Preparation and Characterization of Silymarin-Conjugated Gold Nanoparticles with Enhanced Anti-Fibrotic Therapeutic Effects against Hepatic Fibrosis in Rats: Role of MicroRNAs as Molecular Targets
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Regent
2.2. Preparation and Characterization of Nanoparticles
2.2.1. Preparation of Gold Nanoparticles
2.2.2. Synthesis of Silymarin/Gold Nanoparticles
2.3. Physicochemical Characterizations of Nanoparticles
2.3.1. UV–Visible Spectrophotometry
2.3.2. Dynamic Light Scattering (DLS) and Zeta Potential (ZP) Analysis
2.3.3. Transmission Electron Microscopy (TEM) Analysis
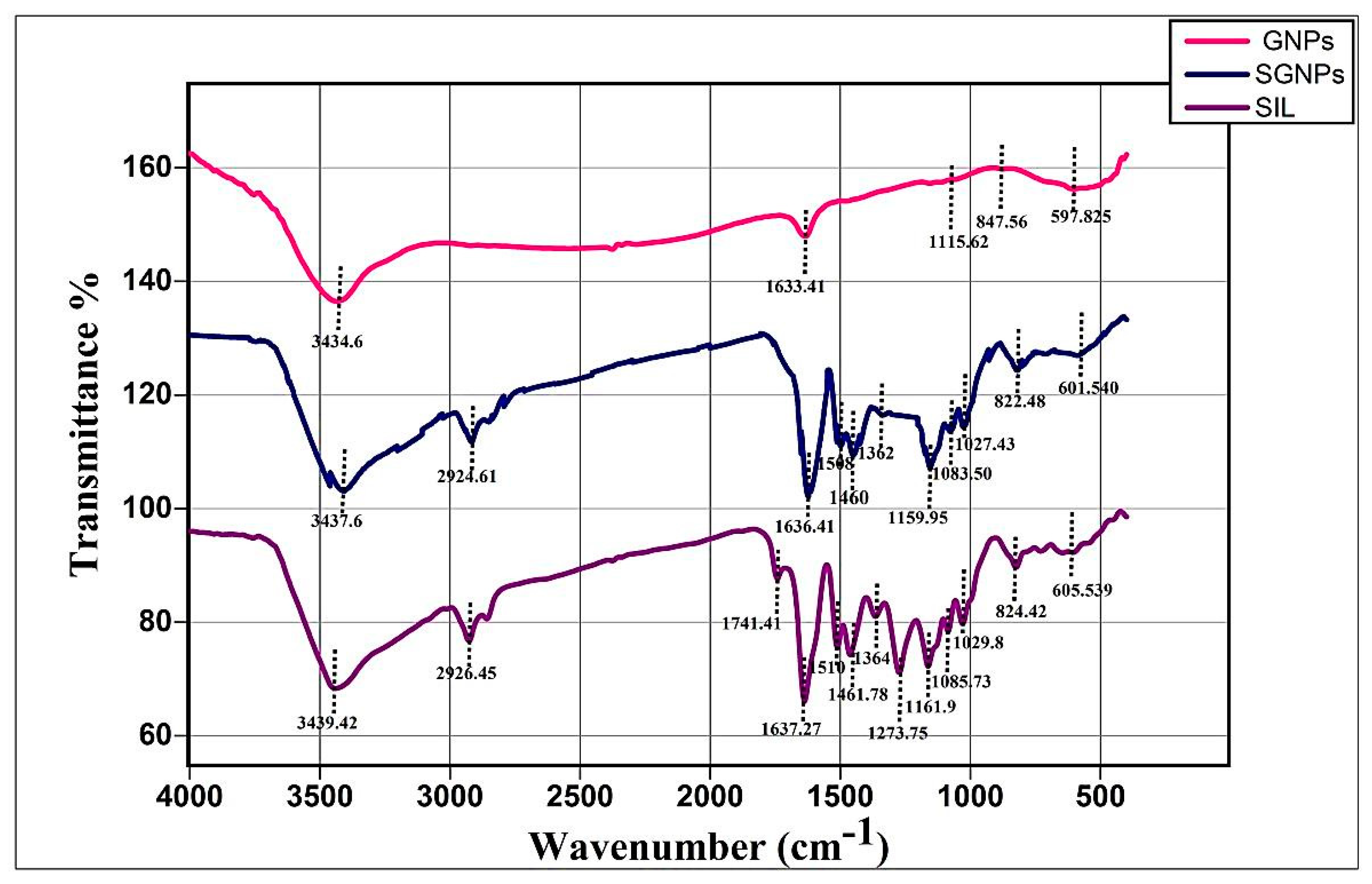
2.3.4. Fourier Transforms Infrared Spectroscopy (FT-IR) Study
2.3.5. Drug Entrapment Efficiency and Loading Capacity
2.3.6. In Vitro Drug Release Study
2.4. In Vivo Animals Studies
2.4.1. Establishment of Liver Fibrosis Model
2.4.2. Experimental Design
2.4.3. Blood Sample Collection and Tissue Preparation
2.4.4. Bioinformatic Study
2.5. Methods
2.5.1. Serum Biomarkers for Liver Function Tests
2.5.2. Malondialdehyde (MDA) as Index of Lipid Peroxidation
2.5.3. ELISA Measurements
2.5.4. Gene Expression Analysis
Assessment of Hepatic microRNAs (miR-22, miR-29c, and miR-219a)
Assessment of Hepatic Expression of TGFβR1, TGFβR2, and COL3A1 Genes
Relative Quantification of Gene Expression
2.6. Histopathology Study
Quantitative Measurements of Fibrosis Area
2.7. Immunohistochemistry (IHC) Study
2.8. Statistical Analysis
3. Results
3.1. Characterization of Gold Nanoparticles (GNPs)
3.1.1. UV–Visible Spectrophotometry
3.1.2. TEM Analysis of GNPs
3.1.3. DLS and Zeta Potential of GNPs
3.2. Characterization of Silymarin-Gold Nanoparticles (SGNPs)
3.2.1. UV–Visible Spectrophotometry
3.2.2. TEM Analysis of SGNPs
3.2.3. DLS and Zeta Potential of SGNPs
3.2.4. FT-IR Study
3.2.5. Drug Entrapment and Loading Capacity of SIL in SGNPs
3.2.6. In Vitro Drug Release Study
3.3. In Vivo Studies
3.3.1. Body Weight Gain, Liver Weight, and Liver Index (%)
3.3.2. Serum Liver Function Markers
3.3.3. Hepatic Redox Parameters and TGFβ-1
3.4. Molecular Analysis
3.4.1. MicroRNAs Expression
3.4.2. Bioinformatics
3.4.3. The Expression of the Target Genes
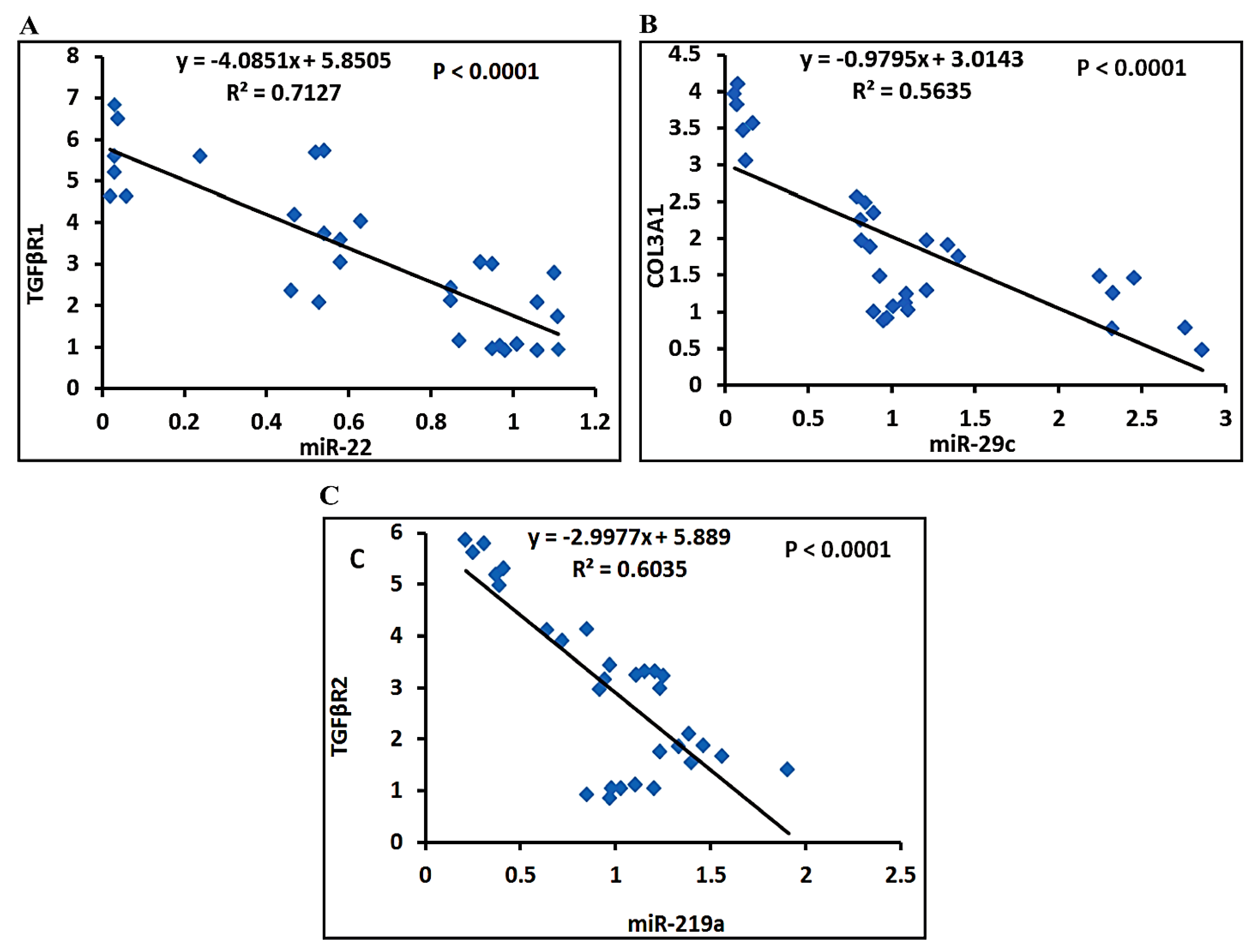
3.4.4. Correlation Studies
3.5. Histopathological Analysis
3.5.1. Liver Morphology
3.5.2. Histopathology of Liver Tissue
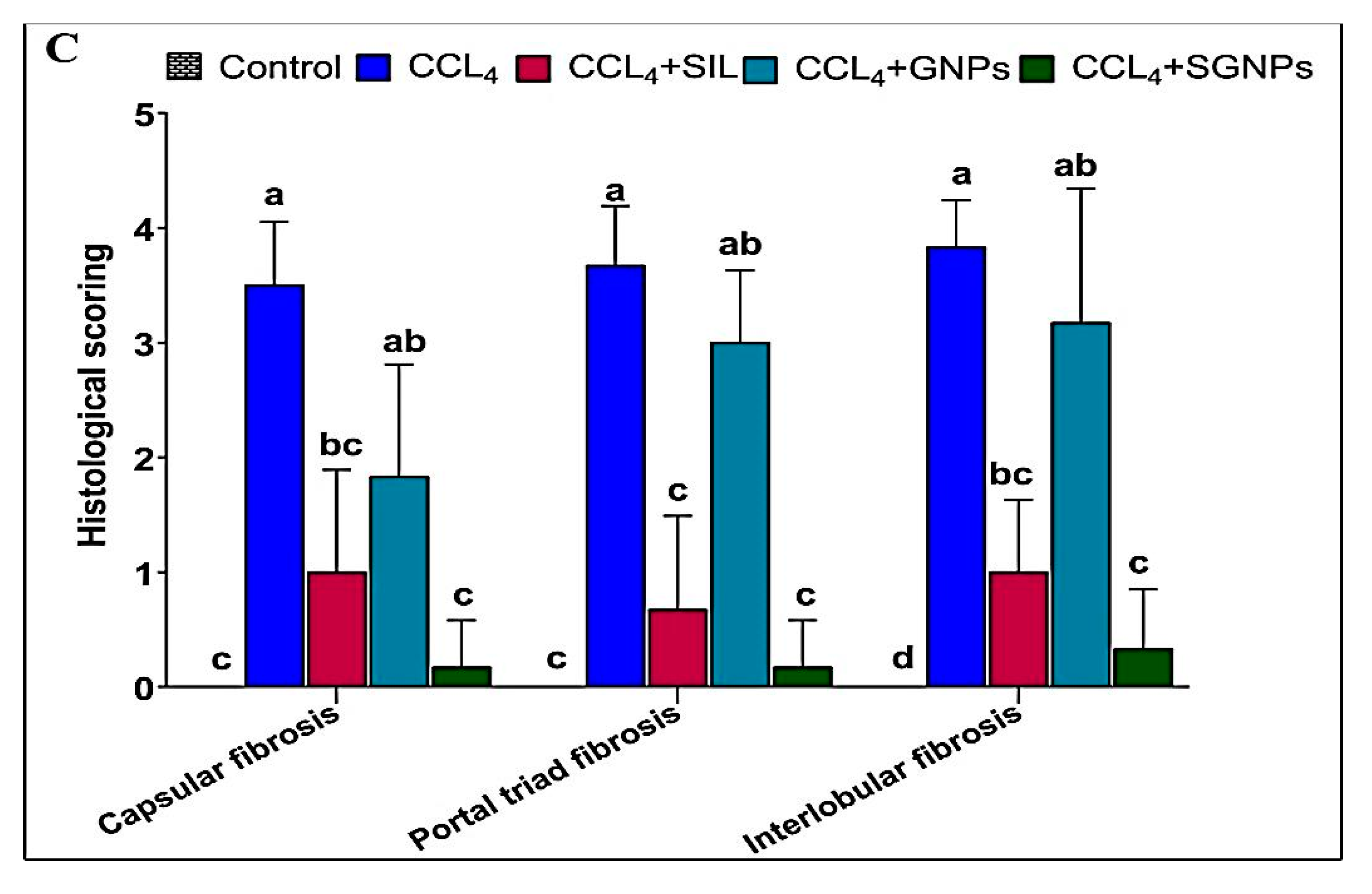
3.5.3. Histological Grading of Fibrosis
3.5.4. Masson’s Trichrome (MT) Staining of Liver Tissue
3.5.5. Immunohistochemistry (IHC) of αSMA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, S.; Zou, L.; Li, L.; Wu, T.J. The protective effect of glycyrrhetinic acid on carbon tetrachloride-induced chronic liver fibrosis in mice via upregulation of Nrf2. PLoS ONE 2013, 8, e53662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collaborators, G. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet J. 2018. [Google Scholar] [CrossRef] [Green Version]
- Moon, A.M.; Singal, A.G.; Tapper, E.B. Contemporary epidemiology of chronic liver disease and cirrhosis. Clin. Gastroenterol. Hepatol. 2020, 18, 2650–2666. [Google Scholar] [CrossRef]
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Sadek, K.; Saleh, E.; Nasr, S.M. Molecular hepatoprotective effects of lipoic acid against carbon tetrachloride-induced liver fibrosis in rats: Hepatoprotection at molecular level. Hum. Exp. Toxicol. 2018, 37, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Jiang, B.; Zheng, B.; Yan, Y.; Wang, J.; Duan, Y.; Li, S.; Yan, L.; Wang, H.; Chen, B.J.P. Heterogenic transplantation of bone marrow-derived rhesus macaque mesenchymal stem cells ameliorates liver fibrosis induced by carbon tetrachloride in mouse. PeerJ 2018, 6, e4336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bataller, R.; Brenner, D.A. Liver fibrosis. J. Clin. Investig. 2005, 115, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Noyan, S.; Cavusoglu, I.; Minbay, F.Z. The effect of vitamin A on CCl4-induced hepatic injuries in rats: A histochemical, immunohistochemical and ultrastructural study. Acta Histochem. 2006, 107, 421–434. [Google Scholar] [CrossRef]
