Machine Learning-Based Identification of Potentially Novel Non-Alcoholic Fatty Liver Disease Biomarkers
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcriptomics
2.1.1. Data Acquisition
2.1.2. Derivation of New Transcriptomics Cohort from Multiple GEO Datasets
2.1.3. Differential Gene Expression (DGE) Analysis
2.1.4. Random Forest-Based Predictions
2.1.5. Downstream Data Analysis
2.2. Lipidomics Data Analysis
2.2.1. Data Acquisition
2.2.2. Data Pre-Processing
2.2.3. Differential Lipid Expression Analysis
2.2.4. Random Forest and ROC Curve Analysis
2.3. Statistical Analysis
2.3.1. Heatmap-Based Visualization of Significant Lipid Features
2.3.2. LIPEA-Based Lipid Pathway Enrichment Analysis
2.3.3. Lipid Network Analysis
3. Results
3.1. Feature Selection for Random Forest Model
3.1.1. Gene Signature Identification
3.1.2. Lipid Signature Identification
3.2. Random Forest Model Performance
3.2.1. Transcriptomic Features Analysis
3.2.2. Lipidomic Features Analysis
3.3. Downstream Analysis
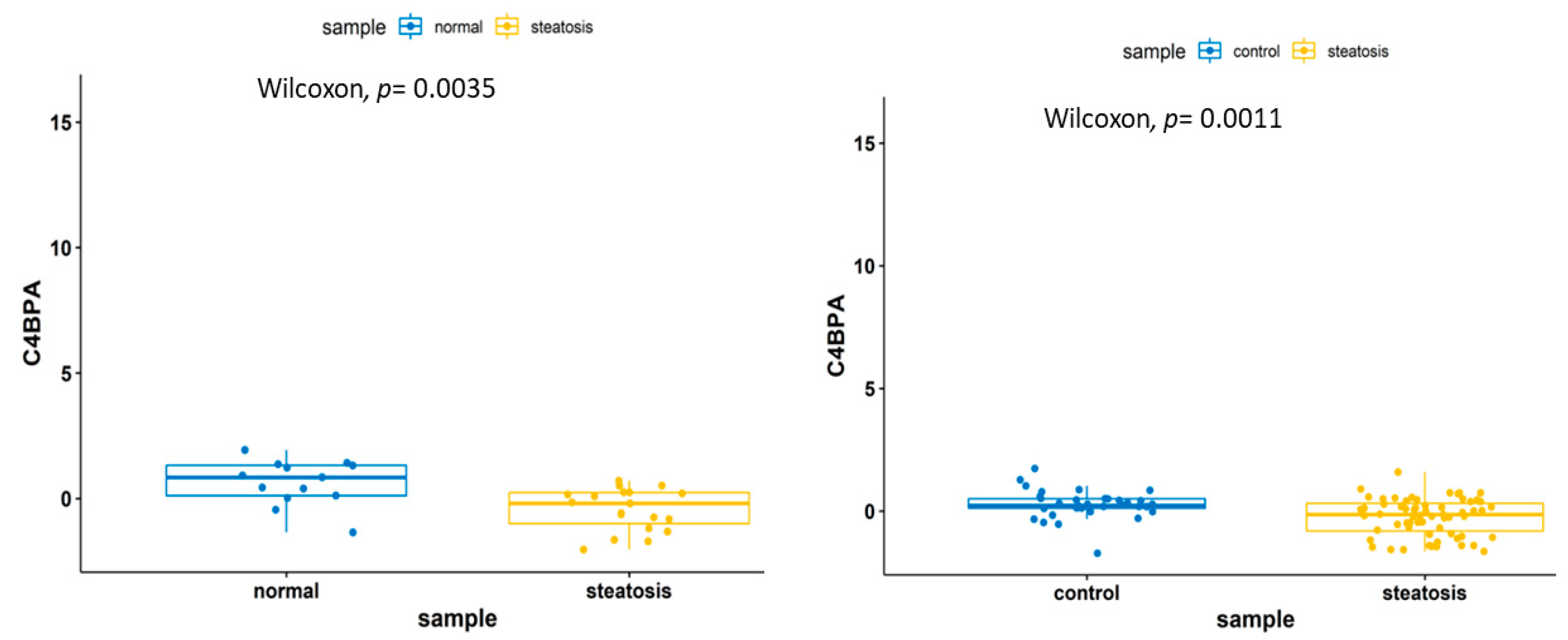
3.3.1. Transcriptomic Feature Study
3.3.2. Lipidomics Feature Study
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, H.Y.; Baik, S.J.; Lee, H.A.; Lee, B.K.; Lee, H.S.; Kim, T.H.; Yoo, K. Relative fat mass at baseline and its early change may be a predictor of incident nonalcoholic fatty liver disease. Sci. Rep. 2020, 10, 17491. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M. Non-alcoholic fatty liver disease—A global public health perspective. J. Hepatol. 2019, 70, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef]
- Tanaka, N.; Kimura, T.; Fujimori, N.; Nagaya, T.; Komatsu, M.; Tanaka, E. Current status, problems, and perspectives of non-alcoholic fatty liver disease research. World J. Gastroenterol. 2019, 25, 163–177. [Google Scholar] [CrossRef]
- Byrne, C.D.; Targher, G. NAFLD: A multisystem disease. J. Hepatol. 2015, 62 (Suppl. S1), S47–S64. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Llauradó, G.; Orešič, M.; Hyötyläinen, T.; Orho-Melander, M.; Yki-Järvinen, H. Circulating triacylglycerol signatures and insulin sensitivity in NAFLD associated with the E167K variant in TM6SF2. J. Hepatol. 2015, 62, 657–663. [Google Scholar] [CrossRef]
