Epigenomics and Lipidomics Integration in Alzheimer Disease: Pathways Involved in Early Stages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants and Samples Collection
2.2. Omics Analysis
2.2.1. Epigenomics
2.2.2. Lipidomics
2.3. Statistical Analysis and Lipidomics-Epigenomics Integration
3. Results
3.1. Participants
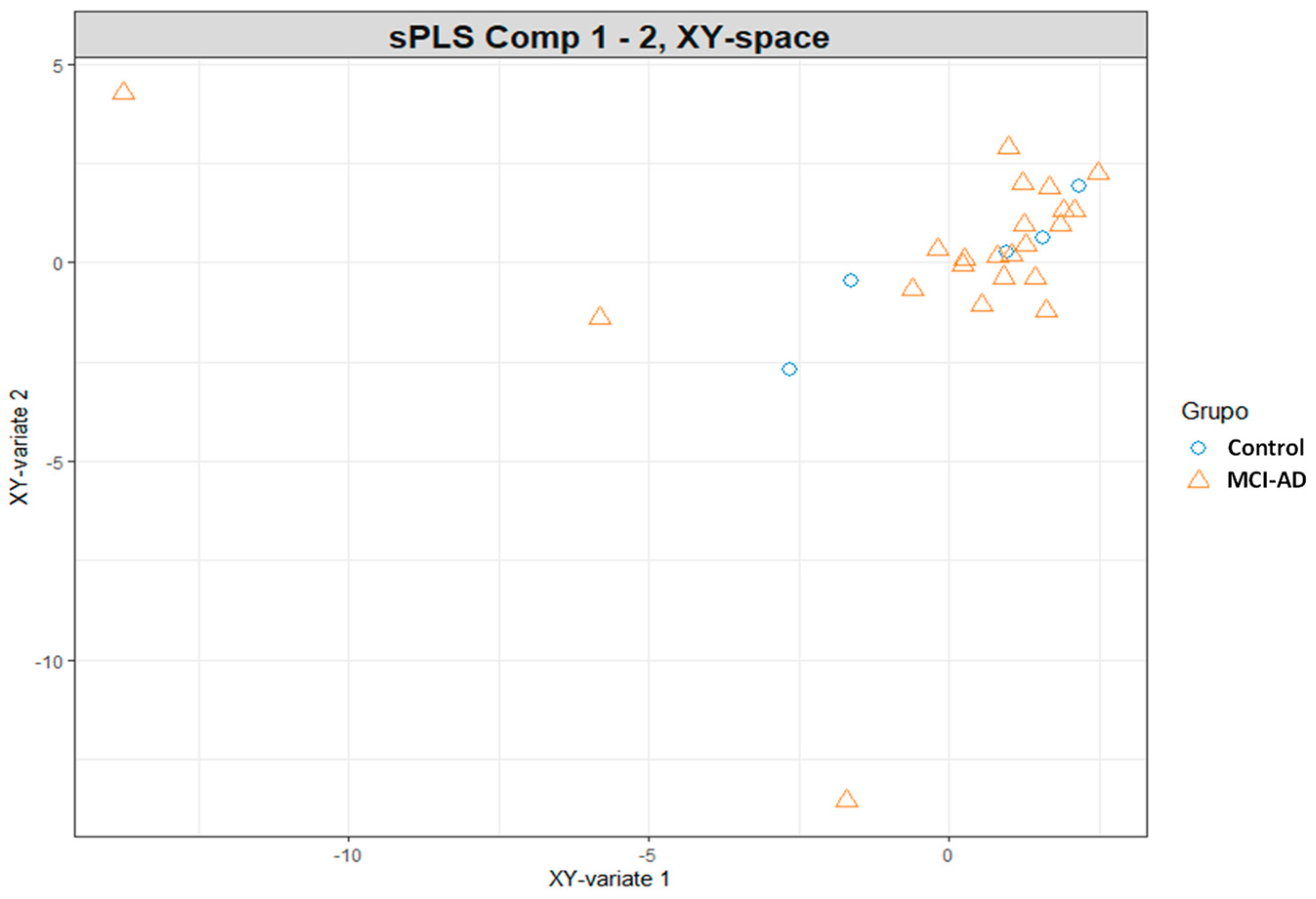
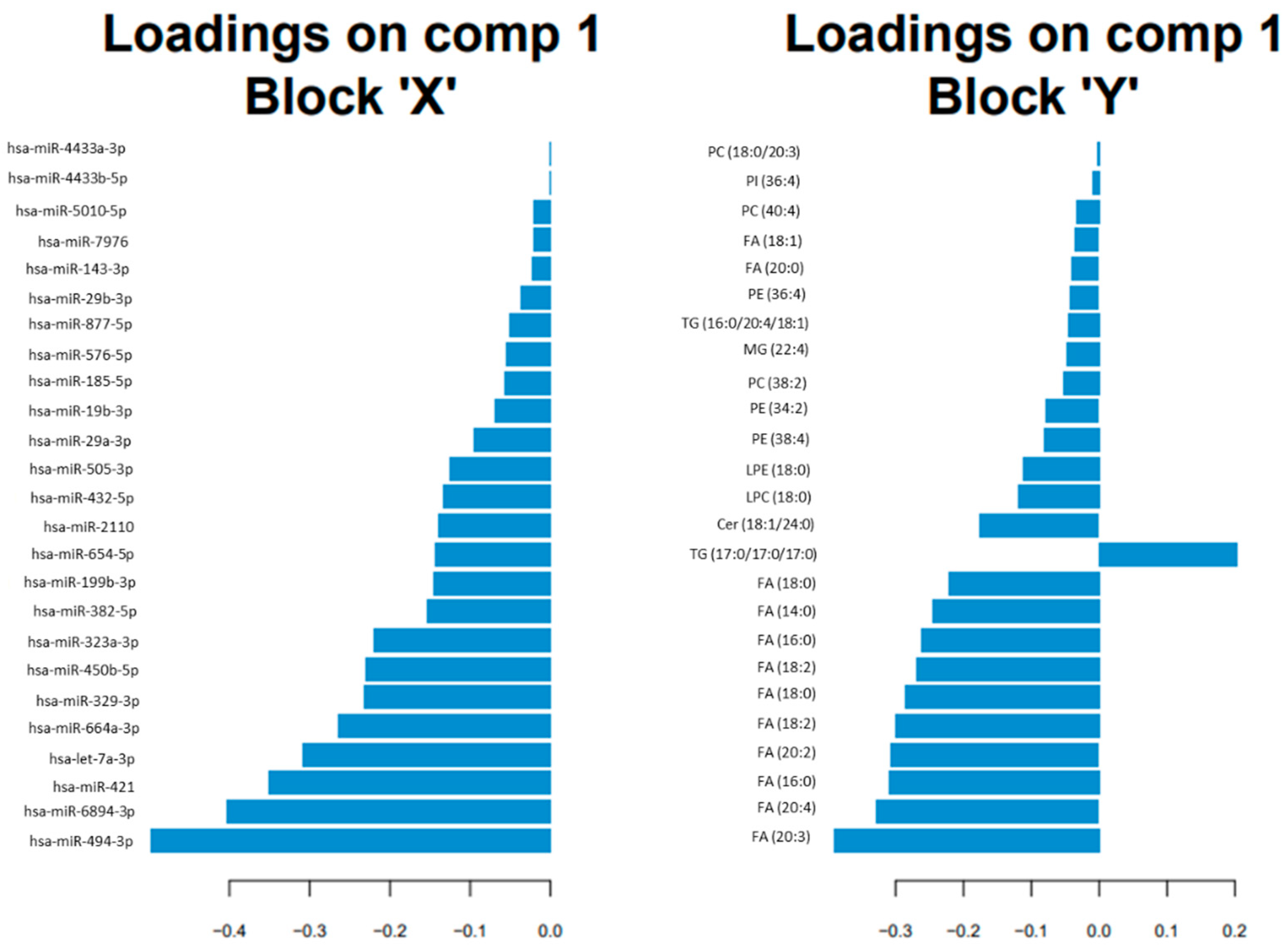
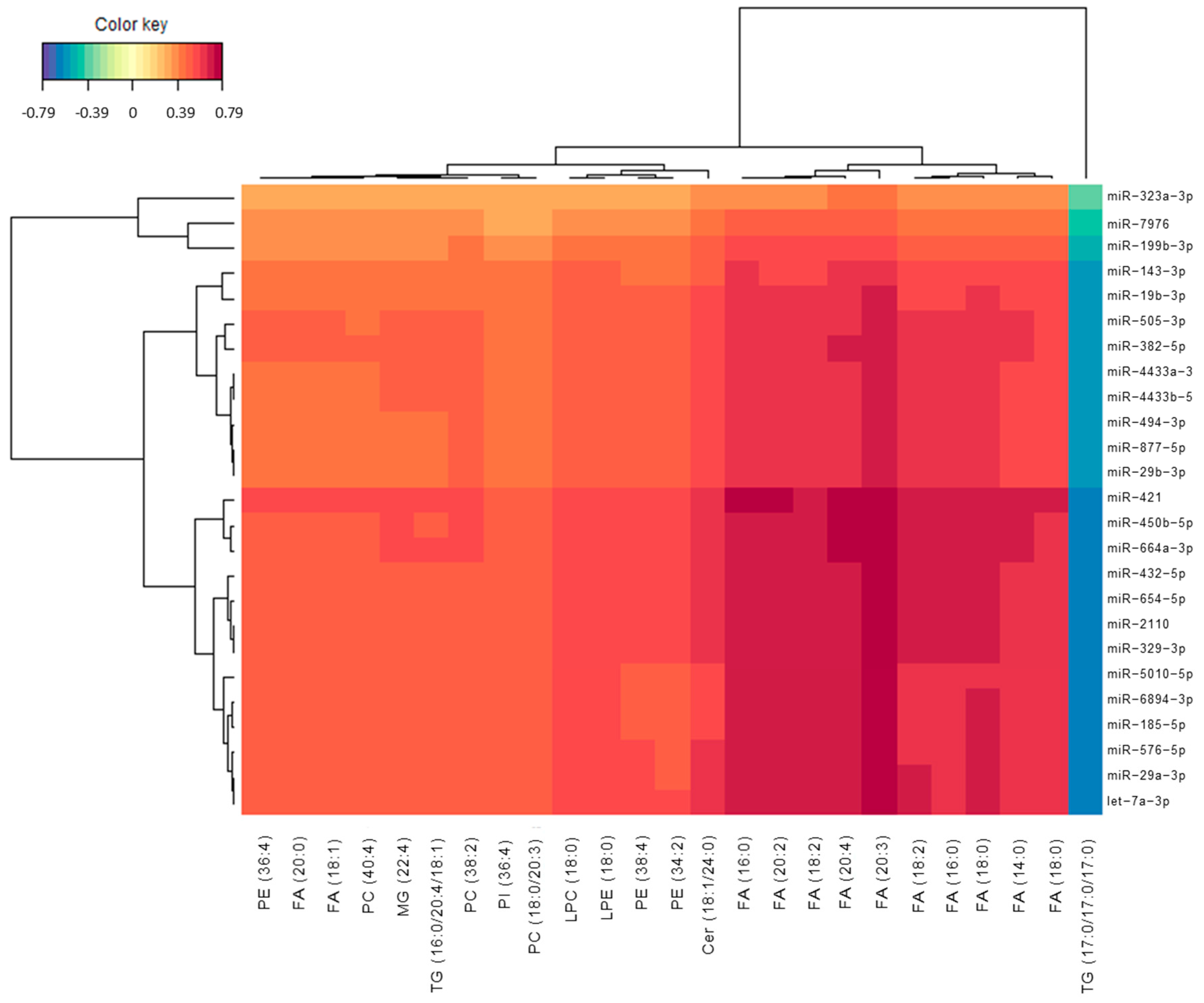
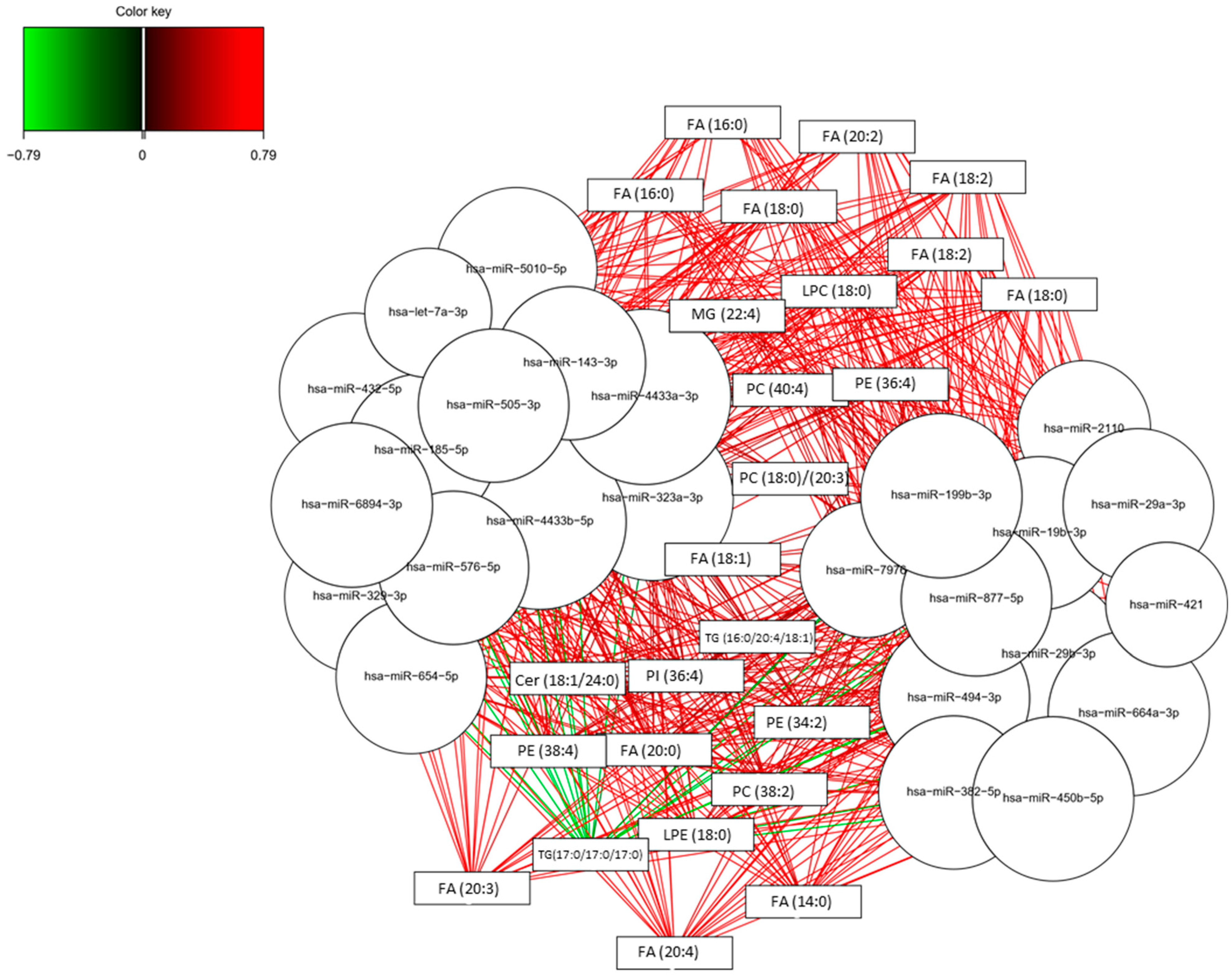
3.2. Omics Integration
3.3. Potential Pathways Involved in AD
3.4. Lipidomics and Epigenomics in AD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alzheimer’s Association. 2020 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2020, 16, 391–460. [Google Scholar] [CrossRef]
- Gong, C.-X.; Liu, F.; Iqbal, K. Multifactorial hypothesis and multi-targets for Alzheimer’s disease. J. Alzheimer’s Dis. 2018, 64, S107–S117. [Google Scholar] [CrossRef]
- Nativio, R.; Lan, Y.; Donahue, G.; Sidoli, S.; Berson, A.; Srinivasan, A.R.; Shcherbakova, O.; Amlie-Wolf, A.; Nie, J.; Cui, X.; et al. An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat. Genet. 2020, 52, 1024–1035. [Google Scholar] [CrossRef] [PubMed]
- Takousis, P.; Sadlon, A.; Schulz, J.; Wohlers, I.; Dobricic, V.; Middleton, L.; Lill, C.M.; Perneczky, R.; Bertram, L. Differential expression of microRNAs in Alzheimer’s disease brain, blood, and cerebrospinal fluid. Alzheimer’s Dement. 2019, 15, 1468–1477. [Google Scholar] [CrossRef] [PubMed]
- Mayo, S.; Benito-León, J.; Peña-Bautista, C.; Baquero, M.; Cháfer-Pericás, C. Recent evidence in epigenomics and proteomics biomarkers for early and minimally invasive diagnosis of Alzheimer’s and Parkinson’s diseases. Curr. Neuropharmacol. 2021, 19, 1273–1303. [Google Scholar] [CrossRef]
- Cannell, I.G.; Kong, Y.W.; Bushell, M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008, 36, 1224–1231. [Google Scholar] [CrossRef] [Green Version]
