The Synergy between Organ-on-a-Chip and Artificial Intelligence for the Study of NAFLD: From Basic Science to Clinical Research
Abstract
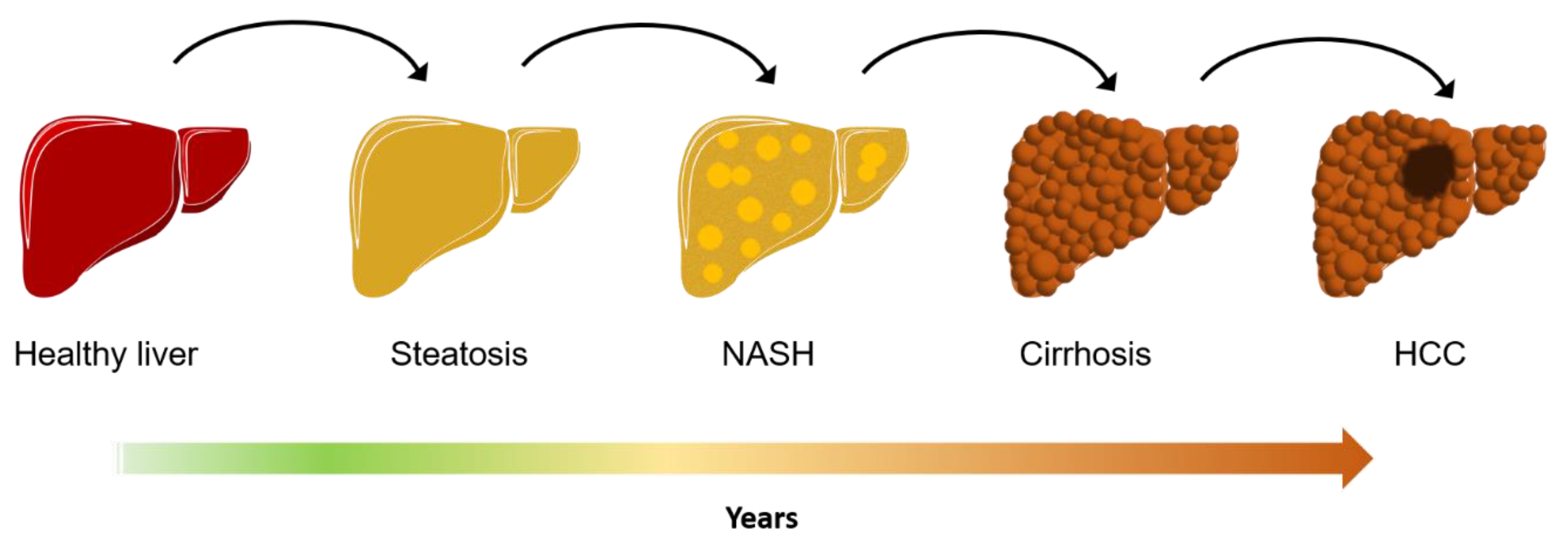
1. Introduction
2. Extra-Hepatic Outcomes: NAFLD as Multiorgan Disease
3. Organ-on-a-Chip 2.0
4. Artificial Intelligence and NAFLD
5. In Vitro NAFLD Features Recognition: The Synergy between OOC and AI
6. Body on a Chip: An Exponential Growth in Complexity
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adams, L.A.; Lymp, J.F.; St. Sauver, J.; Sanderson, S.O.; Lindor, K.D.; Feldstein, A.; Angulo, P. The natural history of nonalcoholic fatty liver disease: A population-based cohort study. Gastroenterology 2005. [Google Scholar] [CrossRef]
- Pang, Q.; Zhang, J.-Y.; Song, S.-D.; Qu, K.; Xu, X.-S.; Liu, S.-S.; Liu, C. Central obesity and nonalcoholic fatty liver disease risk after adjusting for body mass index. World J. Gastroenterol. 2015, 21, 1650–1662. [Google Scholar] [CrossRef]
- Hagström, H.; Nasr, P.; Ekstedt, M.; Hammar, U.; Stål, P.; Hultcrantz, R.; Kechagias, S. Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J. Hepatol. 2017, 67, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease—Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016. [Google Scholar] [CrossRef] [PubMed]
- Lonardo, A.; Leoni, S.; Alswat, K.A.; Fouad, Y. History of Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2020, 21, 5888. [Google Scholar] [CrossRef]
- Marchesini, G.; Day, C.P.; Dufour, J.F.; Canbay, A.; Nobili, V.; Ratziu, V.; Tilg, H.; Roden, M.; Gastaldelli, A.; Yki-Jarvinen, H.; et al. EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J. Hepatol. 2016. [Google Scholar] [CrossRef]
- Rinella, M.E.; Tacke, F.; Sanyal, A.J.; Anstee, Q.M. Report on the AASLD/EASL joint workshop on clinical trial endpoints in NAFLD. J. Hepatol. 2019, 71, 823–833. [Google Scholar] [CrossRef]
- Machado, M.V.; Cortez-Pinto, H. Non-invasive diagnosis of non-alcoholic fatty liver disease. A critical appraisal. J. Hepatol. 2013, 58, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Davison, B.A.; Harrison, S.A.; Cotter, G.; Alkhouri, N.; Sanyal, A.; Edwards, C.; Colca, J.R.; Iwashita, J.; Koch, G.G.; Dittrich, H.C. Suboptimal reliability of liver biopsy evaluation has implications for randomized clinical trials. J. Hepatol. 2020. [Google Scholar] [CrossRef]
- Jensen, V.S.; Tveden-Nyborg, P.; Zacho-Rasmussen, C.; Quaade, M.L.; Ipsen, D.H.; Hvid, H.; Fledelius, C.; Wulff, E.M.; Lykkesfeldt, J. Variation in diagnostic NAFLD/NASH read-outs in paired liver samples from rodent models. J. Pharm. Toxicol. Methods 2020, 101, 106651. [Google Scholar] [CrossRef]
- Fogel, D.B. Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: A review. Contemp. Clin. Trials Commun. 2018, 11, 156–164. [Google Scholar] [CrossRef]
- Freag, M.S.; Namgung, B.; Reyna Fernandez, M.E.; Gherardi, E.; Sengupta, S.; Jang, H.L. Human Nonalcoholic Steatohepatitis on a Chip. Hepatol. Commun. 2020. [Google Scholar] [CrossRef]
