HuR Protein in Hepatocellular Carcinoma: Implications in Development, Prognosis and Treatment
Abstract
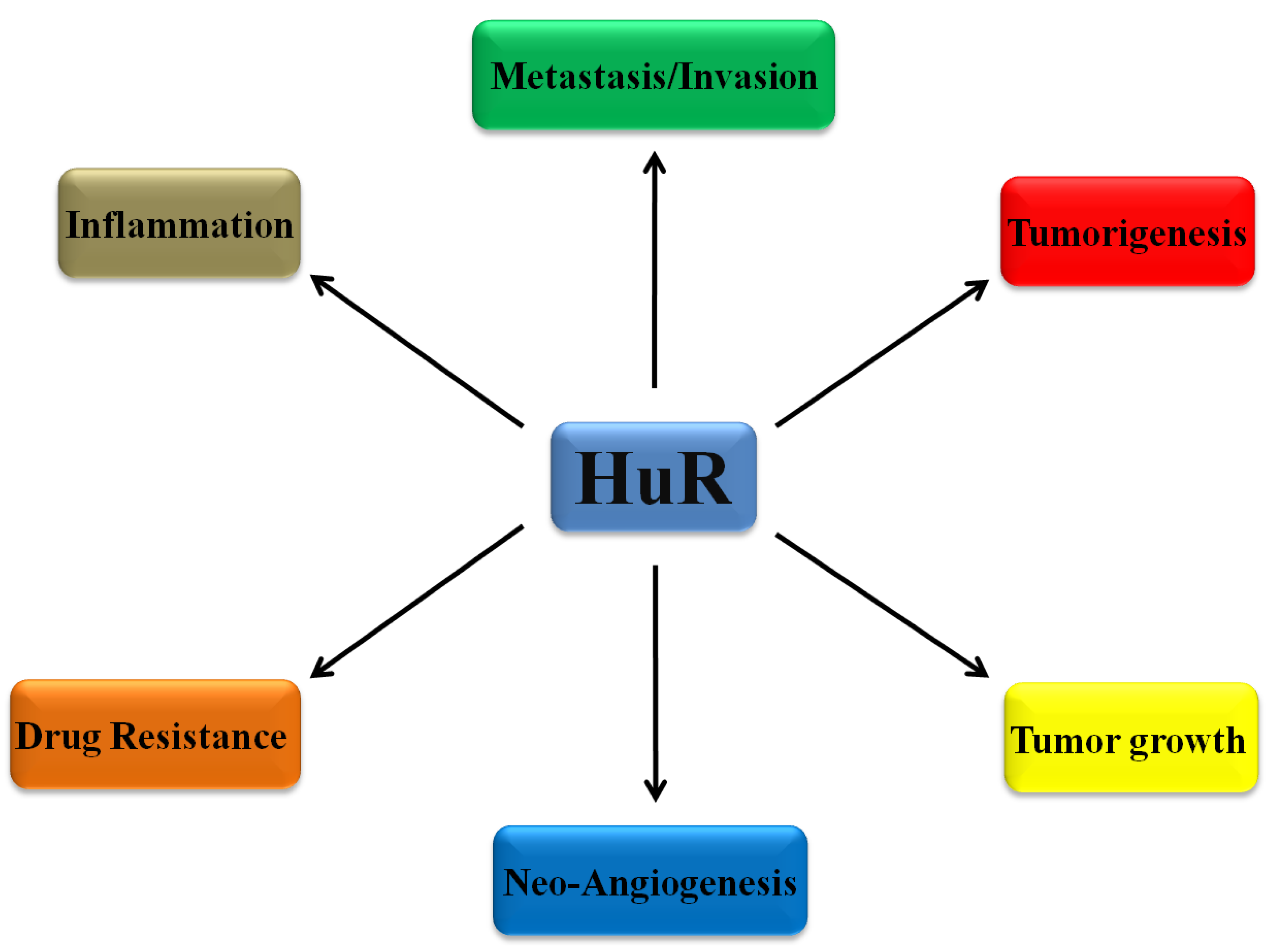
1. Introduction
2. Hepatocellular Carcinoma (HCC)
3. HuR Expression in Normal Liver Tissue and Related Tumor Cell Lines
4. HuR Target Genes and Modulators in Liver Cancer
5. Clinical Significance of HuR Expression
6. HuR Treatment
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ma, W.J.; Cheng, S.; Campbell, C.; Wright, A.; Furneaux, H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996. [Google Scholar] [CrossRef] [PubMed]
- Burd, C.G.; Dreyfuss, G. Conserved structures and diversity of functions of RNA-binding proteins. Science 1994. [Google Scholar] [CrossRef] [PubMed]
- Mazan-Mamczarz, K.; Galbán, S.; De Silanes, I.L.; Martindale, J.L.; Atasoy, U.; Keene, J.D.; Gorospe, M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA 2003, 100, 8354–8359. [Google Scholar] [CrossRef] [PubMed]
- Dolicka, D.; Sobolewski, C.; de Sousa, M.C.; Gjorgjieva, M.; Foti, M. Mrna post-transcriptional regulation by au-rich element-binding proteins in liver inflammation and cancer. Int. J. Mol. Sci. 2020, 21, 6648. [Google Scholar] [CrossRef]
- Fan, X.C.; Steitz, J.A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA 1998, 95, 15293–15298. [Google Scholar] [CrossRef]
- Keene, J.D. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. USA 1999, 96, 5–7. [Google Scholar] [CrossRef]
- Cabilla, J.P.; Nudler, S.I.; Ronchetti, S.A.; Quinteros, F.A.; Lasaga, M.; Duvilanski, B.H. Nitric oxide-sensitive guanylyl cyclase is differentially regulated by nuclear and non-nuclear estrogen pathways in anterior pituitary gland. PLoS ONE 2011. [Google Scholar] [CrossRef]
- Akool, E.-S.; Kleinert, H.; Hamada, F.M.A.; Abdelwahab, M.H.; Forstermann, U.; Pfeilschifter, J.; Eberhardt, W. Nitric Oxide Increases the Decay of Matrix Metalloproteinase 9 mRNA by Inhibiting the Expression of mRNA-Stabilizing Factor HuR. Mol. Cell. Biol. 2003. [Google Scholar] [CrossRef]
- Mazroui, R.; Di Marco, S.; Clair, E.; Von Roretz, C.; Tenenbaum, S.A.; Keene, J.D.; Saleh, M.; Gallouzi, I.E. Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis. J. Cell Biol. 2008. [Google Scholar] [CrossRef]
- Yiakouvaki, A.; Dimitriou, M.; Karakasiliotis, I.; Eftychi, C.; Theocharis, S.; Kontoyiannis, D.L. Myeloid cell expression of the RNA-binding protein HuR protects mice from pathologic inflammation and colorectal carcinogenesis. J. Clin. Investig. 2012. [Google Scholar] [CrossRef]
- Rhee, W.J.; Ni, C.-W.; Zheng, Z.; Chang, K.; Jo, H.; Bao, G. HuR regulates the expression of stress-sensitive genes and mediates inflammatory response in human umbilical vein endothelial cells. Proc. Natl. Acad. Sci. USA 2010. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Modi, Y.; Yarovinsky, T.; Yu, J.; Collinge, M.; Kyriakides, T.; Zhu, Y.; Sessa, W.C.; Pardi, R.; Bender, J.R. Macrophage β 2 integrin-mediated, HuR-dependent stabilization of angiogenic factor-encoding mRNAs in inflammatory angiogenesis. Am. J. Pathol. 2012. [Google Scholar] [CrossRef] [PubMed]
