Using Artificial Neural Network to Discriminate Parkinson’s Disease from Other Parkinsonisms by Focusing on Putamen of Dopamine Transporter SPECT Images
Abstract
:1. Introduction
2. Material and Methods
2.1. Subjects
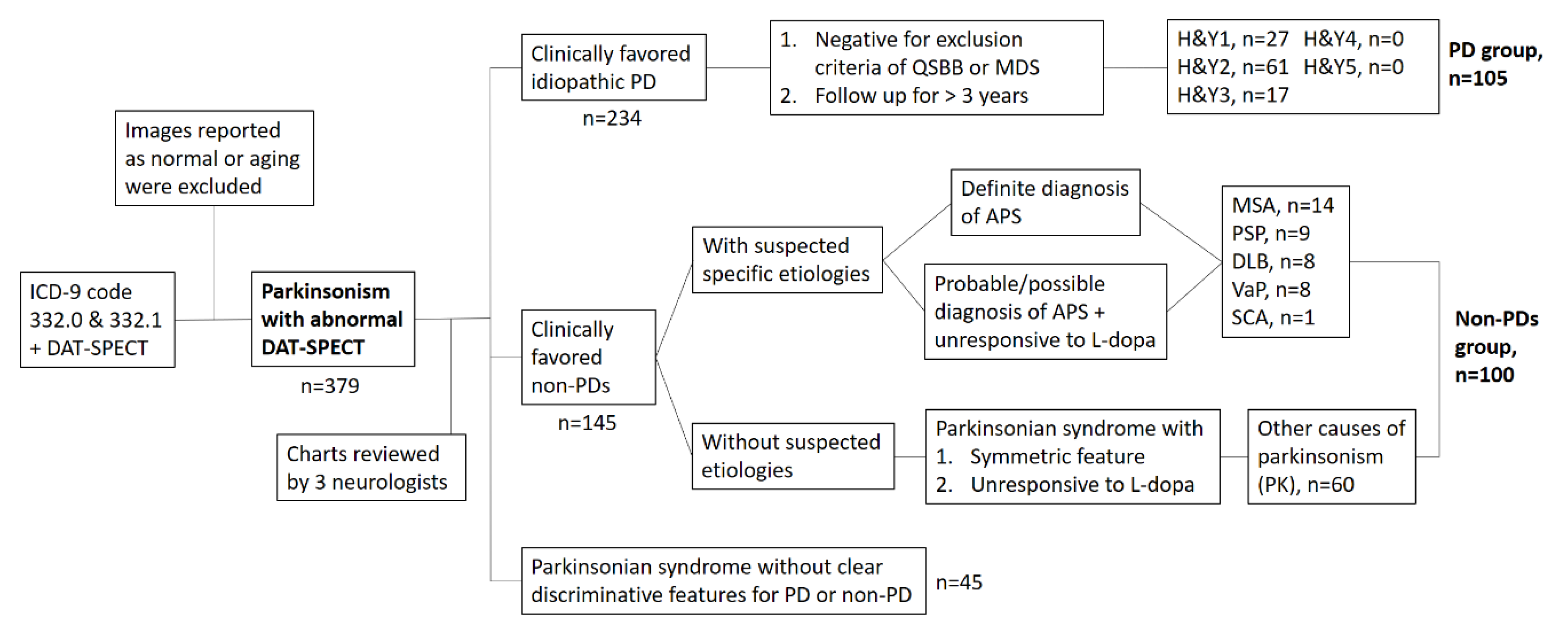
2.1.1. First Set of Images for ANN Training and Validation
2.1.2. Second Set of Images for Testing the ANN Classifier
2.2. Image Processing
2.2.1. Image Pre-Processing
2.2.2. Binary Classification by ANN
2.2.3. Semi-Quantitative Measurements and Machine-Learning Classification
2.2.4. Class-Activation Mapping to Visually Explain the ANN Classifier
3. Results
3.1. Demographic Characteristics
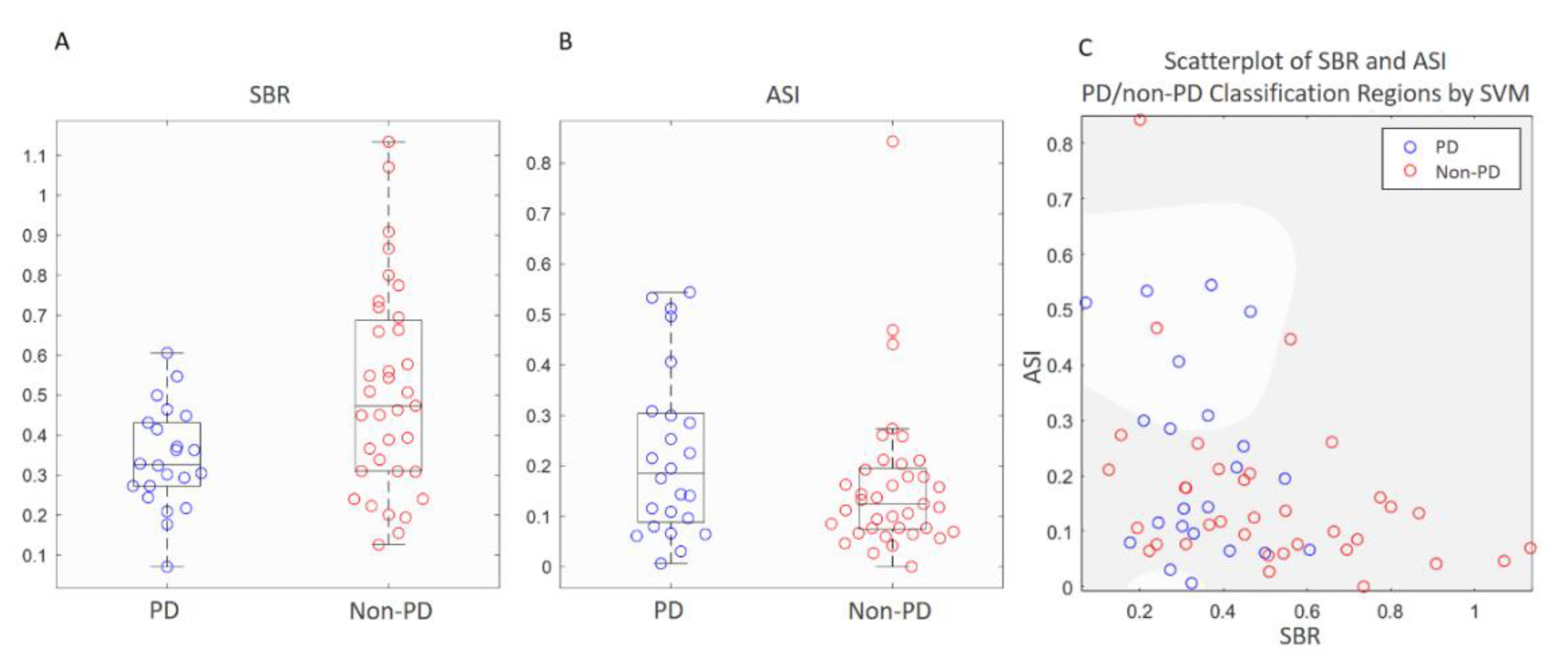
3.2. Comparisons of Semi-Quantitative Measurements and ANN Classifier
3.3. Visualization of Computer-Vision through CAM
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
DAT-SPECT Scan and Reconstruction Protocol
References
- Sardi, S.P.; Simuni, T. New Era in disease modification in Parkinson’s disease: Review of genetically targeted therapeutics. Parkinsonism Relat. Disord. 2019, 59, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.E.; Espay, A.J. Disease Modification in Parkinson’s Disease: Current Approaches, Challenges, and Future Considerations. Mov. Disord. 2018, 33, 660–677. [Google Scholar] [CrossRef] [PubMed]
- Postuma, R.B.; Berg, D.; Stern, M.; Poewe, W.; Olanow, C.W.; Oertel, W.; Obeso, J.; Marek, K.; Litvan, I.; Lang, A.E.; et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov. Disord. 2015, 30, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Marshall, V.L.; Reininger, C.B.; Marquardt, M.; Patterson, J.; Hadley, D.M.; Oertel, W.H.; Benamer, H.T.S.; Kemp, P.; Burn, D.; Tolosa, E.; et al. Parkinson’s disease is overdiagnosed clinically at baseline in diagnostically uncertain cases: A 3-year European multicenter study with repeat [123I]FP-CIT SPECT. Mov. Disord. 2009. [Google Scholar] [CrossRef]
- Berardelli, A.; Wenning, G.K.; Antonini, A.; Berg, D.; Bloem, B.R.; Bonifati, V.; Brooks, D.; Burn, D.J.; Colosimo, C.; Fanciulli, A.; et al. EFNS/MDS-ES/ENS [corrected] recommendations for the diagnosis of Parkinson’s disease. Eur. J. Neurol. 2013, 20, 16–34. [Google Scholar] [CrossRef]