- Jiao, J.; Friedman, S.L.; Aloman, C.J. Hepatic fibrosis. Curr. Opin. Gastroenterol. 2009, 25, 223. [Google Scholar] [CrossRef]
- Arfmann-Knübel, S.; Struck, B.; Genrich, G.; Helm, O.; Sipos, B.; Sebens, S.; Schäfer, H.J. The crosstalk between Nrf2 and TGF-β1 in the epithelial-mesenchymal transition of pancreatic duct epithelial cells. PLoS ONE 2015, 10, e0132978. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.-K.; Pokharel, Y.R.; Lim, S.C.; Han, H.-K.; Ryu, C.S.; Kim, S.K.; Kwak, M.K.; Kang, K.W. Inhibition of liver fibrosis by solubilized coenzyme Q10: Role of Nrf2 activation in inhibiting transforming growth factor-β1 expression. Toxicol. Appl. Pharmacol. 2009, 240, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.-M.; Li, D.; Zhang, C.; Wen, G.-H.; Wu, C.; Xu, Y.-Y.; Kan, Y.; Lu, W.-P.; Ding, H.-Y.; Yang, Y.J.E.; et al. Salvianolic acid B protects against acute and chronic liver injury by inhibiting Smad2C/L phosphorylation. Exp. Ther. Med. 2021, 21, 1. [Google Scholar] [CrossRef] [PubMed]
- Hobert, O.J.S. Gene regulation by transcription factors and microRNAs. Sci. Gov. Rep. 2008, 319, 1785–1786. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, S.J.A. MicroRNAs in fibrosis: Opportunities and challenges. Arthritis Res. Ther. 2016, 18, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, D.; Li, B.; Shao, Q.; Li, F.; Li, Z.; Chen, G. Mir-22 suppresses bmp7 in the development of cirrhosis. Cell. Physiol. Biochem. 2015, 36, 1026–1036. [Google Scholar] [CrossRef]
- Roderburg, C.; Urban, G.W.; Bettermann, K.; Vucur, M.; Zimmermann, H.; Schmidt, S.; Janssen, J.; Koppe, C.; Knolle, P.; Castoldi, M. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatol. Int. 2011, 53, 209–218. [Google Scholar] [CrossRef]
- Ma, L.; Ma, J.; Ou, H.L. MicroRNA219 overexpression serves a protective role during liver fibrosis by targeting tumor growth factor β receptor 2. Mol. Med. Rep. 2019, 19, 1543–1550. [Google Scholar]
- Wang, H.; Zhang, Q.; Wang, B.; Wu, W.; Wei, J.; Li, P.; Huang, R. miR-22 regulates C2C12 myoblast proliferation and differentiation by targeting TGFBR1. Eur. J. Cell Biol. 2018, 97, 257–268. [Google Scholar] [CrossRef]
- Chuang, T.-D.; Khorram, O. Mechanisms underlying aberrant expression of miR-29c in uterine leiomyoma. Fertil. Steril. 2016, 105, 236–245. e231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.; Qie, J.; Fu, Q.; Chen, J.; Jin, Y.; Ding, Z. miR-20a-5p/TGFBR2 axis affects pro-inflammatory macrophages and aggravates liver fibrosis. Front. Oncol. 2020, 10, 107. [Google Scholar] [CrossRef]
- Ferenci, P. Silymarin in the treatment of liver diseases: What is the clinical evidence? Clin. Liver Dis. 2016, 7, 8. [Google Scholar] [CrossRef]
- Javed, S.; Kohli, K.; Ali, M. Reassessing bioavailability of silymarin. Altern. Med. Rev. 2011, 16, 239. [Google Scholar] [PubMed]
- Theodosiou, E.; Purchartová, K.; Stamatis, H.; Kolisis, F.; Křen, V. Bioavailability of silymarin flavonolignans: Drug formulations and biotransformation. Phytochem. Rev. 2014, 13, 1–18. [Google Scholar] [CrossRef]
- Woo, J.S.; Kim, T.-S.; Park, J.-H.; Chi, S. Formulation and biopharmaceutical evaluation of silymarin using SMEDDS. Arch. Pharmacal Res. 2007, 30, 82–89. [Google Scholar] [CrossRef]
- Alipour, M.; reza Bigdeli, M.; Aligholi, H.; Rasoulian, B. Sustained release of silibinin-loaded chitosan nanoparticle induced apoptosis in glioma cells. J. Biomed. Mater. Res. Part A 2020, 108, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Connor, E.E.; Mwamuka, J.; Gole, A.; Murphy, C.J.; Wyatt, M.D. Gold nanoparticles are taken up by human cells but do not cause acute cytotoxicity. Small Rumin. Res. 2005, 1, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, P.; Strano, G.; Zuccarello, L.; Satriano, C. Gold and silver nanoparticles for applications in theranostics. Curr. Top. Med. Chem. 2016, 16, 3069–3102. [Google Scholar] [CrossRef]