- Hébert, S.S.; Horré, K.; Nicolaï, L.; Bergmans, B.; Papadopoulou, A.S.; Delacourte, A.; De Strooper, B. MicroRNA regulation of Alzheimer’s Amyloid precursor protein expression. Neurobiol. Dis. 2009, 33, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.-C.; Ho, P.-C.; Tu, Y.-K.; Jou, I.-M.; Tsai, K.-J. Lipids and Alzheimer’s disease. Int. J. Mol. Sci. 2020, 21, 1505. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Bankov, G.; Kim, M.; Wretlind, A.; Lord, J.; Green, R.; Hodges, A.; Hye, A.; Aarsland, D.; Velayudhan, L.; et al. Integrated lipidomics and proteomics network analysis highlights lipid and immunity pathways associated with Alzheimer’s disease. Transl. Neurodegener. 2020, 9, 36. [Google Scholar] [CrossRef]
- Proitsi, P.; Kim, M.; Whiley, L.; Simmons, A.; Sattlecker, M.; Velayudhan, L.; Lupton, M.K.; Soininen, H.; Kloszewska, I.; Mecocci, P.; et al. Association of blood lipids with Alzheimer’s disease: A comprehensive lipidomics analysis. Alzheimer’s Dement. 2017, 13, 140–151. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Li, A.; Sekiya, M.; Beckmann, N.D.; Quan, X.; Schrode, N.; Fernando, M.B.; Yu, A.; Zhu, L.; Cao, J.; et al. Transformative network modeling of multi-omics data reveals detailed circuits, key regulators, and potential therapeutics for Alzheimer’s disease. Neuron 2021, 109, 257–272.e14. [Google Scholar] [CrossRef] [PubMed]
- Xicota, L.; Ichou, F.; Lejeune, F.-X.; Colsch, B.; Tenenhaus, A.; Leroy, I.; Fontaine, G.; Lhomme, M.; Bertin, H.; Habert, M.-O.; et al. Multi-omics signature of brain amyloid deposition in asymptomatic individuals at-risk for Alzheimer’s disease: The INSIGHT-preAD study. EBioMedicine 2019, 47, 518–528. [Google Scholar] [CrossRef] [Green Version]
- Bihlmeyer, N.A.; Merrill, E.; Lambert, Y.; Srivastava, G.P.; Clark, T.W.; Hyman, B.T.; Das, S. Novel methods for integration and visualization of genomics and genetics data in Alzheimer’s disease. Alzheimer’s Dement. 2019, 15, 788–798. [Google Scholar] [CrossRef] [PubMed]
