Unraveling Autocrine Signaling Pathways through Metabolic Fingerprinting in Serous Ovarian Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture
2.2. Metabolomic Analysis
2.3. Data Processing
2.4. Statistical Analysis
2.5. Principal Component Analysis and Hierarchical Clustering
2.6. Cell Proliferation and InhibitorAnalysis
2.7. 3D-Spheroid Analysis
3. Results
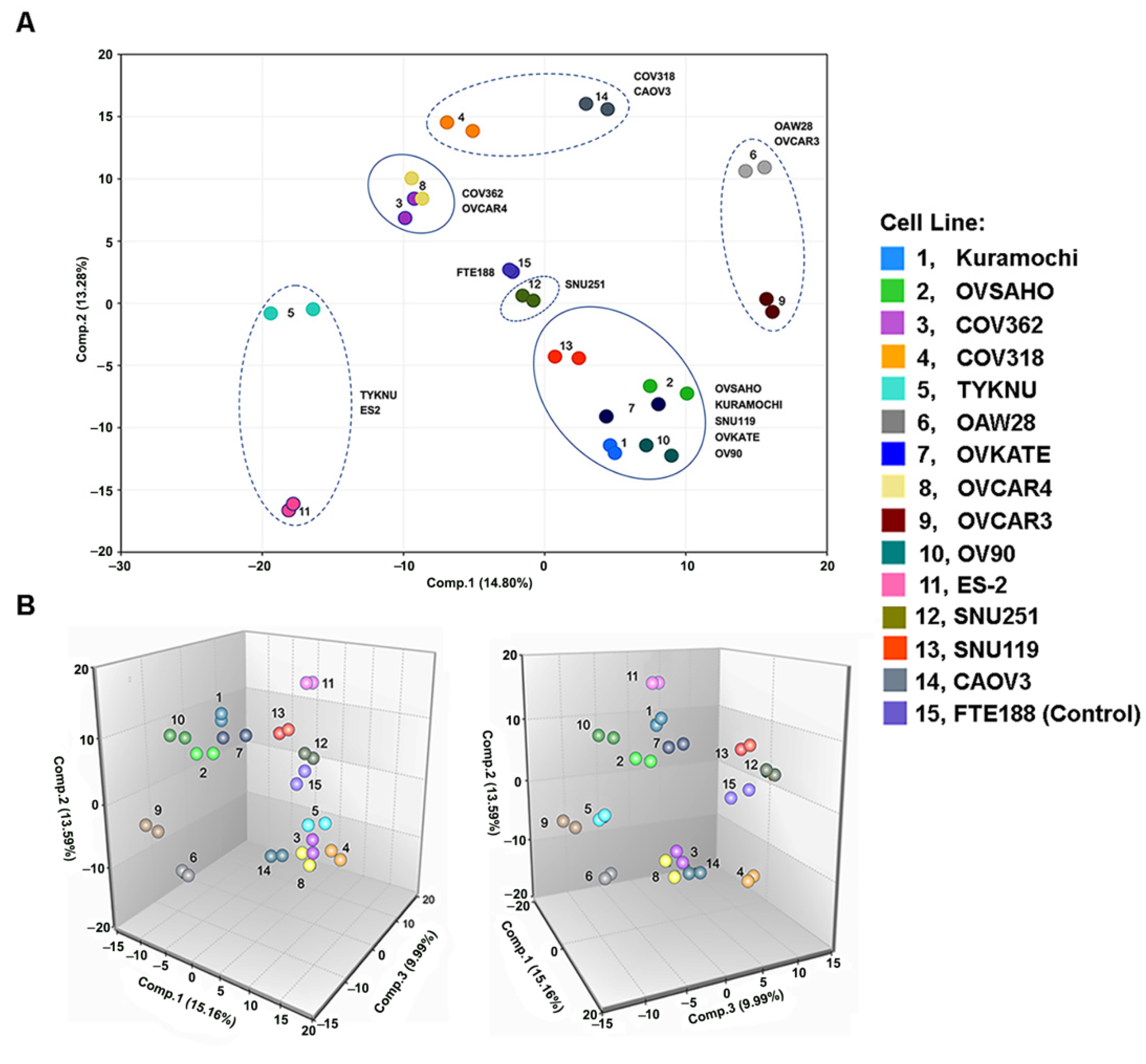
3.1. Metabolomic Heterogeneity and Clustering of HGSOC Cell Lines
3.2. Glycolysis and TCA Metabolism Profile in Ovarian Cancer Cells
3.3. Increased Synthesis and Turnover of Phospholipids in HGSOC Cells
3.4. Increased Amino Acid Metabolism in HGSOC Cells
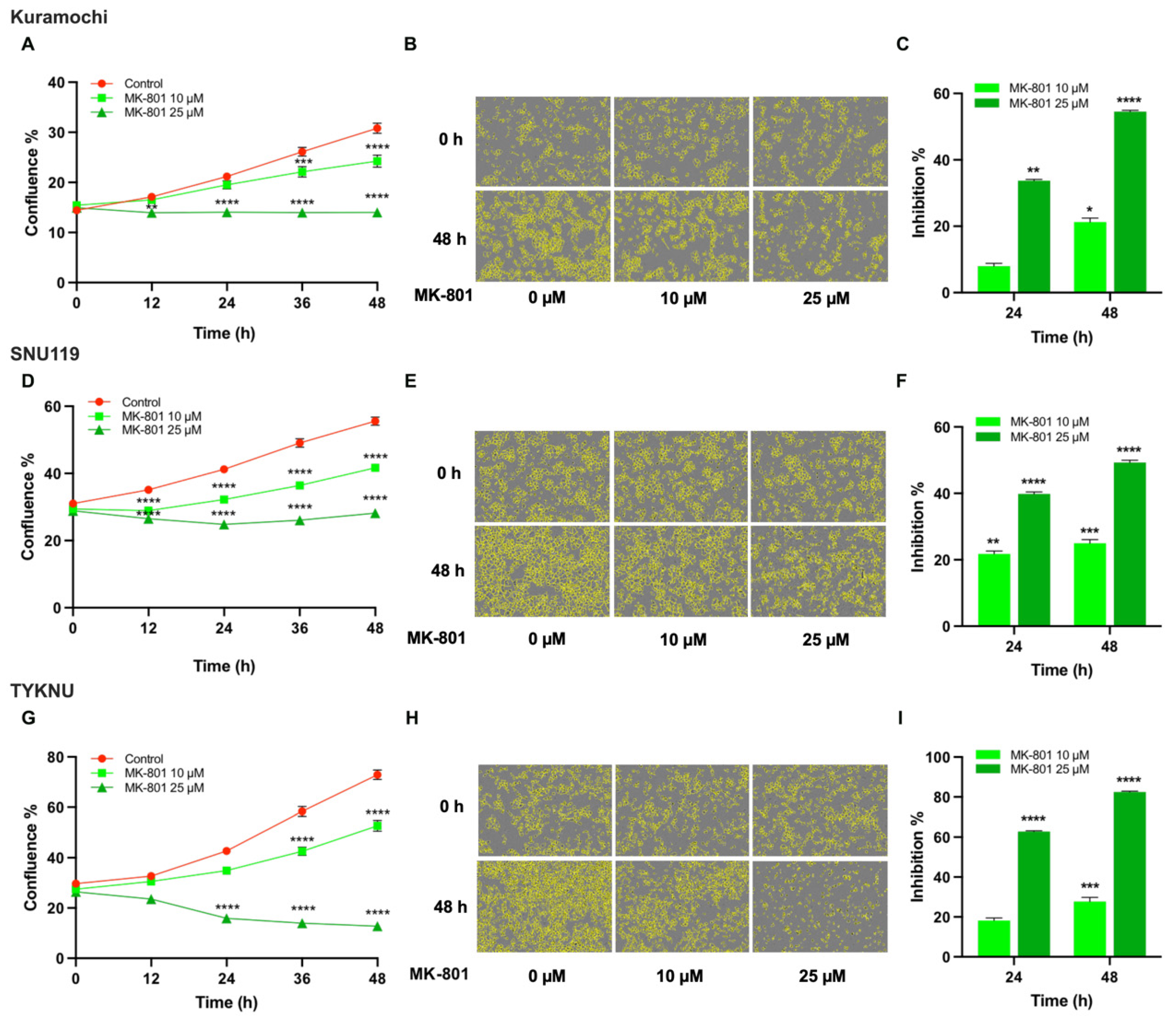
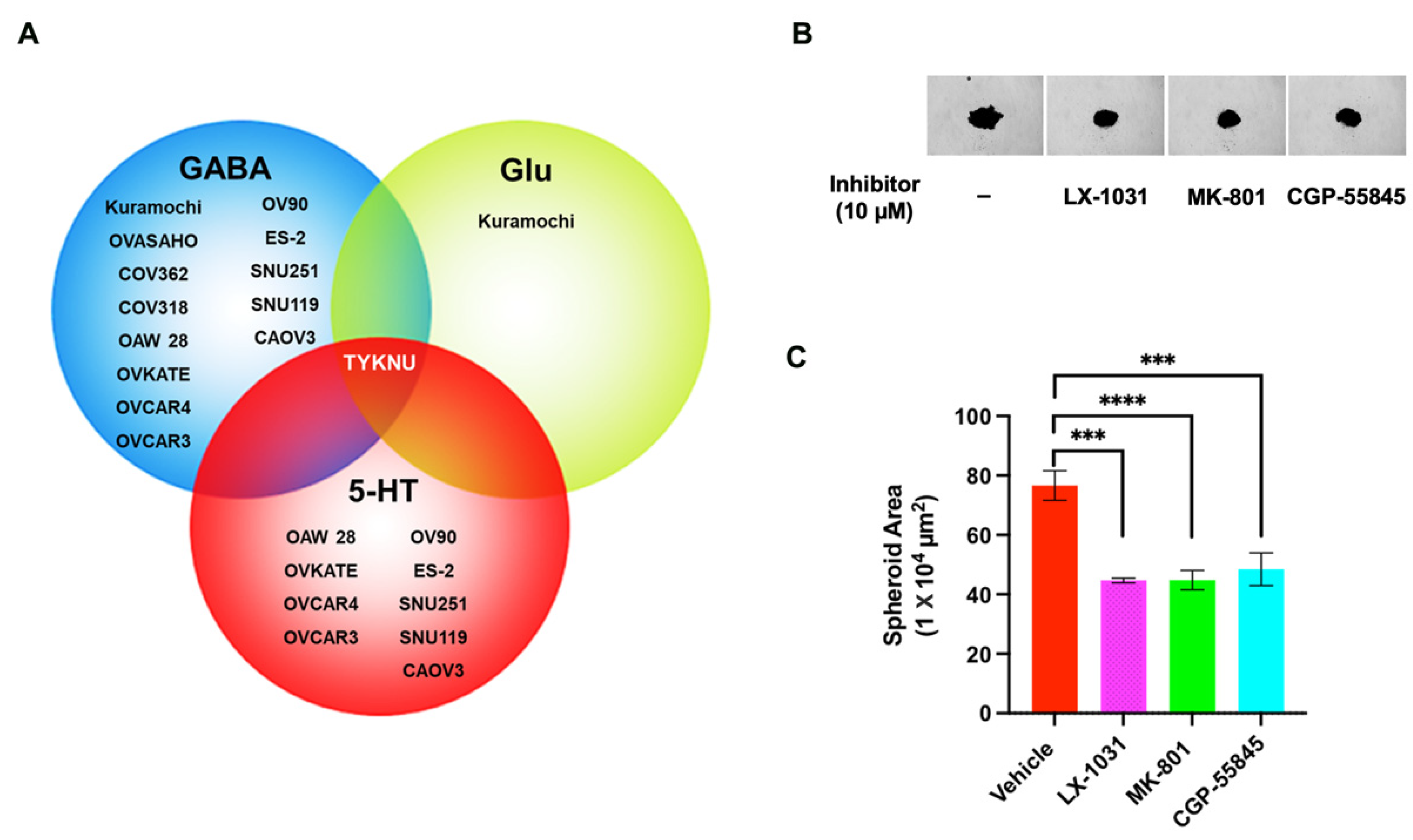
3.5. Targeting the Potential GABA-Mediated Autocrine Signaling in HGSOC Cells
3.6. Targeting the Potential Glu-Mediated Autocrine Signaling in HGSOC Cells
3.7. Evaluating 5-HT-Mediated Autocrine Signaling in HGSOC Cells
3.8. Inhibition of Ovarian Cancer Cell Spheroid Growth by GluR-, GABA-R, and 5-HT Antagonists
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kossai, M.; Leary, A.; Scoazec, J.Y.; Genestie, C. Ovarian Cancer: A Heterogeneous Disease. Pathobiology 2018, 85, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Peng, H.; Qi, X.; Wu, M.; Zhao, X. Targeted therapies in gynecological cancers: A comprehensive review of clinical evidence. Signal Transduct. Target. Ther. 2020, 5, 137. [Google Scholar] [CrossRef] [PubMed]
- Masoodi, T.; Siraj, S.; Siraj, A.K.; Azam, S.; Qadri, Z.; Parvathareddy, S.K.; Tulbah, A.; Al-Dayel, F.; AlHusaini, H.; AlOmar, O.; et al. Genetic heterogeneity and evolutionary history of high-grade ovarian carcinoma and matched distant metastases. Br. J. Cancer 2020, 122, 1219–1230. [Google Scholar] [CrossRef] [Green Version]
- Geistlinger, L.; Oh, S.; Ramos, M.; Schiffer, L.; LaRue, R.S.; Henzler, C.M.; Munro, S.A.; Daughters, C.; Nelson, A.C.; Winterhoff, B.J.; et al. Multiomic Analysis of Subtype Evolution and Heterogeneity in High-Grade Serous Ovarian Carcinoma. Cancer Res. 2020, 80, 4335–4345. [Google Scholar] [CrossRef]
- Kim, J.; DeBerardinis, R.J. Mechanisms and Implications of Metabolic Heterogeneity in Cancer. Cell Metab. 2019, 30, 434–446. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Outschoorn, U.E.; Peiris-Pages, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 2017, 14, 11–31. [Google Scholar] [CrossRef]
- Kosmides, A.K.; Kamisoglu, K.; Calvano, S.E.; Corbett, S.A.; Androulakis, I.P. Metabolomic fingerprinting: Challenges and opportunities. Crit. Rev. Biomed. Eng. 2013, 41, 205–221. [Google Scholar] [CrossRef]
- Clish, C.B. Metabolomics: An emerging but powerful tool for precision medicine. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [Green Version]
