Sp 77.5 in the training phase, AUC = 0.82 Sn 80.0/Sp 77.5 in the validation phase, and AUC = 0.746 Sn 80.0/Sp 77.5 in the external validation phase miR-130a: AUC = 0.742 Sn 82.5/Sp 62.5 in the training phase, AUC = 0.787 Sn 70.0/Sp 80.0 in the validation phase, AUC = 0.697 Sn 70.0/Sp 80.0 in the external validation phase miR-27a + miR-130a: training phase AUC = 0.846 Sn 82.5/Sp 75, validation phase AUC = 0.898, Sn 80.0/Sp 90.0 and external validation phase AUC = 0.801 Sn 80.0/Sp 90.0 | Diagnostic and prognostic |
| Peng et al. (2018) [53] | Exosomal miR-548c-5p | n = 108 CRC | Real-time qPCR | OS: HR 3.40 (95% CI 1.02–11.27, p = 0.046) | Diagnostic and prognostic |
| Jin et al. (2019) [54] | Exosomal miR-21-5p, miR-1246, miR-1229-5p, and miR-96-5p | Drug-resistant CRC cell lines | qRT-PCR | AUC = 0.804, p < 0.05 | Predictive for chemoresistance in advanced CRC |
| Yagi et al. (2019) [55] | Exosomal miR-125b | n = 55 patients with advanced/recurrent CRC treated with mFOLFOX6 | qRT-PCR | PFS: HR 0.71 (95% CI 0.36–0.94, p < 0.041) | Predictive and detection of chemotherapy resistance |
| Wang et al. (2018) [56] | lncRNA H19 | n = 110 paired CRC tissues and para-tumor tissues | qRT-PCR | RFS: log-rank test p < 0.001 High H19: HR 2.383 (95% CI 1.157–4.909, p = 0.018) | Predictive of 5-FU resistance |
| Li et al. (2017) [57] | lncRNA MEG3 | n = 316 CRC | qRT-PCR | AUC = 0.784, Sn 72.86/Sp 61.43 OS: HR 1.390 (95% CI 0.324–2.089, p = 0.007) | Prognostic and promotion of chemosensitivity |
| Sun et al. (2019) [58] | lncRNAs CRNDE, H19, UCA1, and HOTAIR | CRC cell lines (HCT116, HT29, and LoVo) | Gene Expression Profiling Interactive Analysis | HOTAIR OS: HR 1.9, p = 0.0066 DFS: HR 1.8, p = 0.012 | Predictive of treatment sensitivity |
| Tang et al. (2019) [59] | lncRNA GLCC1 | In vitro: Human colorectal cancer cell lines SW1116, SW480, Caco2, LoVo, HT29, RKO, DLD-1, and HCT116 In vivo: BALB/c nude mice | Real-time qPCR | Stabilization of c-Myc after knockdown of lncGLCC1 (p < 0.001) | Prognostic |
| Liu et al. (2016) [60] | Exosomal lncRNA CRNDE-h | n = 468 | qRT-PCR | AUC = 0.892 Sn 70.3/Sp 94.4 | Diagnostic and prognostic |
| Liang et al. (2019) [61] | Exosomal lncRNA RPPH1 | n = 61 CRC | qRT-PCR | OS: HR 2.145 (95% CI 1.450–3.174, p < 0.001) DFS: HR 1.820 (95% CI 1.257–2.637, p = 0.001) | Prognostic, therapeutic, and diagnostic target |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vacante, M.; Ciuni, R.; Basile, F.; Biondi, A. The Liquid Biopsy in the Management of Colorectal Cancer: An Overview. Biomedicines 2020, 8, 308. https://doi.org/10.3390/biomedicines8090308
Vacante M, Ciuni R, Basile F, Biondi A. The Liquid Biopsy in the Management of Colorectal Cancer: An Overview. Biomedicines. 2020; 8(9):308. https://doi.org/10.3390/biomedicines8090308
Chicago/Turabian StyleVacante, Marco, Roberto Ciuni, Francesco Basile, and Antonio Biondi. 2020. "The Liquid Biopsy in the Management of Colorectal Cancer: An Overview" Biomedicines 8, no. 9: 308. https://doi.org/10.3390/biomedicines8090308
APA StyleVacante, M., Ciuni, R., Basile, F., & Biondi, A. (2020). The Liquid Biopsy in the Management of Colorectal Cancer: An Overview. Biomedicines, 8(9), 308. https://doi.org/10.3390/biomedicines8090308