- Shigemizu, D.; Akiyama, S.; Higaki, S.; Sugimoto, T.; Sakurai, T.; Boroevich, K.A.; Sharma, A.; Tsunoda, T.; Ochiya, T.; Niida, S.; et al. Prognosis prediction model for conversion from mild cognitive impairment to Alzheimer’s disease created by integrative analysis of multi-omics data. Alzheimers. Res. Ther. 2020, 12, 145. [Google Scholar] [CrossRef] [PubMed]
- Darst, B.F.; Lu, Q.; Johnson, S.C.; Engelman, C.D. Integrated analysis of genomics, longitudinal metabolomics, and Alzheimer’s risk factors among 1,111 cohort participants. Genet. Epidemiol. 2019, 43, 657–674. [Google Scholar] [CrossRef]
- Hampel, H.; Nisticò, R.; Seyfried, N.T.; Levey, A.I.; Modeste, E.; Lemercier, P.; Baldacci, F.; Toschi, N.; Garaci, F.; Perry, G.; et al. Omics sciences for systems biology in Alzheimer’s disease: State-of-the-art of the evidence. Ageing Res. Rev. 2021, 69, 101346. [Google Scholar] [CrossRef]
- Hedman, Å.K.; Mendelson, M.M.; Marioni, R.E.; Gustafsson, S.; Joehanes, R.; Irvin, M.R.; Zhi, D.; Sandling, J.K.; Yao, C.; Liu, C.; et al. Epigenetic patterns in blood associated with lipid traits predict incident coronary heart disease events and are enriched for results from genome-wide association studies. Circ. Cardiovasc. Genet. 2017, 10, e001487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordevicius, J.; Li, P.; Marshall, L.L.; Killinger, B.A.; Lang, S.; Ensink, E.; Kuhn, N.C.; Cui, W.; Maroof, N.; Lauria, R.; et al. Epigenetic inactivation of the autophagy–lysosomal system in appendix in Parkinson’s disease. Nat. Commun. 2021, 12, 5134. [Google Scholar] [CrossRef] [PubMed]
- Randolph, C.; Tierney, M.C.; Mohr, E.; Chase, T.N. The Repeatable Battery for the Assessment of Neuropsychological Status (RBANS): Preliminary clinical validity. J. Clin. Exp. Neuropsychol. 1998, 20, 310–319. [Google Scholar] [CrossRef]
- Pfeffer, R.I.; Kurosaki, T.T.; Harrah, C.H.; Chance, J.M.; Filos, S. Measurement of functional activities in older adults in the community. J. Gerontol. 1982, 37, 323–329. [Google Scholar] [CrossRef]
- Hughes, C.P.; Berg, L.; Danziger, W.L.; Coben, L.A.; Martin, R.L. A new clinical scale for the staging of dementia. Br. J. Psychiatry 1982, 140, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Kurlowicz, L.; Wallace, M. The mini-mental state examination (MMSE). J. Gerontol. Nurs. 1999, 25, 8–9. [Google Scholar] [CrossRef]