- Puchades-Carrasco, L.; Pineda-Lucena, A. Metabolomics Applications in Precision Medicine: An Oncological Perspective. Curr. Top. Med. Chem. 2017, 17, 2740–2751. [Google Scholar] [CrossRef] [Green Version]
- Vignoli, A.; Risi, E.; McCartney, A.; Migliaccio, I.; Moretti, E.; Malorni, L.; Luchinat, C.; Biganzoli, L.; Tenori, L. Precision Oncology via NMR-Based Metabolomics: A Review on Breast Cancer. Int. J. Mol. Sci. 2021, 22, 4687. [Google Scholar] [CrossRef] [PubMed]
- Robertson-Tessi, M.; Gillies, R.J.; Gatenby, R.A.; Anderson, A.R. Impact of metabolic heterogeneity on tumor growth, invasion, and treatment outcomes. Cancer Res. 2015, 75, 1567–1579. [Google Scholar] [CrossRef] [Green Version]
- Saorin, A.; Di Gregorio, E.; Miolo, G.; Steffan, A.; Corona, G. Emerging Role of Metabolomics in Ovarian Cancer Diagnosis. Metabolites 2020, 10, 419. [Google Scholar] [CrossRef]
- Ha, J.H.; Radhakrishnan, R.; Jayaraman, M.; Yan, M.; Ward, J.D.; Fung, K.M.; Moxley, K.; Sood, A.K.; Isidoro, C.; Mukherjee, P.; et al. LPA Induces Metabolic Reprogramming in Ovarian Cancer via a Pseudohypoxic Response. Cancer Res. 2018, 78, 1923–1934. [Google Scholar] [CrossRef] [Green Version]
- Radhakrishnan, R.; Ha, J.H.; Jayaraman, M.; Liu, J.; Moxley, K.M.; Isidoro, C.; Sood, A.K.; Song, Y.S.; Dhanasekaran, D.N. Ovarian cancer cell-derived lysophosphatidic acid induces glycolytic shift and cancer-associated fibroblast-phenotype in normal and peritumoral fibroblasts. Cancer Lett. 2019, 442, 464–474. [Google Scholar] [CrossRef]
- Ha, J.H.; Gomathinayagam, R.; Yan, M.; Jayaraman, M.; Ramesh, R.; Dhanasekaran, D.N. Determinant role for the gep oncogenes, Galpha12/13, in ovarian cancer cell proliferation and xenograft tumor growth. Genes Cancer 2015, 6, 356–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, A.M.; DeHaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Court, W.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S.A. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef] [Green Version]
- Yi, H.; Talmon, G.; Wang, J. Glutamate in cancers: From metabolism to signaling. J. Biomed. Res. 2019, 34, 260–270. [Google Scholar] [CrossRef]
- Jiang, S.H.; Hu, L.P.; Wang, X.; Li, J.; Zhang, Z.G. Neurotransmitters: Emerging targets in cancer. Oncogene 2020, 39, 503–515. [Google Scholar] [CrossRef]
- Liang, Y.; Li, H.; Gan, Y.; Tu, H. Shedding Light on the Role of Neurotransmitters in the Microenvironment of Pancreatic Cancer. Front. Cell Dev. Biol. 2021, 9, 688953. [Google Scholar] [CrossRef]
- Karmakar, S.; Lal, G. Role of serotonin receptor signaling in cancer cells and anti-tumor immunity. Theranostics 2021, 11, 5296–5312. [Google Scholar] [CrossRef]