- Tolosa, E.; Wenning, G.; Poewe, W. The diagnosis of Parkinson’s disease. Lancet Neurol. 2006, 5, 75–86. [Google Scholar] [CrossRef]
- Huppertz, H.J.; Möller, L.; Südmeyer, M.; Hilker, R.; Hattingen, E.; Egger, K.; Amtage, F.; Respondek, G.; Stamelou, M.; Schnitzler, A.; et al. Differentiation of neurodegenerative parkinsonian syndromes by volumetric magnetic resonance imaging analysis and support vector machine classification. Mov. Disord. 2016. [Google Scholar] [CrossRef]
- Yang, J.; Archer, D.B.; Burciu, R.G.; Muller, M.; Roy, A.; Ofori, E.; Bohnen, N.I.; Albin, R.L.; Vaillancourt, D.E. Multimodal dopaminergic and free-water imaging in Parkinson’s disease. Parkinsonism Relat. Disord. 2019, 62, 10–15. [Google Scholar] [CrossRef]
- Paviour, D.C.; Thornton, J.S.; Lees, A.J.; Jager, H.R. Diffusion-weighted magnetic resonance imaging differentiates Parkinsonian variant of multiple-system atrophy from progressive supranuclear palsy. Mov. Disord. 2007, 22, 68–74. [Google Scholar] [CrossRef]
- Sjostrom, H.; Granberg, T.; Westman, E.; Svenningsson, P. Quantitative susceptibility mapping differentiates between parkinsonian disorders. Parkinsonism Relat. Disord. 2017, 44, 51–57. [Google Scholar] [CrossRef]
- Cheng, Z.; He, N.; Huang, P.; Li, Y.; Tang, R.; Sethi, S.K.; Ghassaban, K.; Yerramsetty, K.K.; Palutla, V.K.; Chen, S.; et al. Imaging the Nigrosome 1 in the substantia nigra using susceptibility weighted imaging and quantitative susceptibility mapping: An application to Parkinson’s disease. NeuroImage Clin. 2020. [Google Scholar] [CrossRef] [PubMed]
- Shinto, A.; Vijayan, K.; Antony, J.; Kamaleshwaran, K.; Kameshwaran, M.; Korde, A.; Samuel, G.; Selvan, A. Correlative 99m Tc-labeled tropane derivative single photon emission computer tomography and clinical assessment in the staging of parkinson disease. World J. Nucl. Med. 2014. [Google Scholar] [CrossRef] [PubMed]
- Eckert, T.; Barnes, A.; Dhawan, V.; Frucht, S.; Gordon, M.F.; Feigin, A.S.; Eidelberg, D. FDG PET in the differential diagnosis of parkinsonian disorders. Neuroimage 2005, 26, 912–921. [Google Scholar] [CrossRef] [PubMed]