- Kulkarni, N.; Alessandrì, L.; Panero, R.; Arigoni, M.; Olivero, M.; Ferrero, G.; Cordero, F.; Beccuti, M.; Calogero, R.A. Reproducible bioinformatics project: A community for reproducible bioinformatics analysis pipelines. BMC Bioinform. 2018, 19, 349. [Google Scholar] [CrossRef] [PubMed]
- González, I.; Cao, K.-A.L.; Davis, M.J.; Déjean, S. Visualising associations between paired ‘omics’ data sets. BioData Min. 2012, 5, 19. [Google Scholar] [CrossRef] [Green Version]
- Murtagh, F.; Legendre, P. Ward’s hierarchical agglomerative clustering method: Which algorithms implement Ward’s criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wu, K.; Bo, T.; Zhou, L.; Gao, L.; Zhou, X.; Chen, W. Integrated microRNA and proteome analysis reveal a regulatory module in hepatic lipid metabolism disorders in mice with subclinical hypothyroidism. Exp. Ther. Med. 2020, 19, 897–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussey, G.S.; Molina, C.P.; Cramer, M.C.; Tyurina, Y.Y.; Tyurin, V.A.; Lee, Y.C.; El-Mossier, S.O.; Murdock, M.H.; Timashev, P.S.; Kagan, V.E.; et al. Lipidomics and RNA sequencing reveal a novel subpopulation of nanovesicle within extracellular matrix biomaterials. Sci. Adv. 2020, 6, eaay4361. [Google Scholar] [CrossRef] [Green Version]
- Alcazar, O.; Hernandez, L.F.; Nakayasu, E.S.; Nicora, C.D.; Ansong, C.; Muehlbauer, M.J.; Bain, J.R.; Myer, C.J.; Bhattacharya, S.K.; Buchwald, P.; et al. Parallel multi-omics in high-risk subjects for the identification of integrated biomarker signatures of type 1 diabetes. Biomolecules 2021, 11, 383. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.S.; Raijas, P.; Ahvenainen, M.; Philips, A.K.; Ukkola-Vuoti, L.; Järvelä, I. Music-listening regulates human microRNA expression. Epigenetics 2021, 16, 554–566. [Google Scholar] [CrossRef]