- Bulutoglu, B.; Rey-Bedón, C.; Kang, Y.B.A.; Mert, S.; Yarmush, M.L.; Usta, O.B. A microfluidic patterned model of non-alcoholic fatty liver disease: Applications to disease progression and zonation. Lab Chip 2019, 19, 3022–3031. [Google Scholar] [CrossRef] [PubMed]
- Gori, M.; Simonelli, M.C.; Giannitelli, S.M.; Businaro, L.; Trombetta, M.; Rainer, A. Investigating Nonalcoholic Fatty Liver Disease in a Liver-on-a-Chip Microfluidic Device. PLoS ONE 2016, 11, e0159729. [Google Scholar] [CrossRef] [PubMed]
- Trietsch, S.J.; Naumovska, E.; Kurek, D.; Setyawati, M.C.; Vormann, M.K.; Wilschut, K.J.; Lanz, H.L.; Nicolas, A.; Ng, C.P.; Joore, J.; et al. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat. Commun. 2017. [Google Scholar] [CrossRef] [PubMed]
- Jordan, N.V.; Bardia, A.; Wittner, B.S.; Benes, C.; Ligorio, M.; Zheng, Y.; Yu, M.; Sundaresan, T.K.; Licausi, J.A.; Desai, R.; et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 2016, 537, 102–106. [Google Scholar] [CrossRef]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Lei, C.; Wu, Y.; Huang, C.-J.; Yasumoto, A.; Jona, M.; Li, W.; Wu, Y.; Yalikun, Y.; Jiang, Y.; et al. Intelligent whole-blood imaging flow cytometry for simple, rapid, and cost-effective drug-susceptibility testing of leukemia. Lab Chip 2019, 19, 2688–2698. [Google Scholar] [CrossRef] [PubMed]
- Isozaki, A.; Harmon, J.; Zhou, Y.; Li, S.; Nakagawa, Y.; Hayashi, M.; Mikami, H.; Lei, C.; Goda, K. AI on a chip. Lab Chip 2020, 20, 3074–3090. [Google Scholar] [CrossRef]
- Targher, G.; Byrne, C.D.; Lonardo, A.; Zoppini, G.; Barbui, C. Non-alcoholic fatty liver disease and risk of incident cardiovascular disease: A meta-analysis. J. Hepatol. 2016. [Google Scholar] [CrossRef]
- Federico, A.; Dallio, M.; Masarone, M.; Persico, M.; Loguercio, C. The epidemiology of non-alcoholic fatty liver disease and its connection with cardiovascular disease: Role of endothelial dysfunction. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4731–4741. [Google Scholar]
- Lonardo, A.; Sookoian, S.; Pirola, C.J.; Targher, G. Non-alcoholic fatty liver disease and risk of cardiovascular disease. Metabolism 2016, 65, 1136–1150. [Google Scholar] [CrossRef]
- Kim, G.A.; Lee, H.C.; Choe, J.; Kim, M.J.; Lee, M.J.; Chang, H.S.; Bae, I.Y.; Kim, H.K.; An, J.; Shim, J.H.; et al. Association between non-alcoholic fatty liver disease and cancer incidence rate. J. Hepatol. 2018. [Google Scholar] [CrossRef]
- Mantovani, A.; Dauriz, M.; Byrne, C.D.; Lonardo, A.; Zoppini, G.; Bonora, E.; Targher, G. Association between nonalcoholic fatty liver disease and colorectal tumours in asymptomatic adults undergoing screening colonoscopy: A systematic review and meta-analysis. Metabolism 2018. [Google Scholar] [CrossRef]
- Shen, H.; Lipka, S.; Kumar, A.; Mustacchia, P. Association between nonalcoholic fatty liver disease and colorectal adenoma: A systemic review and meta-analysis. J. Gastrointest. Oncol. 2014. [Google Scholar] [CrossRef]