- Huang, X.; El-Sayed, M.A. Gold nanoparticles: Optical properties and implementations in cancer diagnosis and photothermal therapy. J. Adv. Res. 2010, 1, 13–28. [Google Scholar] [CrossRef] [Green Version]
- Isoda, K.; Tanaka, A.; Fuzimori, C.; Echigoya, M.; Taira, Y.; Taira, I.; Shimizu, Y.; Akimoto, Y.; Kawakami, H. Toxicity of gold nanoparticles in mice due to nanoparticle/drug interaction induces acute kidney damage. Nanoscale Res. Lett. 2020, 15, 141. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, R.; Raghu, K.J. Biosynthesis and characterization of gold and silver nanoparticles using milk thistle (Silybum marianum) seed extract. J. Nanosci. 2014, 2014, 905404. [Google Scholar] [CrossRef]
- Nambiar, S.; Osei, E.; Fleck, A.; Darko, J.; Mutsaers, A.J.; Wettig, S.J. Synthesis of curcumin-functionalized gold nanoparticles and cytotoxicity studies in human prostate cancer cell line. Appl. Nanosci. 2018, 8, 347–357. [Google Scholar] [CrossRef]
- ElMitwalli, O.S.; Barakat, O.A.; Daoud, R.M.; Akhtar, S.; Henari, F.Z. Green synthesis of gold nanoparticles using cinnamon bark extract, characterization, and fluorescence activity in Au/eosin Y assemblies. J. Nanoparticle Res. 2020, 22, 309. [Google Scholar] [CrossRef]
- McFarland, A.D.; Haynes, C.L.; Mirkin, C.A.; Van Duyne, R.P.; Godwin, H.A. Color my nanoworld. J. Chem. Educ. 2004, 81, 544A. [Google Scholar] [CrossRef]
- Stolarczyk, E.U.; Stolarczyk, K.; Laszcz, M.; Kubiszewski, M.; Les, A.; Michalak, O. Pemetrexed conjugated with gold nanoparticles—Synthesis, characterization and a study of noncovalent interactions. Eur. J. Pharm. Sci. 2017, 109, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Kabir, N.; Ali, H.; Ateeq, M.; Bertino, M.F.; Shah, M.R.; Franzel, L. Silymarin coated gold nanoparticles ameliorates CCl 4-induced hepatic injury and cirrhosis through down regulation of hepatic stellate cells and attenuation of Kupffer cells. RSC Adv. 2014, 4, 9012–9020. [Google Scholar] [CrossRef]
- Sindhu, K.; Rajaram, A.; Sreeram, K.J.; Rajaram, R. Curcumin conjugated gold nanoparticle synthesis and its biocompatibility. RSC Adv. 2014, 4, 1808–1818. [Google Scholar] [CrossRef]
- Safer, A.-M.; Leporatti, S.; Jose, J.; Soliman, M.S. Conjugation of EGCG and chitosan NPs as a novel nano-drug delivery system. Int. J. Nanomed. 2019, 14, 8033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radu, I.C.; Hudita, A.; Zaharia, C.; Stanescu, P.O.; Vasile, E.; Iovu, H.; Stan, M.; Ginghina, O.; Galateanu, B.; Costache, M.; et al. Poly(HydroxyButyrate-co-HydroxyValerate) (PHBHV) Nanocarriers for Silymarin Release as Adjuvant Therapy in Colo-rectal Cancer. Front Pharm. 2017, 8, 508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, A.S.; Lakshmi, B.A.; Kim, S.; Kim, J. Synthesis and characterization of acetyl curcumin-loaded core/shell liposome nanoparticles via an electrospray process for drug delivery, and theranostic applications. Eur. J. Pharm. Biopharm. 2019, 142, 518–530. [Google Scholar] [CrossRef]
- Laouar, A.; Klibet, F.; Bourogaa, E.; Benamara, A.; Boumendjel, A.; Chefrour, A.; Messarah, M. Potential antioxidant properties and hepatoprotective effects of Juniperus phoenicea berries against CCl4 induced hepatic damage in rats. Asian Pac. J. Trop. Med. 2017, 10, 263–269. [Google Scholar] [CrossRef]
- Maksoud, H.A.; Magid, A.D.A.; Mostafa, Y.; Elharrif, M.G.; Sorour, R.I.; Sorour, M.I. Ameliorative effect of liquorice extract versus silymarin in experimentally induced chronic hepatitis: A biochemical and genetical study. Clin. Nutr. Exp. 2019, 23, 69–79. [Google Scholar] [CrossRef] [Green Version]