- Hojati, Z.; Omidi, F.; Dehbashi, M.; Mohammad Soltani, B. The highlighted roles of metabolic and cellular response to stress pathways engaged in circulating hsa-miR-494-3p and hsa-miR-661 in Alzheimer’s disease. Iran. Biomed. J. 2021, 25, 62–67. [Google Scholar] [CrossRef]
- Lv, Z.; Hu, L.; Yang, Y.; Zhang, K.; Sun, Z.; Zhang, J.; Zhang, L.; Hao, Y. Comparative study of microRNA profiling in one Chinese Family with PSEN1 G378E mutation. Metab. Brain Dis. 2018, 33, 1711–1720. [Google Scholar] [CrossRef]
- Gu, H.; Li, L.; Cui, C.; Zhao, Z.; Song, G. Overexpression of let-7a increases neurotoxicity in a PC12 cell model of Alzheimer’s disease via regulating autophagy. Exp. Ther. Med. 2017, 14, 3688–3698. [Google Scholar] [CrossRef] [Green Version]
- Rönnemaa, E.; Zethelius, B.; Vessby, B.; Lannfelt, L.; Byberg, L.; Kilander, L. Serum fatty-acid composition and the risk of Alzheimer’s disease: A longitudinal population-based study. Eur. J. Clin. Nutr. 2012, 66, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Conquer, J.A.; Tierney, M.C.; Zecevic, J.; Bettger, W.J.; Fisher, R.H. Fatty acid analysis of blood plasma of patients with alzheimer’s disease, other types of dementia, and cognitive impairment. Lipids 2000, 35, 1305–1312. [Google Scholar] [CrossRef] [PubMed]
- Goozee, K.; Chatterjee, P.; James, I.; Shen, K.; Sohrabi, H.R.; Asih, P.R.; Dave, P.; Ball, B.; ManYan, C.; Taddei, K.; et al. Alterations in erythrocyte fatty acid composition in preclinical Alzheimer’s disease. Sci. Rep. 2017, 7, 676. [Google Scholar] [CrossRef] [Green Version]
- Johnson, L.A.; Torres, E.R.S.; Impey, S.; Stevens, J.F.; Raber, J. Apolipoprotein E4 and Insulin Resistance Interact to Impair Cognition and Alter the Epigenome and Metabolome. Sci. Rep. 2017, 7, 43701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goedeke, L.; Fernández-Hernando, C. microRNAs: A connection between cholesterol metabolism and neurodegeneration. Neurobiol. Dis. 2014, 72, 48–53. [Google Scholar] [CrossRef] [Green Version]
- Jaouen, F.; Gascon, E. Understanding the role of miR-33 in brain lipid metabolism: Implications for Alzheimer’s disease. J. Neurosci. 2016, 36, 2558–2560. [Google Scholar] [CrossRef]