- Schneider, M.A.; Heeb, L.; Beffinger, M.M.; Pantelyushin, S.; Linecker, M.; Roth, L.; Lehmann, K.; Ungethum, U.; Kobold, S.; Graf, R.; et al. Attenuation of peripheral serotonin inhibits tumor growth and enhances immune checkpoint blockade therapy in murine tumor models. Sci. Transl. Med. 2021, 13, eabc8188. [Google Scholar] [CrossRef] [PubMed]
- Gautam, J.; Banskota, S.; Regmi, S.C.; Ahn, S.; Jeon, Y.H.; Jeong, H.; Kim, S.J.; Nam, T.G.; Jeong, B.S.; Kim, J.A. Tryptophan hydroxylase 1 and 5-HT7 receptor preferentially expressed in triple-negative breast cancer promote cancer progression through autocrine serotonin signaling. Mol. Cancer 2016, 15, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takehara, A.; Hosokawa, M.; Eguchi, H.; Ohigashi, H.; Ishikawa, O.; Nakamura, Y.; Nakagawa, H. Gamma-aminobutyric acid (GABA) stimulates pancreatic cancer growth through overexpressing GABAA receptor pi subunit. Cancer Res. 2007, 67, 9704–9712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.H.; Liu, Y.; Li, Y.D.; Liu, Y.H.; Li, F.; Ju, Q.; Xie, P.L.; Li, G.C. GABA stimulates human hepatocellular carcinoma growth through overexpressed GABAA receptor theta subunit. World J. Gastroenterol. 2012, 18, 2704–2711. [Google Scholar] [CrossRef] [PubMed]
- Briggs, K.J.; Koivunen, P.; Cao, S.; Backus, K.M.; Olenchock, B.A.; Patel, H.; Zhang, Q.; Signoretti, S.; Gerfen, G.J.; Richardson, A.L.; et al. Paracrine Induction of HIF by Glutamate in Breast Cancer: EglN1 Senses Cysteine. Cell 2016, 166, 126–139. [Google Scholar] [CrossRef] [Green Version]
- Nicholson-Guthrie, C.S.; Guthrie, G.D.; Sutton, G.P.; Baenziger, J.C. Urine GABA levels in ovarian cancer patients: Elevated GABA in malignancy. Cancer Lett. 2001, 162, 27–30. [Google Scholar] [CrossRef]
- Maemura, K.; Shiraishi, N.; Sakagami, K.; Kawakami, K.; Inoue, T.; Murano, M.; Watanabe, M.; Otsuki, Y. Proliferative effects of gamma-aminobutyric acid on the gastric cancer cell line are associated with extracellular signal-regulated kinase 1/2 activation. J. Gastroenterol. Hepatol. 2009, 24, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; He, C.; Zhu, Y.; Wang, S.; Li, H.; Zhang, Z.; Jiang, X.; Liu, J. GABABR-Induced EGFR Transactivation Promotes Migration of Human Prostate Cancer Cells. Mol. Pharmacol. 2017, 92, 265–277. [Google Scholar] [CrossRef] [Green Version]
- Stepulak, A.; Rola, R.; Polberg, K.; Ikonomidou, C. Glutamate and its receptors in cancer. J. Neural Transm. 2014, 121, 933–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.J.; Lin, T.Y. mTOR signal pathway and its inhibitors in antitumor therapy: A review. Ai Zheng 2007, 26, 1397–1403. [Google Scholar]
- Paul, S.; Nairn, A.C.; Wang, P.; Lombroso, P.J. NMDA-mediated activation of the tyrosine phosphatase STEP regulates the duration of ERK signaling. Nat. Neurosci. 2003, 6, 34–42. [Google Scholar] [CrossRef]