- Lomonaco, R.; Ortiz-Lopez, C.; Orsak, B.; Webb, A.; Hardies, J.; Darland, C.; Finch, J.; Gastaldelli, A.; Harrison, S.; Tio, F.; et al. Effect of adipose tissue insulin resistance on metabolic parameters and liver histology in obese patients with nonalcoholic fatty liver disease. Hepatology 2012, 55, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Pagano, G.; Pacini, G.; Musso, G.; Gambino, R.; Mecca, F.; Depetris, N.; Cassader, M.; David, E.; Cavallo-Perin, P.; Rizzetto, M. Nonalcoholic steatohepatitis, insulin resistance, and metabolic syndrome: Further evidence for an etiologic association. Hepatology 2002, 35, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.J.; Campbell-Sargent, C.; Mirshahi, F.; Rizzo, W.B.; Contos, M.J.; Sterling, R.K.; Luketic, V.A.; Shiffman, M.L.; Clore, J.N. Nonalcoholic steatohepatitis: Association of insulin resistance and mitochondrial abnormalities. Gastroenterology 2001, 120, 1183–1192. [Google Scholar] [CrossRef]
- Mirmiran, P.; Amirhamidi, Z.; Ejtahed, H.S.; Bahadoran, Z.; Azizi, F. Relationship between diet and non-alcoholic fatty liver disease: A review article. Iran. J. Public Health 2017, 46, 1007–1017. [Google Scholar] [PubMed]
- Maurice, J.; Manousou, P. Non-alcoholic fatty liver disease. Clin. Med. 2018, 18, 245–250. [Google Scholar] [CrossRef]
- Estes, C.; Razavi, H.; Loomba, R.; Younossi, Z.; Sanyal, A.J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018, 67, 123–133. [Google Scholar] [CrossRef]
- Yu, J.; Marsh, S.; Hu, J.; Feng, W.; Wu, C. The pathogenesis of nonalcoholic fatty liver disease: Interplay between diet, gut microbiota, and genetic background. Gastroenterol. Res. Pract. 2016, 2016, 2862173. [Google Scholar] [CrossRef]
- Tilg, H.; Adolph, T.E.; Moschen, A.R. Multiple parallel hits hypothesis in nonalcoholic fatty liver disease: Revisited after a decade. Hepatology 2021, 73, 833–842. [Google Scholar] [CrossRef]
- Fabbrini, E.; Sullivan, S.; Klein, S. Obesity and nonalcoholic fatty liver disease: Biochemical, metabolic, and clinical implications. Hepatology 2010, 51, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the gene expression omnibus (GEO) and bioconductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Aziz, F.; Acharjee, A.; Williams, J.A.; Russ, D.; Bravo-Merodio, L.; Gkoutos, G.V. Biomarker Prioritisation and Power Estimation Using Ensemble Gene Regulatory Network Inference. Int. J. Mol. Sci. 2020, 21, 7886. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Merodio, L.; Acharjee, A.; Russ, D.; Bisht, V.; Williams, J.A.; Tsaprouni, L.G.; Gkoutos, G.V. Translational biomarkers in the era of precision medicine. Adv. Clin. Chem. 2021, 102, 191–232. [Google Scholar]
- Vu, V.Q. Vqv/Ggbiplot: A Biplot Based on Ggplot2. Github. 2015. Available online: http://github.com/vqv/ggbiplot (accessed on 15 March 2021).