- Kumar, S.; Vijayan, M.; Reddy, P.H. MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer’s disease. Hum. Mol. Genet. 2017, 26, 3808–3822. [Google Scholar] [CrossRef]
- Denk, J.; Oberhauser, F.; Kornhuber, J.; Wiltfang, J.; Fassbender, K.; Schroeter, M.L.; Volk, A.E.; Diehl-Schmid, J.; Prudlo, J.; Danek, A.; et al. Specific serum and CSF microRNA profiles distinguish sporadic behavioural variant of frontotemporal dementia compared with Alzheimer patients and cognitively healthy controls. PLoS ONE 2018, 13, e0197329. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, W.; Zan, J.; Wu, C.; Tan, W. Untargeted lipidomics reveals progression of early Alzheimer’s disease in APP/PS1 transgenic mice. Sci. Rep. 2020, 10, 14509. [Google Scholar] [CrossRef] [PubMed]
- Barupal, D.K.; Baillie, R.; Fan, S.; Saykin, A.J.; Meikle, P.J.; Arnold, M.; Nho, K.; Fiehn, O.; Kaddurah-Daouk, R. Sets of coregulated serum lipids are associated with Alzheimer’s disease pathophysiology. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2019, 11, 619–627. [Google Scholar] [CrossRef]
- Ooi, K.-L.M.; Vacy, K.; Boon, W.C. Fatty acids and beyond: Age and Alzheimer’s disease related changes in lipids reveal the neuro-nutraceutical potential of lipids in cognition. Neurochem. Int. 2021, 149, 105143. [Google Scholar] [CrossRef] [PubMed]
- Braga, A.A.; Bortolin, R.H.; Graciano-Saldarriaga, M.E.; Hirata, T.D.; Cerda, A.; de Freitas, R.C.; Lin-Wang, H.T.; Borges, J.B.; França, J.I.; Masi, L.N.; et al. High serum miR-421 is associated with metabolic dysregulation and inflammation in patients with metabolic syndrome. Epigenomics 2021, 13, 423–436. [Google Scholar] [CrossRef] [PubMed]
- He, P.-P.; Jiang, T.; OuYang, X.-P.; Liang, Y.-Q.; Zou, J.-Q.; Wang, Y.; Shen, Q.-Q.; Liao, L.; Zheng, X.-L. Lipoprotein lipase: Biosynthesis, regulatory factors, and its role in atherosclerosis and other diseases. Clin. Chim. Acta 2018, 480, 126–137. [Google Scholar] [CrossRef] [PubMed]