- Deutsch, S.I.; Tang, A.H.; Burket, J.A.; Benson, A.D. NMDA receptors on the surface of cancer cells: Target for chemotherapy? Biomed. Pharmacother. 2014, 68, 493–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- North, W.G.; Liu, F.; Tian, R.; Abbasi, H.; Akerman, B. NMDA receptors are expressed in human ovarian cancer tissues and human ovarian cancer cell lines. Clin. Pharmacol. 2015, 7, 111–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, E.H.; Kemp, J.A.; Priestley, T.; Knight, A.R.; Woodruff, G.N.; Iversen, L.L. The anticonvulsant MK-801 is a potent N-methyl-D-aspartate antagonist. Proc. Natl. Acad. Sci. USA 1986, 83, 7104–7108. [Google Scholar] [CrossRef] [Green Version]
- Henriksen, R.; Dizeyi, N.; Abrahamsson, P.A. Expression of serotonin receptors 5-HT1A, 5-HT1B, 5-HT2B and 5-HT4 in ovary and in ovarian tumours. Anticancer Res. 2012, 32, 1361–1366. [Google Scholar]
- Ye, D.; Xu, H.; Tang, Q.; Xia, H.; Zhang, C.; Bi, F. The role of 5-HT metabolism in cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188618. [Google Scholar] [CrossRef]
- Camilleri, M. LX-1031, a tryptophan 5-hydroxylase inhibitor that reduces 5-HT levels for the potential treatment of irritable bowel syndrome. IDrugs 2010, 13, 921–928. [Google Scholar] [PubMed]
- Brown, P.M.; Drossman, D.A.; Wood, A.J.; Cline, G.A.; Frazier, K.S.; Jackson, J.I.; Bronner, J.; Freiman, J.; Zambrowicz, B.; Sands, A.; et al. The tryptophan hydroxylase inhibitor LX1031 shows clinical benefit in patients with nonconstipating irritable bowel syndrome. Gastroenterology 2011, 141, 507–516. [Google Scholar] [CrossRef] [Green Version]
- Lagies, S.; Schlimpert, M.; Neumann, S.; Waldin, A.; Kammerer, B.; Borner, C.; Peintner, L. Cells grown in three-dimensional spheroids mirror in vivo metabolic response of epithelial cells. Commun. Biol. 2020, 3, 246. [Google Scholar] [CrossRef] [PubMed]
- Bialkowska, K.; Komorowski, P.; Bryszewska, M.; Milowska, K. Spheroids as a Type of Three-Dimensional Cell Cultures-Examples of Methods of Preparation and the Most Important Application. Int. J. Mol. Sci. 2020, 21, 6225. [Google Scholar] [CrossRef]
- Zhang, Z.; Huang, K.; Gu, C.; Zhao, L.; Wang, N.; Wang, X.; Zhao, D.; Zhang, C.; Lu, Y.; Meng, Y. Molecular Subtyping of Serous Ovarian Cancer Based on Multi-omics Data. Sci. Rep. 2016, 6, 26001. [Google Scholar] [CrossRef]
- Testa, U.; Petrucci, E.; Pasquini, L.; Castelli, G.; Pelosi, E. Ovarian Cancers: Genetic Abnormalities, Tumor Heterogeneity and Progression, Clonal Evolution and Cancer Stem Cells. Medicines 2018, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Natanzon, Y.; Goode, E.L.; Cunningham, J.M. Epigenetics in ovarian cancer. Semin. Cancer Biol. 2018, 51, 160–169. [Google Scholar] [CrossRef]