- Kandimalla, R.; Dash, S.; Bhowal, A.C.; Kalita, S.; Talukdar, N.C.; Kundu, S.; Kotoky, J. Glycogen-gold nanohybrid escalates the potency of silymarin. Int. J. Nanomed. 2017, 12, 7025–7038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Draper, H.; Hadley, M. Malondialdehyde determination as index of lipid Peroxidation. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1990; Volume 186, pp. 421–431. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Suvarna, K.S.; Layton, C.; Bancroft, J.D. Bancroft’s Theory and Practice of Histological Techniques E-Book; Elsevier Health Sciences: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Wu, Y.C.; Huang, H.H.; Wu, Y.J.; Manousakas, I.; Yang, C.C.; Kuo, S.M. Therapeutic and Protective Effects of Liposomal Encapsulation of Astaxanthin in Mice with Alcoholic Liver Fibrosis. Int. J. Mol. Sci. 2019, 20, 4057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Luo, J.; Li, L.; Cheng, M.; Huang, N.; Liu, J.; Waalkes, M.P. The collagenolytic effects of the traditional Chinese medicine preparation, Han-Dan-Gan-Le, contribute to reversal of chemical-induced liver fibrosis in rats. Life Sci. 2003, 72, 1563–1571. [Google Scholar] [CrossRef]
- Hsu, S.-M.; Raine, L.; Fanger, H. A comparative study of the peroxidase-antiperoxidase method and an avidin-biotin complex method for studying polypeptide hormones with radioimmunoassay antibodies. Am. J. Clin. Pathol. 1981, 75, 734–738. [Google Scholar] [CrossRef] [PubMed]
- Gabius, H.-J.; Gabius, S. Lectins and Glycobiology; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Narayanan, K.B.; Sakthivel, N. Phytosynthesis of gold nanoparticles using leaf extract of Coleus amboinicus Lour. Mater. Charact. 2010, 61, 1232–1238. [Google Scholar] [CrossRef]
- Jayaseelan, C.; Ramkumar, R.; Rahuman, A.A.; Perumal, P. Green synthesis of gold nanoparticles using seed aqueous extract of Abelmoschus esculentus and its antifungal activity. Ind. Crop. Prod. 2013, 45, 423–429. [Google Scholar] [CrossRef]
- Mohandas, A.; Deepthi, S.; Biswas, R.; Jayakumar, R. Chitosan based metallic nanocomposite scaffolds as antimicrobial wound dressings. Bioact. Mater. 2018, 3, 267–277. [Google Scholar] [CrossRef]
- Katas, H.; Lim, C.S.; Azlan, A.Y.H.N.; Buang, F.; Busra, M.F.M. Antibacterial activity of biosynthesized gold nanoparticles using biomolecules from Lignosus rhinocerotis and chitosan. Saudi Pharm. J. 2019, 27, 283–292. [Google Scholar] [CrossRef]
- Sulaiman, G.M.; Waheeb, H.M.; Jabir, M.S.; Khazaal, S.H.; Dewir, Y.H.; Naidoo, Y. Hesperidin loaded on gold nanoparticles as a drug delivery system for a successful biocompatible, anti-cancer, anti-inflammatory and phagocytosis inducer model. Sci. Rep. Ist. Super. Sanita 2020, 10, 9362. [Google Scholar] [CrossRef] [PubMed]
- Srinath, B.; Rai, V.R. Biosynthesis of highly monodispersed, spherical gold nanoparticles of size 4–10 nm from spent cultures of Klebsiella pneumoniae. 3 Biotech 2015, 5, 671–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, M.M.; Fouad, S.A.; Elshoky, H.A.; Mohammed, G.M.; Salaheldin, T.A. Antibacterial effect of gold nanoparticles against Corynebacterium pseudotuberculosis. Int. J. Vet. Sci. Med. 2017, 5, 23–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pongsuchart, M.; Danladkaew, C.; Khomvarn, T.; Sereemaspun, A. Effect of glutathione-stabilized gold nanoparticles in 3T3 fibroblast cell. In Proceedings of the International Conference on Clean and Green Energy IPCBEE, International Conference on Clean and Green Energy IPCBEE, Hong Kong, China, 5–7 January 2012. [Google Scholar]
- Zhang, H.; Liu, G.; Zeng, X.; Wu, Y.; Yang, C.; Mei, L.; Wang, Z.; Huang, L. Fabrication of genistein-loaded biodegradable TPGS-b-PCL nanoparticles for improved therapeutic effects in cervical cancer cells. Int. J. Nanomed. 2015, 10, 2461. [Google Scholar]
- Forest, V.; Pourchez, J. Preferential binding of positive nanoparticles on cell membranes is due to electrostatic interactions: A too simplistic explanation that does not take into account the nanoparticle protein corona. Mater. Sci. Eng. 2017, 70, 889–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begum, N.A.; Mondal, S.; Basu, S.; Laskar, R.A.; Mandal, D. Biogenic synthesis of Au and Ag nanoparticles using aqueous solutions of Black Tea leaf extracts. Biointerfaces 2009, 71, 113–118. [Google Scholar] [CrossRef]