| Test or Biomarker | Participant Group | |
|---|---|---|
| Control | MCI-AD | |
| CDR * | 0–0.5 | 0.5–1 |
| MMSE * | ≥27 | <27 |
| RBANS.DM * | ≥85 | <85 |
| FAQ | <9 | >9 |
| Neuroimaging Structural (NMR-TAC) | Normal | Altered |
| CSF amyloid β42 (pg mL−1) | ≥700 | <700 |
| CSF t-Tau (pg mL−1) | <350 | ≥350 |
| CSF p-Tau (pg mL−1) | <85 | ≥85 |
| Variables | Healthy Group (n = 5) | MCI-AD Group (n = 22) |
|---|---|---|
| Age (years, median (IQR)) | 68 (68, 72) | 72 (69, 74) |
| Gender (female, n (%)) | 2 (40%) | 12 (54.5%) |
| CSF amyloid β-42 (pg mL−1, median (IQR)) | 1346.74 (930, 1421) | 517.16 (453.86, 634.45) |
| CSF amyloid β-42/amyloid β-40 (median, IQR) | 0.1 (0.09, 0.11) | 0.05 (0.05, 0.05) |
| CSF t-Tau (pg mL−1, median (IQR)) | 240 (238, 276) | 566 (450, 780) |
| CSF p-Tau (pg mL−1, median (IQR)) | 35 (35, 40) | 81 (64.5, 107) |
| CSF NfL (pg mL−1, median (IQR)) | 826.94 (791, 847.7) | 1428.68 (1123.24, 1555.91) |
| CSF t-Tau/amyloid β-42 (median (IQR)) | 0.2 (0.19, 0.25) | 0.99 (0.79, 1.32) |
| CDR (score, median (IQR)) | 0 (0–0.5) | 0.5 (0–1) |
| MMSE (score, median (IQR)) | 29 (29, 30) | 24 (23, 26) |
| RBANS_DM (score, median (IQR)) | 100 (98, 110) | 44 (40, 64) |
| FAQ (score, median (IQR)) | 1 (0, 2) | 7 (4, 9) |
| miRNA | Target Genes |
|---|---|
| hsa-miR-494-3p | ELOVL3 (ELOVL fatty acid elongase 3) |
| ELOVL5 (ELOVL fatty acid elongase 5) | |
| hsa-miR-6894-3p | |
| hsa-miR-421 | ARV1 (ARV1 homolog, fatty acid homeostasis modulator) |
| FAR1 (fatty acyl-CoA reductase 1) | |
| ELOVL2 (ELOVL fatty acid elongase 2) | |
| hsa-let-7a-3p | ELOVL2 (ELOVL fatty acid elongase 2) |
| FA2H (fatty acid 2-hydroxylase) | |
| ELOVL7 (ELOVL fatty acid elongase 7) | |
| hsa-miR-664a-3p | FAR1 (fatty acyl-CoA reductase 1) |
| ELOVL4 (ELOVL fatty acid elongase 4) | |
| ELOVL7 ELOVL fatty acid elongase 7 | |
| ELOVL5 ELOVL fatty acid elongase 5 | |
| hsa-miR-329-3p | |
| hsa-miR-450b-5p | ELOVL6 (ELOVL fatty acid elongase 6) |
| hsa-miR-323a-3p | |
| hsa-miR-382-5p | |
| hsa-miR-199b-3p | |
| hsa-miR-654-5p | FADS6 (fatty acid desaturase 6) |