- Garraux, G.; Phillips, C.; Schrouff, J.; Kreisler, A.; Lemaire, C.; Degueldre, C.; Delcour, C.; Hustinx, R.; Luxen, A.; Destée, A.; et al. Multiclass classification of FDG PET scans for the distinction between Parkinson’s disease and atypical parkinsonian syndromes. NeuroImage Clin. 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenning, G.K.; Shephard, B.; Hawkes, C.; Petruckevitch, A.; Lees, A.; Quinn, N. Olfactory function in atypical parkinsonian syndromes. Acta Neurol. Scand. 1995, 91, 247–250. [Google Scholar] [CrossRef]
- Goldstein, D.S.; Holmes, C.; Bentho, O.; Sato, T.; Moak, J.; Sharabi, Y.; Imrich, R.; Conant, S.; Eldadah, B.A. Biomarkers to detect central dopamine deficiency and distinguish Parkinson disease from multiple system atrophy. Parkinsonism Relat. Disord. 2008, 14, 600–607. [Google Scholar] [CrossRef] [Green Version]
- Skowronek, C.; Zange, L.; Lipp, A. Cardiac 123I-MIBG Scintigraphy in Neurodegenerative Parkinson Syndromes: Performance and Pitfalls in Clinical Practice. Front. Neurol. 2019, 10, 152. [Google Scholar] [CrossRef] [Green Version]
- Augimeri, A.; Cherubini, A.; Cascini, G.L.; Galea, D.; Caligiuri, M.E.; Barbagallo, G.; Arabia, G.; Quattrone, A. CADA—computer-aided DaTSCAN analysis. EJNMMI Phys. 2016. [Google Scholar] [CrossRef] [Green Version]
- Nicastro, N.; Wegrzyk, J.; Preti, M.G.; Fleury, V.; Van de Ville, D.; Garibotto, V.; Burkhard, P.R. Classification of degenerative parkinsonism subtypes by support-vector-machine analysis and striatal (123)I-FP-CIT indices. J. Neurol. 2019, 266, 1771–1781. [Google Scholar] [CrossRef] [Green Version]
- Badoud, S.; Van De Ville, D.; Nicastro, N.; Garibotto, V.; Burkhard, P.R.; Haller, S. Discriminating among degenerative parkinsonisms using advanced 123I-ioflupane SPECT analyses. NeuroImage Clin. 2016. [Google Scholar] [CrossRef]
- Joling, M.; Vriend, C.; van der Zande, J.J.; Lemstra, A.W.; van den Heuvel, O.A.; Booij, J.; Berendse, H.W. Lower 123I-FP-CIT binding to the striatal dopamine transporter, but not to the extrastriatal serotonin transporter, in Parkinson’s disease compared with dementia with Lewy bodies. NeuroImage Clin. 2018. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.; van Ginneken, B.; Sanchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaccaro, M.G.; Sarica, A.; Quattrone, A.; Chiriaco, C.; Salsone, M.; Morelli, M.; Quattrone, A. Neuropsychological assessment could distinguish among different clinical phenotypes of progressive supranuclear palsy: A Machine Learning approach. J. Neuropsychol. 2020. [Google Scholar] [CrossRef]