- Islan, G.A.; Das, S.; Cacicedo, M.L.; Halder, A.; Mukherjee, A.; Cuestas, M.L.; Roy, P.; Castro, G.R.; Mukherjee, A. Silybin-conjugated gold nanoparticles for antimicrobial chemotherapy against Gram-negative bacteria. J. Drug Deliv. Sci. Technol. 2019, 53, 101181. [Google Scholar] [CrossRef]
- Das, S.; Roy, P.; Auddy, R.G.; Mukherjee, A. Silymarin nanoparticle prevents paracetamol-induced hepatotoxicity. Int. J. Nanomed. 2011, 6, 1291. [Google Scholar]
- Shamaila, S.; Zafar, N.; Riaz, S.; Sharif, R.; Nazir, J.; Naseem, S. Gold nanoparticles: An efficient antimicrobial agent against enteric bacterial human pathogen. Nanomaterials 2016, 6, 71. [Google Scholar] [CrossRef] [Green Version]
- Thimmaraju, M.K.; Mondal, P.; Venu, K.; Padmaja, B.; Babu, G.S.; Dinesh Kumar, R.; Ravi Kumar, K. Carbon tetrachloride, alcohol and ranitidine induced hepatotoxicity and its protection by bark extracts of Bassia Latifolia in Wister rats. J. Herbs Spices Med. Plants 2020, 26, 275–290. [Google Scholar] [CrossRef]
- Tamayo, R.P. Is cirrhosis of the liver experimentally produced by CC14 an adequate model of human cirrhosis? Hepatol. Int. 1983, 3, 112–120. [Google Scholar] [CrossRef]
- Muriel, P. Peroxidation of lipids and liver damage. In Oxidants, Antioxidants, and Free Radicals; Taylor & Francis Group: Boca Raton, FL, USA, 1997; pp. 237–257. [Google Scholar]
- Dutta, S.; Chakraborty, A.K.; Dey, P.; Kar, P.; Guha, P.; Sen, S.; Kumar, A.; Sen, A.; Chaudhuri, T.K. Amelioration of CCl4 induced liver injury in swiss albino mice by antioxidant rich leaf extract of Croton bonplandianus Baill. PLoS ONE 2018, 13, e0196411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naz, I.; Khan, M.R.; Zai, J.A.; Batool, R.; Zahra, Z.; Tahir, A. Pilea umbrosa ameliorate CCl4 induced hepatic injuries by regulating endoplasmic reticulum stress, pro-inflammatory and fibrosis genes in rat. Environ. Health Prev. Med. 2020, 25, 53. [Google Scholar] [CrossRef]
- Poli, G. Pathogenesis of liver fibrosis: Role of oxidative stress. Mol. Asp. Med. 2000, 21, 49–98. [Google Scholar] [CrossRef]
- Devaraj, E.; Roy, A.; Veeraragavan, G.R.; Magesh, A.; Sleeba, A.V.; Arivarasu, L.; Parasuraman, B.M. β-Sitosterol attenuates carbon tetrachloride–induced oxidative stress and chronic liver injury in rats. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2020, 393, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Wahhab, M.A.; Aljawish, A.; El-Nekeety, A.A.; Abdel-Aziem, S.H.; Hassan, N.S. Chitosan nanoparticles plus quercetin suppress the oxidative stress, modulate DNA fragmentation and gene expression in the kidney of rats fed ochratoxin A-contaminated diet. Food Chem. Toxicol. 2017, 99, 209–221. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, H.; Cao, Y.; Li, Y.; Sun, S.; Zhang, J.; Zhang, G. Schisandrin B attenuates CCl4-induced liver fibrosis in rats by regulation of Nrf2-ARE and TGF-β/Smad signaling pathways. Drug Des. Devel. Ther. 2017, 11, 2179. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.-J.; Tao, H.; Huang, C.; Li, J. Nuclear erythroid 2-related factor 2: A novel potential therapeutic target for liver fibrosis. Food Chem. Toxicol. 2013, 59, 421–427. [Google Scholar] [CrossRef]
- Xu, W.; Hellerbrand, C.; Koehler, U.A.; Bugnon, P.; Kan, Y.-W.; Werner, S.; Beyer, T.A. The Nrf2 transcription factor protects from toxin-induced liver injury and fibrosis. Lab. Investig. 2008, 88, 1068–1078. [Google Scholar] [CrossRef]
- Huang, W.; Li, L.; Tian, X.; Yan, J.; Yang, X.; Wang, X.; Liao, G.; Qiu, G. Astragalus and Paeoniae radix rubra extract inhibits liver fibrosis by modulating the transforming growth factorβ/Smad pathway in rats. Mol. Med. Rep. 2015, 11, 805–814. [Google Scholar] [CrossRef] [Green Version]