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Arendt, B.M.; Comelli, E.M.; Ma, D.W.; Lou, W.; Teterina, A.; Kim, T.; Fung, S.K.; Wong, D.K.; McGilvray, I.; Fischer, S.E.; et al. Altered hepatic gene expression in nonalcoholic fatty liver disease is associated with lower hepatic n-3 and n-6 polyunsaturated fatty acids. Hepatology 2015, 61, 1565–1578. [Google Scholar] [CrossRef] [PubMed]
- Kriss, M.; Golden-Mason, L.; Kaplan, J.; Mirshahi, F.; Setiawan, V.W.; Sanyal, A.J.; Rosen, H.R. Increased hepatic and circulating chemokine and osteopontin expression occurs early in human NAFLD development. PLoS ONE 2020, 15, e0236353. [Google Scholar] [CrossRef]
- Du Plessis, J.; Van Pelt, J.; Korf, H.; Mathieu, C.; Van der Schueren, B.; Lannoo, M.; Oyen, T.; Topal, B.; Fetter, G.; Nayler, S.; et al. Association of Adipose Tissue Inflammation With Histologic Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology 2015, 149, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Frades, I.; Andreasson, E.; Mato, J.M.; Alexandersson, E.; Matthiesen, R.; Martínez-Chantar, M.L. Integrative genomic signatures of hepatocellular carcinoma derived from nonalcoholic Fatty liver disease. PLoS ONE 2015, 10, e0124544. [Google Scholar] [CrossRef]
- Starmann, J.; Fälth, M.; Spindelböck, W.; Lanz, K.L.; Lackner, C.; Zatloukal, K.; Trauner, M.; Sültmann, H. Gene expression profiling unravels cancer-related hepatic molecular signatures in steatohepatitis but not in steatosis. PLoS ONE 2012, 7, e46584. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77–2105. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic. Acids Res. 2016, 44(W1), W90–W97. [Google Scholar] [CrossRef]
- Wei T, S.V. R Package “Corrplot”: Visualization of a Correlation Matrix. GitHub. 2017. Available online: https://github.com/taiyun/corrplot (accessed on 15 March 2021).
- Sanders, F.W.B.; Acharjee, A.; Walker, C.; Marney, L.; Roberts, L.D.; Imamura, F.; Jenkins, B.; Case, J.; Ray, S.; Virtue, S.; et al. Hepatic steatosis risk is partly driven by increased de novo lipogenesis following carbohydrate consumption. Genome. Biol. 2018, 19, 79. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics For Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 15 March 2021).
- Kassambara, A. Rstatix: Pipe-Friendly Framework for Basic Statistical Tests; R Package Version 0.7.0. 2021. Available online: https://CRAN.R-project.org/package=rstatix (accessed on 15 March 2021).
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Acevedo, A. LIPEA: Lipid Pathway Enrichment Analysis. bioRxiv 2018. [Google Scholar] [CrossRef]
- Epskamp, S. Qgraph: Network visualizations of relationships in psychometric data. J. Stat. Softw. 2012, 48, 1–8. [Google Scholar] [CrossRef]
- Wang, R.; Wang, X.; Zhuang, L. Gene expression profiling reveals key genes and pathways related to the development of non-alcoholic fatty liver disease. Ann. Hepatol. 2016, 15, 190–199. [Google Scholar] [PubMed]
- Niederreiter, L.; Tilg, H. Cytokines and fatty liver diseases. Liver Res. 2018, 2, 14–20. [Google Scholar] [CrossRef]
- Tomizawa, M.; Kawanabe, Y.; Shinozaki, F.; Sato, S.; Motoyoshi, Y.; Sugiyama, T.; Yamamoto, S.; Sueishi, M. Triglyceride is strongly associated with nonalcoholic fatty liver disease among markers of hyperlipidemia and diabetes. Biomed. Rep. 2014, 2, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Perakakis, N.; Stefanakis, K.; Mantzoros, C.S. The role of omics in the pathophysiology, diagnosis and treatment of non-alcoholic fatty liver disease. Metab. Clin. Exp. 2020, 111, 154320. [Google Scholar] [CrossRef] [PubMed]
- Kosmalski, M.; Mokros, Ł.; Kuna, P.; Witusik, A.; Pietras, T. Changes in the immune system—The key to diagnostics and therapy of patients with non-alcoholic fatty liver disease. Cent. Eur. J. Immunol. 2018, 43, 231–239. [Google Scholar] [CrossRef]