| ELOVL1 (ELOVL fatty acid elongase 1) | |
| hsa-miR-2110 | ELOVL4 (ELOVL fatty acid elongase 4) |
| hsa-miR-432-5p | |
| hsa-miR-505-3p | ELOVL4 (ELOVL fatty acid elongase 4) |
| hsa-miR-29a-3p | ELOVL4 (ELOVL fatty acid elongase 4) |
| hsa-miR-19b-3p | ELOVL5 (ELOVL fatty acid elongase 5) |
| hsa-miR-185-5p | ELOVL4 (ELOVL fatty acid elongase 4) |
| ELOVL2 (ELOVL fatty acid elongase 2) | |
| FAR1 (fatty acyl-CoA reductase 1) | |
| hsa-miR-576-5p | FAR2 (fatty acyl-CoA reductase 2) |
| hsa-miR-877-5p | |
| hsa-miR-29b-3p | ELOVL4 (ELOVL fatty acid elongase 4) |
| hsa-miR-143-3p | FADS6 (fatty acid desaturase 6) |
| FAR1 (fatty acyl-CoA reductase 1) | |
| hsa-miR-7976 | |
| hsa-miR-5010-5p | |
| hsa-miR-4433b-5p | |
| hsa-miR-4433a-3p | FABP7 (fatty acid binding protein 7) |
| ELOVL4 (ELOVL fatty acid elongase 4) | |
| ELOVL2 (ELOVL fatty acid elongase 2) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peña-Bautista, C.; Álvarez-Sánchez, L.; Cañada-Martínez, A.J.; Baquero, M.; Cháfer-Pericás, C. Epigenomics and Lipidomics Integration in Alzheimer Disease: Pathways Involved in Early Stages. Biomedicines 2021, 9, 1812. https://doi.org/10.3390/biomedicines9121812
Peña-Bautista C, Álvarez-Sánchez L, Cañada-Martínez AJ, Baquero M, Cháfer-Pericás C. Epigenomics and Lipidomics Integration in Alzheimer Disease: Pathways Involved in Early Stages. Biomedicines. 2021; 9(12):1812. https://doi.org/10.3390/biomedicines9121812
Chicago/Turabian StylePeña-Bautista, Carmen, Lourdes Álvarez-Sánchez, Antonio José Cañada-Martínez, Miguel Baquero, and Consuelo Cháfer-Pericás. 2021. "Epigenomics and Lipidomics Integration in Alzheimer Disease: Pathways Involved in Early Stages" Biomedicines 9, no. 12: 1812. https://doi.org/10.3390/biomedicines9121812
APA StylePeña-Bautista, C., Álvarez-Sánchez, L., Cañada-Martínez, A. J., Baquero, M., & Cháfer-Pericás, C. (2021). Epigenomics and Lipidomics Integration in Alzheimer Disease: Pathways Involved in Early Stages. Biomedicines, 9(12), 1812. https://doi.org/10.3390/biomedicines9121812