- Supriono, S.; Nugraheni, A.; Kalim, H.; Eko, M.H. The Effect of Curcumin on Regression of Liver Fibrosis through Decreased Expression of Transforming Growth Factor-β1 (TGF-β1). Indones. Biomed. J. 2019, 11, 52–58. [Google Scholar] [CrossRef]
- Niu, L.; Cui, X.; Qi, Y.; Xie, D.; Wu, Q.; Chen, X.; Ge, J.; Liu, Z. Involvement of TGF-β1/Smad3 signaling in carbon tetrachloride-induced acute liver injury in mice. PLoS ONE 2016, 11, e0156090. [Google Scholar] [CrossRef]
- Gressner, A.; Weiskirchen, R.J. Modern pathogenetic concepts of liver fibrosis suggest stellate cells and TGF-β as major players and therapeutic targets. J. Cell. Mol. Med. 2006, 10, 76–99. [Google Scholar] [CrossRef] [Green Version]
- Tu, X.; Zhang, H.; Zhang, J.; Zhao, S.; Zheng, X.; Zhang, Z.; Zhu, J.; Chen, J.; Dong, L.; Zang, J. MicroRNA-101 suppresses liver fibrosis by targeting the TGF β signalling pathway. J. Pathol. 2014, 234, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Chen, B.; Fan, X.; Li, G.; Dong, P.; Zheng, J. Epigenetically-regulated microRNA-9-5p suppresses the activation of hepatic stellate cells via TGFBR1 and TGFBR2. Cell. Physiol. Biochem. 2017, 43, 2242–2252. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Benz, F.; Luedde, T.; Roderburg, C. The role of miRNAs in the regulation of inflammatory processes during hepatofibrogenesis. Hepatobiliary Surg. Nutr. 2015, 4, 24. [Google Scholar] [PubMed]
- Gjorgjieva, M.; Sobolewski, C.; Ay, A.-S.; Abegg, D.; Correia de Sousa, M.; Portius, D.; Berthou, F.; Fournier, M.; Maeder, C.; Rantakari, P. Genetic ablation of MiR-22 fosters diet-induced obesity and NAFLD development. J. Pers. Med. 2020, 10, 170. [Google Scholar] [CrossRef] [PubMed]
- Roulot, D.; Sevcsik, A.M.; Coste, T.; Strosberg, A.D.; Marullo, S. Role of transforming growth factor β type II receptor in hepatic fibrosis: Studies of human chronic hepatitis C and experimental fibrosis in rats. Hepatol. Int. 1999, 29, 1730–1738. [Google Scholar] [CrossRef]
- Hong, Y.; Cao, H.; Wang, Q.; Ye, J.; Sui, L.; Feng, J.; Cai, X.; Song, H.; Zhang, X.; Chen, X. MiR-22 may suppress fibrogenesis by targeting TGFβR I in cardiac fibroblasts. Cell. Physiol. Biochem. 2016, 40, 1345–1353. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Q.; Wang, Q.; Zhao, X.; Zhao, P.; Geng, T.; Gong, D. Role of miR29c in goose fatty liver is mediated by its target genes that are involved in energy homeostasis and cell growth. BMC Vet. Res. 2018, 14, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, T.F.; Cohen-Gould, L.; Factor, S.M.; Eghbali, M.; Blumenfeld, O.O. Structure and function of connective tissue in cardiac muscle: Collagen types I and III in endomysial struts and pericellular fibers. Scanning Microsc. 1988, 2, 33. [Google Scholar]
- Hafez, M.M.; Hamed, S.S.; El-Khadragy, M.F.; Hassan, Z.K.; Al Rejaie, S.S.; Sayed-Ahmed, M.M.; Al-Harbi, N.O.; Al-Hosaini, K.A.; Al-Harbi, M.M.; Alhoshani, A.R.; et al. Effect of ginseng extract on the TGF-β1 signaling pathway in CCl 4-induced liver fibrosis in rats. BMC Complement. Altern. Med. 2017, 17, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leti, F.; Malenica, I.; Doshi, M.; Courtright, A.; Van Keuren-Jensen, K.; Legendre, C.; Still, C.D.; Gerhard, G.S.; DiStefano, J.K. High-throughput sequencing reveals altered expression of hepatic microRNAs in nonalcoholic fatty liver disease–related fibrosis. Transl. Res. 2015, 166, 304–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, R.; Zhou, J.; Wang, R.; Sun, R.; Feng, D.; Wang, Z.; Zhao, Y.; Lv, L.; Tian, X.; Yao, J.; et al. Protocatechuic acid-mediated miR-219a-5p activation inhibits the p66shc oxidant pathway to alleviate alcoholic liver injury. Oxid. Med. Cell. Longev. 2019, 2019, 3527809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohseni, R.; Karimi, J.; Tavilani, H.; Khodadadi, I.; Hashemnia, M. Carvacrol ameliorates the progression of liver fibrosis through targeting of Hippo and TGF-β signaling pathways in carbon tetrachloride (CCl4)-induced liver fibrosis in rats. Immunopharmacol. Immunotoxicol. 2019, 41, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Santibañez, J.F.; Quintanilla, M.; Bernabeu, C. TGF-β/TGF-β receptor system and its role in physiological and pathological conditions. Clin. Sci. (Lond.) 2011, 121, 233–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansal, M.B.; Chamroonkul, N. Antifibrotics in liver disease: Are we getting closer to clinical use? Hepatol. Int. 2019, 13, 25–39. [Google Scholar] [CrossRef]