- Dunkelberger, J.R.; Song, W.C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010, 20, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Luque, A.; Serrano, I.; Ripoll, E.; Malta, C.; Gomà, M.; Blom, A.M.; Grinyó, J.M.; de Córdoba, S.R.; Torras, J.; Aran, J.M. Noncanonical immunomodulatory activity of complement regulator C4BP(β-) limits the development of lupus nephritis. Kidney Int. 2020, 97, 551–566. [Google Scholar] [CrossRef]
- Martin, M.; Gottsäter, A.; Nilsson, P.M.; Mollnes, T.E.; Lindblad, B.; Blom, A.M. Complement activation and plasma levels of C4b-binding protein in critical limb ischemia patients. J. Vasc. Surg. 2009, 50, 100–106. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Varghese, P.M.; Murugaiah, V.; Beirag, N.; Temperton, N.; Khan, H.A.; Alrokayan, S.H.; Al-Ahdal, M.N.; Nal, B.; Al-Mohanna, F.A.; Sim, R.B.; et al. C4b binding protein acts as an innate immune effector against influenza a virus. Front. Immunol. 2021, 11, 585361. [Google Scholar] [CrossRef]
- Rodriguez de Cordoba, S.; Sanchez-Corral, P.; Rey-Campos, J. Structure of the gene coding for the alpha polypeptide chain of the human complement component C4b-binding protein. J. Exp. Med. 1991, 173, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Bettoni, S.; Shaughnessy, J.; Maziarz, K.; Ermert, D.; Gulati, S.; Zheng, B.; Mörgelin, M.; Jacobsson, S.; Riesbeck, K.; Unemo, M.; et al. C4BP-IgM protein as a therapeutic approach to treat Neisseria gonorrhoeae infections. JCI Insight 2019, 4, e131886. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Yuan, R.; Zhang, Y.; Geng, S.; Li, L. Tollip deficiency alters atherosclerosis and steatosis by disrupting lipophagy. J Am. Heart Assoc. 2017, 6, e004078. [Google Scholar] [CrossRef] [PubMed]
- Mirea, A.M.; Tack, C.J.; Chavakis, T.; Joosten, L.A.B.; Toonen, E.J.M. IL-1 family cytokine pathways underlying NAFLD: Towards new treatment strategies. Trends Mol. Med. 2018, 24, 458–471. [Google Scholar] [CrossRef]
- Phieler, J.; Garcia-Martin, R.; Lambris, J.D.; Chavakis, T. The role of the complement system in metabolic organs and metabolic diseases. Semin. Immunol. 2013, 25, 47–53. [Google Scholar] [CrossRef]
- Okrój, M.; Blom, A.M. Chapter 24—C4b-binding protein. In The Complement FactsBook, 2nd ed.; Barnum, S., Schein, T., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 251–259. [Google Scholar]
- Moreno-Navarrete, J.M.; Fernández-Real, J.M. The complement system is dysfunctional in metabolic disease: Evidences in plasma and adipose tissue from obese and insulin resistant subjects. Semin. Cell Dev. Biol. 2019, 85, 164–172. [Google Scholar] [CrossRef]
- Rawal, N.; Rajagopalan, R.; Salvi, V.P. Stringent regulation of complement lectin pathway C3/C5 convertase by C4b-binding protein (C4BP). Mol. Immunol. 2009, 46, 2902–2910. [Google Scholar] [CrossRef]
- Rensen, S.S.; Slaats, Y.; Driessen, A.; Peutz-Kootstra, C.J.; Nijhuis, J.; Steffensen, R.; Greve, J.W.; Buurman, W.A. Activation of the complement system in human nonalcoholic fatty liver disease. Hepatology 2009, 50, 1809–1817. [Google Scholar] [CrossRef] [PubMed]
- Reca, R.; Wysoczynski, M.; Yan, J.; Lambris, J.D.; Ratajczak, M.Z. The role of third complement component (C3) in homing of hematopoietic stem/progenitor cells into bone marrow. Adv. Exp. Med. Biol. 2006, 586, 35–51. [Google Scholar]
- Saleh, J.; Wahab, R.A.; Farhan, H.; Al-Amri, I.; Cianflone, K. Plasma levels of acylation-stimulating protein are strongly predicted by waist/hip ratio and correlate with decreased LDL size in men. ISRN Obes. 2013, 2013, 342802. [Google Scholar] [CrossRef] [PubMed]