- De Silanes, I.L.; Lal, A.; Gorospe, M. HuR: Post-Transcriptional Paths to Malignancy. RNA Biol. 2005. [Google Scholar] [CrossRef] [PubMed]
- Kotta-Loizou, I.; Giaginis, C.; Theocharis, S. Clinical significance of HuR expression in human malignancy. Med. Oncol. 2014, 31, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.M.; Steitz, J.A. HuR and mRNA stability. Cell. Mol. Life Sci. 2001, 58, 266–277. [Google Scholar] [CrossRef]
- Levidou, G.; Kotta-Loizou, I.; Tasoulas, J.; Papadopoulos, T.; Theocharis, S. Clinical significance and biological role of HuR in head and neck carcinomas. Dis. Markers 2018, 2018, 4020937. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Lal, A.; Hyeon, H.K.; Gorospe, M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle 2007, 6, 1288–1292. [Google Scholar] [CrossRef]
- Giaginis, C.; Sampani, A.; Kotta-Loizou, I.; Giannopoulou, I.; Danas, E.; Politi, E.; Tsourouflis, G.; Kouraklis, G.; Patsouris, E.; Keramopoulos, A.; et al. Elevated Hu-Antigen Receptor (HuR) Expression is Associated with Tumor Aggressiveness and Poor Prognosis but not with COX-2 Expression in Invasive Breast Carcinoma Patients. Pathol. Oncol. Res. 2018, 24, 631–640. [Google Scholar] [CrossRef]
- Pons, F.; Varela, M.; Llovet, J.M. Staging systems in hepatocellular carcinoma. HPB 2005. [Google Scholar] [CrossRef]
- Llovet, J.M.; Burroughs, A.; Bruix, J. Hepatocellular carcinoma. Lancet 2003. [Google Scholar] [CrossRef]
- Buendia, M.-A. Genetics of hepatocellular carcinoma. Semin. Cancer Biol. 2000. [Google Scholar] [CrossRef]
- Thorgeirsson, S.S.; Grisham, J.W. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002, 31, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, S.; Tao, Y. Regulating tumor suppressor genes: Post-translational modifications. Signal Transduct. Target. Ther. 2020, 5, 90. [Google Scholar] [CrossRef]
- Embade, N.; Fernández-Ramos, D.; Varela-Rey, M.; Beraza, N.; Sini, M.; de Juan, V.G.; Woodhoo, A.; Martínez-López, N.; Rodríguez-Iruretagoyena, B.; Bustamante, F.J.; et al. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology 2012. [Google Scholar] [CrossRef]
- Zhu, H.; Berkova, Z.; Mathur, R.; Sehgal, L.; Khashab, T.; Tao, R.-H.; Ao, X.; Feng, L.; Sabichi, A.L.; Blechacz, B.; et al. HuR Suppresses Fas Expression and Correlates with Patient Outcome in Liver Cancer. Mol. Cancer Res. 2015. [Google Scholar] [CrossRef]
- MartíNez-LóPez, N.; Varela-Rey, M.; FernáNdez-Ramos, D.; Woodhoo, A.; VáZquez-Chantada, M.; Embade, N.; Espinosa-Hevia, L.; Bustamante, F.J.; Parada, L.A.; Rodriguez, M.S.; et al. Activation of LKB1-Akt pathway independent of phosphoinositide 3-kinase plays a critical role in the proliferation of hepatocellular carcinoma from nonalcoholic steatohepatitis. Hepatology 2010. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Chantada, M.; Fernández-Ramos, D.; Embade, N.; Martínez-Lopez, N.; Varela-Rey, M.; Woodhoo, A.; Luka, Z.; Wagner, C.; Anglim, P.P.; Finnell, R.H.; et al. HuR/Methyl-HuR and AUF1 Regulate the MAT Expressed During Liver Proliferation, Differentiation, and Carcinogenesis. Gastroenterology 2010. [Google Scholar] [CrossRef]