- Choi, H.; Ha, S.; Im, H.J.; Paek, S.H.; Lee, D.S. Refining diagnosis of Parkinson’s disease with deep learning-based interpretation of dopamine transporter imaging. NeuroImage Clin. 2017, 16, 586–594. [Google Scholar] [CrossRef]
- Taylor, J.C.; Fenner, J.W. Comparison of machine learning and semi-quantification algorithms for (I123)FP-CIT classification: The beginning of the end for semi-quantification? EJNMMI Phys. 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage 2006, 31, 1116–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, T.F.; Vese, L.A. Active contours without edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef] [Green Version]
- Iizuka, T.; Fukasawa, M.; Kameyama, M. Deep-learning-based imaging-classification identified cingulate island sign in dementia with Lewy bodies. Sci. Rep. 2019, 9, 8944. [Google Scholar] [CrossRef] [Green Version]
- Vlaar, A.M.; van Kroonenburgh, M.J.; Kessels, A.G.; Weber, W.E. Meta-analysis of the literature on diagnostic accuracy of SPECT in parkinsonian syndromes. BMC Neurol. 2007, 7, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joling, M.; Vriend, C.; Raijmakers, P.G.H.M.; van der Zande, J.J.; Lemstra, A.W.; Berendse, H.W.; Booij, J.; van den Heuvel, O.A. Striatal DAT and extrastriatal SERT binding in early-stage Parkinson’s disease and dementia with Lewy bodies, compared with healthy controls: An 123 I-FP-CIT SPECT study. NeuroImage Clin. 2019. [Google Scholar] [CrossRef]
- Swanson, R.L.; Newberg, A.B.; Acton, P.D.; Siderowf, A.; Wintering, N.; Alavi, A.; Mozley, P.D.; Plossl, K.; Udeshi, M.; Hurtig, H. Differences in [99mTc]TRODAT-1 SPECT binding to dopamine transporters in patients with multiple system atrophy and Parkinson’s disease. Eur. J. Nucl. Med. Mol. Imaging 2005, 32, 302–307. [Google Scholar] [CrossRef] [PubMed]


| Data | Training/Validation Set (n = 205) | Test Set (n = 57) | ||||
|---|---|---|---|---|---|---|