- Ebrahimi, H.; Naderian, M.; Sohrabpour, A.A. New concepts on reversibility and targeting of liver fibrosis; a review article. Middle East J. Dig. Dis. 2018, 10, 133. [Google Scholar] [CrossRef]
- Gu, X.; Manautou, J.E. Molecular mechanisms underlying chemical liver injury. Expert Rev. Mol. Med. 2012, 14. [Google Scholar] [CrossRef] [Green Version]
- Clichici, S.; David, L.; Moldovan, B.; Baldea, I.; Olteanu, D.; Filip, M.; Nagy, A.; Luca, V.; Crivii, C.; Mircea, P.; et al. Hepatoprotective effects of silymarin coated gold nanoparticles in experimental cholestasis. Mater. Sci. Eng. 2020, 115, 111117. [Google Scholar] [CrossRef]
- Staroverov, S.; Kozlov, S.; Fomin, A.; Gabalov, K.; Volkov, A.; Domnitsky, I.; Dykman, L.; Guliy, O. Synthesis of Silymarin−Gold Nanoparticle Conjugate and Analysis of its Liver-Protecting Activity. Curr. Pharm. Biotechnol. 2021, 22, 2001–2007. [Google Scholar] [CrossRef]
- Gillessen, A.; Schmidt, H.H. Silymarin as supportive treatment in liver diseases: A narrative review. Adv. Ther. 2020, 37, 1279–1301. [Google Scholar] [CrossRef] [Green Version]
- Stratton, M.; Ramachandran, A.; Camacho, E.J.M.; Patil, S.; Waris, G.; Grice, K.A. JAnti-fibrotic activity of gold and platinum complexes–Au (I) compounds as a new class of anti-fibrotic agents. J. Inorg. Biochem. 2020, 206, 111023. [Google Scholar] [CrossRef]
- Dkhil, M.A.; Bauomy, A.A.; Diab, M.S.; Al-Quraishy, S.J. Antioxidant and hepatoprotective role of gold nanoparticles against murine hepatic schistosomiasis. Int. J. Nanomed. 2015, 10, 7467. [Google Scholar]
- de Carvalho, T.G.; Garcia, V.B.; de Araújo, A.A.; da Silva Gasparotto, L.H.; Silva, H.; Guerra, G.C.B.; de Castro Miguel, E.; de Carvalho Leitão, R.F.; da Silva Costa, D.V.; Cruz, L.J. Spherical neutral gold nanoparticles improve anti-inflammatory response, oxidative stress and fibrosis in alcohol-methamphetamine-induced liver injury in rats. Int. J. Pharm. 2018, 548, 1–14. [Google Scholar] [CrossRef]
- Wang, L.; Huang, Q.-H.; Li, Y.-X.; Huang, Y.-F.; Xie, J.-H.; Xu, L.-Q.; Dou, Y.-X.; Su, Z.-R.; Zeng, H.-F.; Chen, J. Protective effects of silymarin on triptolide-induced acute hepatotoxicity in rats. Mol. Med. Rep. 2018, 17, 789–800. [Google Scholar] [CrossRef]
- Javanshir, R.; Honarmand, M.; Hosseini, M.; Hemmati, M. Anti-dyslipidemic properties of green gold nanoparticle: Improvement in oxidative antioxidative balance and associated atherogenicity and insulin resistance. Clin. Phytoscience 2020, 6, 74. [Google Scholar] [CrossRef]
- Teksoy, O.; Sahinturk, V.; Cengiz, M.; İnal, B.; Ayhancı, A. The Protective Effects of Silymarin on Thioacetamide-Induced Liver Damage: Measurement of miR-122, miR-192, and miR-194 Levels. Appl. Biochem. Biotechnol. 2020, 191, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Yang, S.-G.; Kim, J.M.; Lee, J.-W.; Kim, Y.S.; Lee, J.I. Silymarin suppresses hepatic stellate cell activation in a dietary rat model of non-alcoholic steatohepatitis: Analysis of isolated hepatic stellate cells. Int. J. Mol. Med. 2012, 30, 473–479. [Google Scholar] [CrossRef] [Green Version]
- Meng, S.; Yang, F.; Wang, Y.; Qin, Y.; Xian, H.; Che, H.; Wang, L. Silymarin ameliorates diabetic cardiomyopathy via inhibiting TGF-β1/Smad signaling. Cell Biol. Int. Rep. 2019, 43, 65–72. [Google Scholar] [CrossRef] [Green Version]