- Cai, J.; Mao, Z.; Hwang, J.J.; Lu, S.C. Differential expression of methionine adenosyltransferase genes influences the rate of growth of human hepatocellular carcinoma cells. Cancer Res. 1998, 58, 1444–1450. [Google Scholar]
- Marras, V.; Massarelli, G.; Daino, L.; Pinna, G.; Bennati, S.; Carta, M.; Seddaiu, M.A.; Feo, F. Chemoprevention of Rat Liver Carcinogenesis by 5-Adenosyl-L-methionine: A Long-Term Study. Cancer Res. 1992, 52, 4979–4986. [Google Scholar]
- Woodhoo, A.; Iruarrizaga-Lejarreta, M.; Beraza, N.; Garcia-Rodriguez, J.L.; Embade, N.; Fernandez-Ramos, D.; Matinez-Lopez, N.; Gutierre, V.; Arteta, B.; Caballeria, J.; et al. HuR contributes to Hepatic Stellate Cell activation and liver fibrosis. Hepatology 2012. [Google Scholar] [CrossRef]
- Stevenson, L.F.; Sparks, A.; Allende-Vega, N.; Xirodimas, D.P.; Lane, D.P.; Saville, M.K. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; An, P.; Xie, E.; Wu, Q.; Fang, X.; Gao, H.; Zhang, Z.; Li, Y.; Wang, X.; Zhang, J.; et al. Characterization of ferroptosis in murine models of hemochromatosis. Hepatology 2017. [Google Scholar] [CrossRef] [PubMed]
- Ji, E.; Kim, C.; Kang, H.; Ahn, S.; Jung, M.; Hong, Y.; Tak, H.; Lee, S.; Kim, W.; Lee, E.K. RNA Binding Protein HuR Promotes Autophagosome Formation by Regulating Expression of Autophagy-Related Proteins 5, 12, and 16 in Human Hepatocellular Carcinoma Cells. Mol. Cell. Biol. 2019, 39. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Sanders, A.J.; Ye, L.; Jiang, W.G. HuR, a key post-transcriptional regulator, and its implication in progression of breast cancer. Histol. Histopathol. 2010, 25, 1331–1340. [Google Scholar] [PubMed]
- Abdelmohsen, K.; Gorospe, M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA 2010, 1, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Govindaraju, S.; Lee, B.S. Adaptive and maladaptive expression of the mRNA regulatory protein HuR. World J. Biol. Chem. 2013. [Google Scholar] [CrossRef]
- Guo, J.; Lv, J.; Chang, S.; Chen, Z.; Lu, W.; Xu, C.; Liu, M.; Pang, X. Inhibiting cytoplasmic accumulation of HuR synergizes genotoxic agents in urothelial carcinoma of the bladder. Oncotarget 2016. [Google Scholar] [CrossRef]
- Zhang, D.-Y.; Zou, X.-J.; Cao, C.-H.; Zhang, T.; Lei, L.; Qi, X.-L.; Liu, L.; Wu, D.-H. Identification and Functional Characterization of Long Non-coding RNA MIR22HG as a Tumor Suppressor for Hepatocellular Carcinoma. Theranostics 2018, 8, 3751–3765. [Google Scholar] [CrossRef]
- Wu, X.; Gardashova, G.; Lan, L.; Han, S.; Zhong, C.; Marquez, R.T.; Wei, L.; Wood, S.; Roy, S.; Gowthaman, R.; et al. Targeting the interaction between RNA-binding protein HuR and FOXQ1 suppresses breast cancer invasion and metastasis. Commun. Biol. 2020, 3, 1–16. [Google Scholar] [CrossRef]
- Denkert, C.; Koch, I.; Von Keyserlingk, N.; Noske, A.; Niesporek, S.; Dietel, M.; Weichert, W. Expression of the ELAV-like protein HuR in human colon cancer: Association with tumor stage and cyclooxygenase-2. Mod. Pathol. 2006, 19, 1261–1269. [Google Scholar] [CrossRef]