- Liu, X.; Wu, A.; Wang, X.; Liu, Y.; Xu, Y.; Liu, G.; Liu, L. Identification of metabolism-associated molecular subtype in ovarian cancer. J. Cell. Mol. Med. 2021, 25, 9617–9626. [Google Scholar] [CrossRef] [PubMed]
- Bodelon, C.; Killian, J.K.; Sampson, J.N.; Anderson, W.F.; Matsuno, R.; Brinton, L.A.; Lissowska, J.; Anglesio, M.S.; Bowtell, D.D.L.; Doherty, J.A.; et al. Molecular Classification of Epithelial Ovarian Cancer Based on Methylation Profiling: Evidence for Survival Heterogeneity. Clin. Cancer Res. 2019, 25, 5937–5946. [Google Scholar] [CrossRef] [PubMed]
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013, 4, 2126. [Google Scholar] [CrossRef]
- Beaufort, C.M.; Helmijr, J.C.; Piskorz, A.M.; Hoogstraat, M.; Ruigrok-Ritstier, K.; Besselink, N.; Murtaza, M.; van, I.W.F.; Heine, A.A.; Smid, M.; et al. Ovarian cancer cell line panel (OCCP): Clinical importance of in vitro morphological subtypes. PLoS ONE 2014, 9, e103988. [Google Scholar] [CrossRef]
- Phadngam, S.; Castiglioni, A.; Ferraresi, A.; Morani, F.; Follo, C.; Isidoro, C. PTEN dephosphorylates AKT to prevent the expression of GLUT1 on plasmamembrane and to limit glucose consumption in cancer cells. Oncotarget 2016, 7, 84999–85020. [Google Scholar] [CrossRef] [Green Version]
- Jacome Sanz, D.; Raivola, J.; Karvonen, H.; Arjama, M.; Barker, H.; Murumagi, A.; Ungureanu, D. Evaluating Targeted Therapies in Ovarian Cancer Metabolism: Novel Role for PCSK9 and Second Generation mTOR Inhibitors. Cancers 2021, 13, 3727. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kwon, J.; Lee, K.A. Bcl2l10 induces metabolic alterations in ovarian cancer cells by regulating the TCA cycle enzymes SDHD and IDH1. Oncol. Rep. 2021, 45, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Qian, Y.; Yu, J.; Wong, C.C. Metabolic rewiring in the promotion of cancer metastasis: Mechanisms and therapeutic implications. Oncogene 2020, 39, 6139–6156. [Google Scholar] [CrossRef]
- Xintaropoulou, C.; Ward, C.; Wise, A.; Marston, H.; Turnbull, A.; Langdon, S.P. A comparative analysis of inhibitors of the glycolysis pathway in breast and ovarian cancer cell line models. Oncotarget 2015, 6, 25677–25695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, N.M.; Mucka, P.; Kern, J.G.; Feng, H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 2018, 9, 216–237. [Google Scholar] [CrossRef]
- Cheng, M.; Bhujwalla, Z.M.; Glunde, K. Targeting Phospholipid Metabolism in Cancer. Front. Oncol. 2016, 6, 266. [Google Scholar] [CrossRef] [Green Version]
- Bandu, R.; Mok, H.J.; Kim, K.P. Phospholipids as cancer biomarkers: Mass spectrometry-based analysis. Mass Spectrom. Rev. 2018, 37, 107–138. [Google Scholar] [CrossRef] [PubMed]