| Group | PD | Non-PD | p Value | PD | Non-PD | p Value |
| Age (years) (mean ± SD) | 65.4 ± 10.2 | 66.6 ± 12.8 | 0.44 | 70.3 ± 9.8 | 70.6 ± 13.4 | 0.93 |
| Gender (F/M) | 52/53 | 45/55 | 0.51 | 8/14 | 12/23 | 0.87 |
| Mean disease duration (years) (IQR) | 2.32 (2) | 1.89 (1) | 0.27 | 2.57 (2.5) | 3.56 (3) | 0.34 |
| Classifier | SVM | ANN | |
|---|---|---|---|
| Learning Method | Machine Learning | Deep Learning | |
| Input data | SBR & ASI | Whole-brain image | SR image |
| Accuracy | 68.4% | 68.4% | 86.0% |
| Sensitivity | 31.8% | 81.8% | 81.8% |
| Specificity | 91.4% | 60.0% | 88.6% |
| Predicted Positive (Classified as PD) | Predicted Negative (Classified as non-PD) | ||
|---|---|---|---|
| Actual positive (PDs = 22) | TP 18 | FN 4 | Sensitivity (recall) 0.818 |
| Actual negative (non-PDs = 35) | FP 4 | TN 31 | Specificity 0.886 |
| Precision 0.818 | Negative Predictive value 0.886 | Accuracy 0.860 | |
| F1 score: 2 × (precision × recall)/(precision + recall) = 0.818 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chien, C.-Y.; Hsu, S.-W.; Lee, T.-L.; Sung, P.-S.; Lin, C.-C. Using Artificial Neural Network to Discriminate Parkinson’s Disease from Other Parkinsonisms by Focusing on Putamen of Dopamine Transporter SPECT Images. Biomedicines 2021, 9, 12. https://doi.org/10.3390/biomedicines9010012
Chien C-Y, Hsu S-W, Lee T-L, Sung P-S, Lin C-C. Using Artificial Neural Network to Discriminate Parkinson’s Disease from Other Parkinsonisms by Focusing on Putamen of Dopamine Transporter SPECT Images. Biomedicines. 2021; 9(1):12. https://doi.org/10.3390/biomedicines9010012
Chicago/Turabian StyleChien, Chung-Yao, Szu-Wei Hsu, Tsung-Lin Lee, Pi-Shan Sung, and Chou-Ching Lin. 2021. "Using Artificial Neural Network to Discriminate Parkinson’s Disease from Other Parkinsonisms by Focusing on Putamen of Dopamine Transporter SPECT Images" Biomedicines 9, no. 1: 12. https://doi.org/10.3390/biomedicines9010012
APA StyleChien, C.-Y., Hsu, S.-W., Lee, T.-L., Sung, P.-S., & Lin, C.-C. (2021). Using Artificial Neural Network to Discriminate Parkinson’s Disease from Other Parkinsonisms by Focusing on Putamen of Dopamine Transporter SPECT Images. Biomedicines, 9(1), 12. https://doi.org/10.3390/biomedicines9010012