- Song, X.; Shi, X.; Li, W.; Zhang, F.; Cai, Z. The RNA-Binding Protein HuR in Digestive System Tumors. Biomed Res. Int. 2020, 2020, 9656051. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guo, Y.; Chu, H.; Guan, Y.; Bi, J.; Wang, B. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int. J. Mol. Sci. 2013, 14, 10015–10041. [Google Scholar] [CrossRef]
- Lauriola, L.; Granone, P.; Ramella, S.; Lanza, P.; Ranelletti, F.O. Expression of the RNA-binding protein HuR and its clinical significance in human stage I and II lung adenocarcinoma. Histol. Histopathol. 2012, 27, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Oba, A.; Ban, D.; Kirimura, S.; Akahoshi, K.; Mitsunori, Y.; Matsumura, S.; Ochiai, T.; Kudo, A.; Tanaka, S.; Minoru, T. Clinical application of the biomarkers for the selection of adjuvant chemotherapy in pancreatic ductal adenocarcinoma. J. Hepatobiliary Pancreat. Sci. 2016, 23, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Filippova, N.; Yang, X.; Wang, Y.; Gillespie, G.Y.; Langford, C.; King, P.H.; Wheeler, C.; Nabors, L.B. The RNA-binding protein HuR promotes glioma growth and treatment resistance. Mol. Cancer Res. 2011, 9, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, J. HuR stabilizes TFAM mRNA in an ATM/p38-dependent manner in ionizing irradiated cancer cells. Cancer Sci. 2018, 109, 2446–2457. [Google Scholar] [CrossRef] [PubMed]
- Levy, N.S.; Chung, S.; Furneaux, H.; Levy, A.P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 1998, 273, 6417–6423. [Google Scholar] [CrossRef] [PubMed]
- Suswam, E.A.; Nabors, L.B.; Huang, Y.; Yang, X.; King, P.H. IL-1β induces stabilization of IL-8 mRNA in malignant breast cancer cells via the 3′ untranslated region: Involvement of divergent RNA-binding factors HuR, KSRP and TIAR. Int. J. Cancer 2005, 113, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wu, K.; Li, Y.; Sun, R.; Li, X. Human antigen R: A potential therapeutic target for liver diseases. Pharmacol. Res. 2020, 155, 104684. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Bhattacharya, A.; Ivanov, D.N. Identification of Small-Molecule Inhibitors of the HuR/RNA Interaction using a fluorescence polarization screening assay followed by NMR validation. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Aotani, Y.; Saitoh, Y. Structure Determination of MS-444; a New Myosin Light Chain Kinase Inhibitor. J. Antibiot. 1995, 48, 952–953. [Google Scholar] [CrossRef] [PubMed]
- Meisner, N.C.; Hintersteiner, M.; Mueller, K.; Bauer, R.; Seifert, J.M.; Naegeli, H.U.; Ottl, J.; Oberer, L.; Guenat, C.; Moss, S.; et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat. Chem. Biol. 2007, 3, 508–515. [Google Scholar] [CrossRef]
- Blanco, F.F.; Preet, R.; Aguado, A.; Vishwakarma, V.; Stevens, L.E.; Vyas, A.; Padhye, S.; Xu, L.; Weir, S.J.; Anant, S.; et al. Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis. Oncotarget 2016, 7, 74043–74058. [Google Scholar] [CrossRef] [PubMed]