- Kopecka, J.; Trouillas, P.; Gasparovic, A.C.; Gazzano, E.; Assaraf, Y.G.; Riganti, C. Phospholipids and cholesterol: Inducers of cancer multidrug resistance and therapeutic targets. Drug Resist. Updat. 2020, 49, 100670. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G.; Sellers, Z.P.; Bujko, K.; Kakar, S.S.; Kucia, M.; Ratajczak, M.Z. Novel pleiotropic effects of bioactive phospholipids in human lung cancer metastasis. Oncotarget 2017, 8, 58247–58263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.H.; Santidrian, A.F.; LeBoeuf, S.E.; Kurczy, M.E.; Rattray, N.J.W.; Rattray, Z.; Warth, B.; Ritland, M.; Hoang, L.T.; Loriot, C.; et al. Metabolomics guided pathway analysis reveals link between cancer metastasis, cholesterol sulfate, and phospholipids. Cancer Metab. 2017, 5, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.R.; Yung, M.M.H.; Xuan, Y.; Zhan, S.; Leung, L.L.; Liang, R.R.; Leung, T.H.Y.; Yang, H.; Xu, D.; Sharma, R.; et al. Targeting of lipid metabolism with a metabolic inhibitor cocktail eradicates peritoneal metastases in ovarian cancer cells. Commun. Biol. 2019, 2, 281. [Google Scholar] [CrossRef]
- Ji, Z.; Shen, Y.; Feng, X.; Kong, Y.; Shao, Y.; Meng, J.; Zhang, X.; Yang, G. Deregulation of Lipid Metabolism: The Critical Factors in Ovarian Cancer. Front. Oncol. 2020, 10, 593017. [Google Scholar] [CrossRef]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, L.; Sandhu, J.K.; Harper, M.E.; Cuperlovic-Culf, M. Role of Glutathione in Cancer: From Mechanisms to Therapies. Biomolecules 2020, 10, 1429. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, X.; Cheng, C.; Yu, W.; Yi, P. Metabolism of Amino Acids in Cancer. Front. Cell Dev. Biol. 2020, 8, 603837. [Google Scholar] [CrossRef]
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino acids in cancer. Exp. Mol. Med. 2020, 52, 15–30. [Google Scholar] [CrossRef]
- Endicott, M.; Jones, M.; Hull, J. Amino acid metabolism as a therapeutic target in cancer: A review. Amino Acids 2021, 53, 1169–1179. [Google Scholar] [CrossRef]
- Scalise, M.; Console, L.; Rovella, F.; Galluccio, M.; Pochini, L.; Indiveri, C. Membrane Transporters for Amino Acids as Players of Cancer Metabolic Rewiring. Cells 2020, 9, 2028. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.; Pereira, C.; Medeiros, R. ASCT2 and LAT1 Contribution to the Hallmarks of Cancer: From a Molecular Perspective to Clinical Translation. Cancers 2021, 13, 203. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Soga, T. Amino acid transporters as emerging therapeutic targets in cancer. Cancer Sci. 2021, 112, 2958–2965. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Moss, T.; Mangala, L.S.; Marini, J.; Zhao, H.; Wahlig, S.; Armaiz-Pena, G.; Jiang, D.; Achreja, A.; Win, J.; et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol. Syst. Biol. 2014, 10, 728. [Google Scholar] [CrossRef]