- Kawano, Y.; Cohen, D.E. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J. Gastroenterol. 2013, 48, 434–441. [Google Scholar] [CrossRef]
- Eguchi, Y.; Hyogo, H.; Ono, M.; Mizuta, T.; Ono, N.; Fujimoto, K.; Chayama, K.; Saibara, T. Prevalence and associated metabolic factors of nonalcoholic fatty liver disease in the general population from 2009 to 2010 in Japan: A multicenter large retrospective study. J. Gastroenterol. 2012, 47, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Arvind, A.; Osganian, S.A.; Cohen, D.E.; Corey, K.E. Lipid and Lipoprotein Metabolism in Liver Disease; MDText.com, Inc.: Endotext South Dartmouth, MA, USA, 2000. [Google Scholar]
- Morigny, P.; Houssier, M.; Mouisel, E.; Langin, D. Adipocyte lipolysis and insulin resistance. Biochimie 2016, 125, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Cignarelli, A.; Genchi, V.A.; Perrini, S.; Natalicchio, A.; Laviola, L.; Giorgino, F. Insulin and insulin receptors in adipose tissue development. Int. J. Mol. Sci. 2019, 20, 759. [Google Scholar] [CrossRef]









| S.No | GEO | Number of Samples | Number of Features | Platform | Reference |
|---|---|---|---|---|---|
| 1 | GSE89632 | Control (n = 24) vs. Steatosis (n = 20) | 29,377 | Illumina HumanHT-12 WG-DASL V4.0 R2 expression beadchip | [22] |
| 2 | GSE151158 | Control (n = 21) vs. Steatosis (n =23) | 618 | NanoString Human Immunology v2 Code Set (NS_Immunology_v2_C2328+PLS_Golden_1_C5164) | [23] |
| 3 | GSE58979 | Control (n = 0) vs. Steatosis (n = 17) | 49,395 | Affymetrix Human Gene Expression Array | [24] |
| 4 | GSE63067 | Control (n = 7) vs. Steatosis (n = 2) | 54,675 | [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | [25] |
| 5 | GSE33814 | Control (n = 13) vs. Steatosis (n = 19) | 48,803 | Illumina HumanWG-6 v3.0 expression beadchip | [26] |
| Pathway Name | Pathway Lipids | Converted Lipids (Number) | Converted Lipids (Percentage) | Converted Lipids (List) | p-Value |
|---|---|---|---|---|---|
| Long-term depression | 3 | 2 | 50.00 | C00165, C00641 | 0.0147783 |
| Regulation of lipolysis in adipocytes | 6 | 2 | 50.00 | C00165, C00422 | 0.0147783 |
| Glycerolipid metabolism | 15 | 2 | 50.00 | C00422, C00641 | 0.0421456 |
| Insulin resistance | 4 | 2 | 50.00 | C00165, C00422 | 0.0421456 |
| Fat digestion and absorption | 8 | 2 | 50.00 | C00165, C00422 | 0.0800387 |
| Rap1 signaling pathway | 1 | 1 | 25.00 | C00165 | 0.137931 |
| Chemokines signaling pathway | 2 | 1 | 25.00 | C00165 | 0.137931 |
| Ras signaling pathway | 2 | 1 | 25.00 | C00165 | 0.137931 |
| MAPK signaling pathway | 1 | 1 | 25.00 | C00165 | 0.137931 |
| NF-kappa B signaling pathway | 1 | 1 | 25.00 | C00165 | 0.137931 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shafiha, R.; Bahcivanci, B.; Gkoutos, G.V.; Acharjee, A. Machine Learning-Based Identification of Potentially Novel Non-Alcoholic Fatty Liver Disease Biomarkers. Biomedicines 2021, 9, 1636. https://doi.org/10.3390/biomedicines9111636
Shafiha R, Bahcivanci B, Gkoutos GV, Acharjee A. Machine Learning-Based Identification of Potentially Novel Non-Alcoholic Fatty Liver Disease Biomarkers. Biomedicines. 2021; 9(11):1636. https://doi.org/10.3390/biomedicines9111636
Chicago/Turabian StyleShafiha, Roshan, Basak Bahcivanci, Georgios V. Gkoutos, and Animesh Acharjee. 2021. "Machine Learning-Based Identification of Potentially Novel Non-Alcoholic Fatty Liver Disease Biomarkers" Biomedicines 9, no. 11: 1636. https://doi.org/10.3390/biomedicines9111636
APA StyleShafiha, R., Bahcivanci, B., Gkoutos, G. V., & Acharjee, A. (2021). Machine Learning-Based Identification of Potentially Novel Non-Alcoholic Fatty Liver Disease Biomarkers. Biomedicines, 9(11), 1636. https://doi.org/10.3390/biomedicines9111636