- Lang, M.; Berry, D.; Passecker, K.; Mesteri, I.; Bhuju, S.; Ebner, F.; Sedlyarov, V.; Evstatiev, R.; Dammann, K.; Loy, A.; et al. HuR small-molecule inhibitor elicits differential effects in adenomatosis polyposis and colorectal carcinogenesis. Cancer Res. 2017, 77, 2424–2438. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan, R.; Mehta, M.; Ahmed, R.; Roy, S.; Xu, L.; Aubé, J.; Chen, A.; Zhao, Y.D.; Herman, T.; Ramesh, R.; et al. HuR-targeted small molecule inhibitor exhibits cytotoxicity towards human lung cancer cells. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan, R.; Babu, A.; Amreddy, N.; Srivastava, A.; Chen, A.; Zhao, Y.D.; Kompella, U.B.; Munshi, A.; Ramesh, R. Tumor-targeted nanoparticle delivery of HuR siRNA inhibits lung tumor growth in vitro and in vivo by disrupting the oncogenic activity of the RNA-binding protein HuR. Mol. Cancer Ther. 2017, 16, 1470–1486. [Google Scholar] [CrossRef]
- Allegri, L.; Baldan, F.; Roy, S.; Aubé, J.; Russo, D.; Filetti, S.; Damante, G. The HuR CMLD-2 inhibitor exhibits antitumor effects via MAD2 downregulation in thyroid cancer cells. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Giaginis, C.; Alexandrou, P.; Tsoukalas, N.; Sfiniadakis, I.; Kavantzas, N.; Agapitos, E.; Patsouris, E.; Theocharis, S. Hu-antigen receptor (HuR) and cyclooxygenase-2 (COX-2) expression in human non-small-cell lung carcinoma: Associations with clinicopathological parameters, tumor proliferative capacity and patients’ survival. Tumor Biol. 2014, 36, 315–327. [Google Scholar] [CrossRef]
- Giaginis, C.; Alexandrou, P.; Delladetsima, I.; Karavokyros, I.; Danas, E.; Giagini, A.; Patsouris, E.; Theocharis, S. Clinical Significance of Hu-Antigen Receptor (HuR) and Cyclooxygenase-2 (COX-2) Expression in Human Malignant and Benign Thyroid Lesions. Pathol. Oncol. Res. 2016, 22, 189–196. [Google Scholar] [CrossRef]
- Doller, A.; Badawi, A.; Schmid, T.; Brauß, T.; Pleli, T.; Zu Heringdorf, D.M.; Piiper, A.; Pfeilschifter, J.; Eberhardt, W. The cytoskeletal inhibitors latrunculin A and blebbistatin exert antitumorigenic properties in human hepatocellular carcinoma cells by interfering with intracellular HuR trafficking. Exp. Cell Res. 2015, 330, 66–80. [Google Scholar] [CrossRef]
- Eberhardt, W.; Badawi, A.; Biyanee, A.; Pfeilschifter, J. Cytoskeleton-dependent transport as a potential target for interfering with post-transcriptional HuR mRNA regulons. Front. Pharmacol. 2016, 7, 251. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-Y.; Chung, T.-W.; Choi, H.-J.; Lee, C.H.; Eun, J.S.; Han, Y.T.; Choi, J.-Y.; Kim, S.-Y.; Han, C.-W.; Jeong, H.-S.; et al. A novel cantharidin analog N-ABenzylcantharidinamide reduces the expression of MMP-9 and invasive potentials of Hep3B via inhibiting cytosolic translocation of HuR. Biochem. Biophys. Res. Commun. 2014, 447, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Mittelstadt, M.L.; Patel, R.C. AP-1 mediated transcriptional repression of matrix metalloproteinase-9 by recruitment of histone deacetylase 1 in response to interferon β. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.; Amreddy, N.; Babu, A.; Munshi, A.; Ramesh, R. Combinatorial nanoparticle delivery of siRNA and antineoplastics for lung cancer treatment. In Methods in Molecular Medicine; Humana Press Inc.: Clifton, NJ, USA, 2019; Volume 1974, pp. 265–290. [Google Scholar]