- Kanbara, K.; Otsuki, Y.; Watanabe, M.; Yokoe, S.; Mori, Y.; Asahi, M.; Neo, M. GABAB receptor regulates proliferation in the high-grade chondrosarcoma cell line OUMS-27 via apoptotic pathways. BMC Cancer 2018, 18, 263. [Google Scholar] [CrossRef] [Green Version]
- Shu, Q.; Liu, J.; Liu, X.; Zhao, S.; Li, H.; Tan, Y.; Xu, J. GABAB R/GSK-3beta/NF-kappaB signaling pathway regulates the proliferation of colorectal cancer cells. Cancer Med. 2016, 5, 1259–1267. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.; Li, P.; Xue, Y.; Tian, H.; Zhou, T.; Zhang, G. Glutamate receptor, ionotropic, Nmethyl Daspartateassociated protein 1 promotes colorectal cancer cell proliferation and metastasis, and is negatively regulated by miR2963p. Mol. Med. Rep. 2021, 24, 1–11. [Google Scholar] [CrossRef]
- Corsi, L.; Mescola, A.; Alessandrini, A. Glutamate Receptors and Glioblastoma Multiforme: An Old “Route” for New Perspectives. Int. J. Mol. Sci. 2019, 20, 1796. [Google Scholar] [CrossRef] [Green Version]
- Du, S.; Sung, Y.S.; Wey, M.; Wang, Y.; Alatrash, N.; Berthod, A.; MacDonnell, F.M.; Armstrong, D.W. Roles of N-methyl-D-aspartate receptors and D-amino acids in cancer cell viability. Mol. Biol. Rep. 2020, 47, 6749–6758. [Google Scholar] [CrossRef]
- Balakrishna, P.; George, S.; Hatoum, H.; Mukherjee, S. Serotonin Pathway in Cancer. Int. J. Mol. Sci. 2021, 22, 1268. [Google Scholar] [CrossRef]
- Sarrouilhe, D.; Mesnil, M. Serotonin and human cancer: A critical view. Biochimie 2019, 161, 46–50. [Google Scholar] [CrossRef]
- Ngo, D.H.; Vo, T.S. An Updated Review on Pharmaceutical Properties of Gamma-Aminobutyric Acid. Molecules 2019, 24, 2678. [Google Scholar] [CrossRef] [Green Version]
- Durant, C.F.; Paterson, L.M.; Turton, S.; Wilson, S.J.; Myers, J.F.M.; Muthukumaraswamy, S.; Venkataraman, A.; Mick, I.; Paterson, S.; Jones, T.; et al. Using Baclofen to Explore GABA-B Receptor Function in Alcohol Dependence: Insights From Pharmacokinetic and Pharmacodynamic Measures. Front. Psychiatry 2018, 9, 664. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Gao, Y.; Lin, S.; Zou, X.; Zhu, Y.; Chen, X.; Wan, H.; Zhu, H. Effects of activating GABAB1 receptor on proliferation, migration, invasion and epithelial-mesenchymal transition of ovarian cancer cells. J. Ovarian Res. 2020, 13, 126. [Google Scholar] [CrossRef]