- Anestopoulos, I.; Sfakianos, A.P.; Franco, R.; Chlichlia, K.; Panayiotidis, M.I.; Kroll, D.J.; Pappa, A. A novel role of silibinin as a putative epigenetic modulator in human prostate carcinoma. Mol. Cells 2017, 22, 62. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Shen, N.; Yang, Y.; Yu, H.; Xu, S.; Yang, Y.-W.; Liu, S.; Meguellati, K.; Yan, F. Targeting epigenetic pathway with gold nanoparticles for acute myeloid leukemia therapy. Biomaterials 2018, 167, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Maccora, D.; Dini, V.; Battocchio, C.; Fratoddi, I.; Cartoni, A.; Rotili, D.; Castagnola, M.; Faccini, R.; Bruno, I.; Scotognella, T. Gold nanoparticles and nanorods in nuclear medicine: A mini review. Appl. Sci. 2019, 9, 3232. [Google Scholar] [CrossRef] [Green Version]













| Gene | Sequence | |
|---|---|---|
| COL3A1 | Forward | 5′-AAC GGA GCT CCT GGC CCC AT-3′ |
| Reverse | 5′-ATT GCC TCG AGC ACC TGC GG-3′ | |
| TGFβR1 | Forward | 5′-GCT GAC ATC TAT GCA ATG GG-3′ |
| Reverse | 5′-ATA TTT GGC CTT AAC TTC TGT TC-3′ | |
| TGFβR2 | Forward | 5′-CCA GGG CAT CCA GAT CGT GTG-3′ |
| Reverse | 5′-TAG TGT TCA GGG AGC CGT CTT-3′ | |
| GAPDH | Forward | 5′-GGG TGT GAA CCA CGA GAA ATA-3′ |
| Reverse | 5′-AGT TGT CAT GGA TGA CCT T-3′ |
| Silymarin Concentration | |||
|---|---|---|---|
| Parameters | 0.482 mg/mL | 1 mg/mL | 1.2 mg/mL |
| Entrapment efficiency | 90% | 95% | 96% |
| Drug loading capacity | 19% | 34% | 38.69% |
| Groups | Parameters | ||
|---|---|---|---|
| Body Weight Gain (g) | Liver Weight (g) | Liver Index (%) | |
| Control | 101.50 a ± 11.24 | 9.08 c ± 0.64 | 2.59 d ± 0.21 |
| CCl4 | −44.14 c ± 10.07 | 13.22 a ± 0.71 | 6.41 a ± 0.26 |
| CCl4+SIL | 52.83 b ± 5.49 | 11.25 b ± 0.23 | 3.70 c ± 0.11 |
| CCl4+GNPs | −30 c ± 9.92 | 11.50 b ± 0.48 | 5.16 b ± 0.32 |
| CCl4+SGNPs | 93.67 a ± 5.47 | 9.24 c ± 0.52 | 2.68 d ± 0.11 |
| Groups | Parameters | ||||
|---|---|---|---|---|---|
| AST (U/L) | ALT (U/L) | ALP (U/L) | Total Bilirubin (mg/dL) | Albumin (g/dL) | |
| Control | 111.5 d ± 12.44 | 43.33 d ± 7.42 | 101.3 d ± 13.79 | 0.40 c ± 0.10 | 4.11 a ± 0.21 |
| CCl4 | 195.5 a ± 17.76 | 130.8 a ± 13.70 | 252.2 a ± 19.62 | 1.15 a ± 0.30 | 3.14 c ± 0.13 |
| CCl4+SIL | 134.3 bcd ± 12.75 | 61.17 c ± 9.02 | 148.7 c ± 16.72 | 0.75 b ± 0.21 | 3.49 bc ± 0.24 |
| CCl4+GNPs | 159.0 b ± 15.10 | 104.7 b ± 8.91 | 189.0 b ± 16.42 | 0.83 b ± 0.12 | 3.27 bc ± 0.27 |
| CCl4+SGNPs | 125.0 cd ± 10.14 | 51.8 cd ± 6.77 | 133.5 c ± 12.58 | 0.67 bc ± 0.10 | 3.60 b ± 0.08 |
| Groups | Parameters | ||
|---|---|---|---|
| MDA (nmol/g Tissues) | NRF2 (pg/mg Protein) | TGF-β1 (ng/mg Protein) | |
| Control | 11.68 c ± 2.46 | 109.4 a ± 9.48 | 7.24 d ± 2.04 |
| CCl4 | 25.75 a ± 4.43 | 56.97 c ± 8.31 | 28.44 a ± 3.04 |
| CCl4+SIL | 17.30 b ± 2.13 | 68.88 bc ± 11.77 | 19.12 c ± 1.91 |
| CCl4+GNPs | 18.11 b ± 3.81 | 59.27 bc ± 7.60 | 23.40 b ± 2.53 |
| CCl4+SGNPs | 14.48 bc ± 2.06 | 76.30 b ± 14.95 | 18.05 c ± 1.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullah, A.S.; El Sayed, I.E.T.; El-Torgoman, A.M.A.; Alghamdi, N.A.; Ullah, S.; Wageh, S.; Kamel, M.A. Preparation and Characterization of Silymarin-Conjugated Gold Nanoparticles with Enhanced Anti-Fibrotic Therapeutic Effects against Hepatic Fibrosis in Rats: Role of MicroRNAs as Molecular Targets. Biomedicines 2021, 9, 1767. https://doi.org/10.3390/biomedicines9121767
Abdullah AS, El Sayed IET, El-Torgoman AMA, Alghamdi NA, Ullah S, Wageh S, Kamel MA. Preparation and Characterization of Silymarin-Conjugated Gold Nanoparticles with Enhanced Anti-Fibrotic Therapeutic Effects against Hepatic Fibrosis in Rats: Role of MicroRNAs as Molecular Targets. Biomedicines. 2021; 9(12):1767. https://doi.org/10.3390/biomedicines9121767
Chicago/Turabian StyleAbdullah, Abdullah Saad, Ibrahim El Tantawy El Sayed, Abdel Moneim A. El-Torgoman, Noweir Ahmad Alghamdi, Sami Ullah, S. Wageh, and Maher A. Kamel. 2021. "Preparation and Characterization of Silymarin-Conjugated Gold Nanoparticles with Enhanced Anti-Fibrotic Therapeutic Effects against Hepatic Fibrosis in Rats: Role of MicroRNAs as Molecular Targets" Biomedicines 9, no. 12: 1767. https://doi.org/10.3390/biomedicines9121767