- Muralidharan, R.; Babu, A.; Amreddy, N.; Basalingappa, K.; Mehta, M.; Chen, A.; Zhao, Y.D.; Kompella, U.B.; Munshi, A.; Ramesh, R. Folate receptor-targeted nanoparticle delivery of HuR-RNAi suppresses lung cancer cell proliferation and migration. J. Nanobiotechnol. 2016, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.Q.; Wang, Y.; Liu, D.G. Cancer cell growth suppression by a 62nt AU-rich RNA from C/EBPβ 3’UTR through competitive binding with HuR. Biochem. Biophys. Res. Commun. 2012, 426, 122–128. [Google Scholar] [CrossRef] [PubMed]

| mRNA Target | Function of mRNA Target | Result of HuR Intervention | REF |
|---|---|---|---|
| MAT2A | catalyse the synthesis of S-adenosylmethionine (SAMe) |
| [27] |
| Mdm2 | cell cycle regulator |
| [24] |
| cyclin A and cyclin D1 | cell cycle regulator |
| [24] |
| caspase 3 | involved in apoptosis |
| [24] |
| HAUSP | along with Mdm2 modulate p53 function |
| [26] |
| Fas | involved in apoptosis |
| [25] |
| PDGF and TGFb | hepatic stellate cell activators | promotion of liver fibrosis | [30] |
| Compound | Function | REF |
|---|---|---|
| MS-444 | Inhibits the cytoplasmic translocation of HuR, | [50,53] |
| N-Benzylcantharidinamide | Inhibits the cytoplasmic translocation of HuR, | [62] |
| Latrunculin A | Inhibits the cytoplasmic translocation of HuR, | [60,61] |
| Blebbistatin | Inhibits the cytoplasmic translocation of HuR, | [60,61] |
| HuR-FNP | Targets HuR’s mRNA and regulates its expression. | [65] |
| CMLD2 | Inhibits HuR binding to target mRNAs | [57] |
| KH3 | Inhibits HuR binding to target mRNAs | [39] |
| Short RNA with AU-rich elements, obtained from C/EBPβ 3′UTR | Inhibits HuR binding to target mRNAs | [66] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papatheofani, V.; Levidou, G.; Sarantis, P.; Koustas, E.; Karamouzis, M.V.; Pergaris, A.; Kouraklis, G.; Theocharis, S. HuR Protein in Hepatocellular Carcinoma: Implications in Development, Prognosis and Treatment. Biomedicines 2021, 9, 119. https://doi.org/10.3390/biomedicines9020119
Papatheofani V, Levidou G, Sarantis P, Koustas E, Karamouzis MV, Pergaris A, Kouraklis G, Theocharis S. HuR Protein in Hepatocellular Carcinoma: Implications in Development, Prognosis and Treatment. Biomedicines. 2021; 9(2):119. https://doi.org/10.3390/biomedicines9020119
Chicago/Turabian StylePapatheofani, Vasiliki, Georgia Levidou, Panagiotis Sarantis, Evangelos Koustas, Michalis V. Karamouzis, Alexandros Pergaris, Gregorios Kouraklis, and Stamatios Theocharis. 2021. "HuR Protein in Hepatocellular Carcinoma: Implications in Development, Prognosis and Treatment" Biomedicines 9, no. 2: 119. https://doi.org/10.3390/biomedicines9020119
APA StylePapatheofani, V., Levidou, G., Sarantis, P., Koustas, E., Karamouzis, M. V., Pergaris, A., Kouraklis, G., & Theocharis, S. (2021). HuR Protein in Hepatocellular Carcinoma: Implications in Development, Prognosis and Treatment. Biomedicines, 9(2), 119. https://doi.org/10.3390/biomedicines9020119