- Neman, J.; Termini, J.; Wilczynski, S.; Vaidehi, N.; Choy, C.; Kowolik, C.M.; Li, H.; Hambrecht, A.C.; Roberts, E.; Jandial, R. Human breast cancer metastases to the brain display GABAergic properties in the neural niche. Proc. Natl. Acad. Sci. USA 2014, 111, 984–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, R.A.; Watt, M.J. Unsuspected Protumorigenic Signaling Role for the Oncometabolite GABA in Advanced Prostate Cancer. Cancer Res. 2019, 79, 4580–4581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatami, F.; Payab, M.; Sarvari, M.; Gilany, K.; Larijani, B.; Arjmand, B.; Tavangar, S.M. Oncometabolites as biomarkers in thyroid cancer: A systematic review. Cancer Manag. Res. 2019, 11, 1829–1841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, R.R.J.; Patel, K.; Putnam, W.C.; Kapur, P.; Rakheja, D. Oncometabolites: A New Paradigm for Oncology, Metabolism, and the Clinical Laboratory. Clin. Chem 2017, 63, 1812–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Predescu, D.V.; Cretoiu, S.M.; Cretoiu, D.; Pavelescu, L.A.; Suciu, N.; Radu, B.M.; Voinea, S.C. G Protein-Coupled Receptors (GPCRs)-Mediated Calcium Signaling in Ovarian Cancer: Focus on GPCRs activated by Neurotransmitters and Inflammation-Associated Molecules. Int. J. Mol. Sci. 2019, 20, 5568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, H.Y.; Yang, S.D.; Ju, W.; Ahn, J.H. Aberrant epigenetic regulation of GABRP associates with aggressive phenotype of ovarian cancer. Exp. Mol. Med. 2017, 49, e335. [Google Scholar] [CrossRef]
- Lee, J.Y.; Yang, C.; Lim, W.; Song, G. Methiothepin Suppresses Human Ovarian Cancer Cell Growth by Repressing Mitochondrion-Mediated Metabolism and Inhibiting Angiogenesis In Vivo. Pharmaceutics 2020, 12, 686. [Google Scholar] [CrossRef] [PubMed]
- Zink, K.E.; Dean, M.; Burdette, J.E.; Sanchez, L.M. Imaging Mass Spectrometry Reveals Crosstalk between the Fallopian Tube and the Ovary that Drives Primary Metastasis of Ovarian Cancer. ACS Cent. Sci. 2018, 4, 1360–1370. [Google Scholar] [CrossRef] [PubMed]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha, J.H.; Jayaraman, M.; Nadhan, R.; Kashyap, S.; Mukherjee, P.; Isidoro, C.; Song, Y.S.; Dhanasekaran, D.N. Unraveling Autocrine Signaling Pathways through Metabolic Fingerprinting in Serous Ovarian Cancer Cells. Biomedicines 2021, 9, 1927. https://doi.org/10.3390/biomedicines9121927
Ha JH, Jayaraman M, Nadhan R, Kashyap S, Mukherjee P, Isidoro C, Song YS, Dhanasekaran DN. Unraveling Autocrine Signaling Pathways through Metabolic Fingerprinting in Serous Ovarian Cancer Cells. Biomedicines. 2021; 9(12):1927. https://doi.org/10.3390/biomedicines9121927
Chicago/Turabian StyleHa, Ji Hee, Muralidharan Jayaraman, Revathy Nadhan, Srishti Kashyap, Priyabrata Mukherjee, Ciro Isidoro, Yong Sang Song, and Danny N. Dhanasekaran. 2021. "Unraveling Autocrine Signaling Pathways through Metabolic Fingerprinting in Serous Ovarian Cancer Cells" Biomedicines 9, no. 12: 1927. https://doi.org/10.3390/biomedicines9121927