- Nseir, W.; Abu-Rahmeh, Z.; Tsipis, A.; Mograbi, J.; Mahamid, M. Relationship between non-alcoholic fatty liver disease and breast cancer. Isr. Med. Assoc. J. 2017, 19, 242–245. [Google Scholar] [PubMed]
- Campbell, P.T.; Deka, A.; Jacobs, E.J.; Newton, C.C.; Hildebrand, J.S.; McCullough, M.L.; Limburg, P.J.; Gapstur, S.M. Prospective study reveals associations between colorectal cancer and type 2 diabetes mellitus or insulin use in men. Gastroenterology 2010. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, A.E.; Goodwin, P.J.; Chlebowski, R.T.; Pan, K.; Stambolic, V.; Dowling, R.J.O. Association of obesity-related metabolic disruptions with cancer risk and outcome. J. Clin. Oncol. 2016, 34, 4249–4255. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Lee, S.E.; Lee, Y.B.; Jun, J.E.; Ahn, J.; Bae, J.C.; Jin, S.M.; Hur, K.Y.; Jee, J.H.; Lee, M.K.; et al. Relationship Between Relative Skeletal Muscle Mass and Nonalcoholic Fatty Liver Disease: A 7-Year Longitudinal Study. Hepatology 2018. [Google Scholar] [CrossRef]
- Montano-Loza, A.J.; Meza-Junco, J.; Prado, C.M.M.; Lieffers, J.R.; Baracos, V.E.; Bain, V.G.; Sawyer, M.B. Muscle Wasting Is Associated With Mortality in Patients With Cirrhosis. Clin. Gastroenterol. Hepatol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Tsien, C.; Garber, A.; Narayanan, A.; Shah, S.N.; Barnes, D.; Eghtesad, B.; Fung, J.; Mccullough, A.J.; Dasarathy, S. Post-liver transplantation sarcopenia in cirrhosis: A prospective evaluation. J. Gastroenterol. Hepatol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Marcuccilli, M.; Chonchol, M. NAFLD and chronic kidney disease. Int. J. Mol. Sci. 2016, 17, 562. [Google Scholar] [CrossRef] [PubMed]
- Targher, G.; Byrne, C.D. Non-alcoholic fatty liver disease: An emerging driving force in chronic kidney disease. Nat. Rev. Nephrol. 2017, 13, 297–310. [Google Scholar] [CrossRef] [PubMed]
- El Azeem, H.A.; Khalek, E.S.A.; El-Akabawy, H.; Naeim, H.; Khalik, H.A.; Alfifi, A.A. Association between nonalcoholic fatty liver disease and the incidence of cardiovascular and renal events. J. Saudi Hear. Assoc. 2013. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, G.; Zelber-Sagi, S.; Preis, S.R.; Beiser, A.S.; DeCarli, C.; Speliotes, E.K.; Satizabal, C.L.; Vasan, R.S.; Seshadri, S. Association of nonalcoholic fatty liver disease with lower brain volume in healthy middle-aged adults in the Framingham Study. JAMA Neurol. 2018. [Google Scholar] [CrossRef]
- Fargion, S.; Porzio, M.; Fracanzani, A.L. Nonalcoholic fatty liver disease and vascular disease: State-of-the-art. World J. Gastroenterol. 2014, 20, 13306. [Google Scholar] [CrossRef]
- Lombardi, R.; Fargion, S.; Fracanzani, A.L. Brain involvement in non-alcoholic fatty liver disease (NAFLD): A systematic review. Dig. Liver Dis. 2019, 51, 1214–1222. [Google Scholar] [CrossRef]
- Hadjihambi, A.; De Chiara, F.; Hosford, P.S.; Habtetion, A.; Karagiannis, A.; Davies, N.; Gourine, A.V.; Jalan, R. Ammonia mediates cortical hemichannel dysfunction in rodent models of chronic liver disease. Hepatology 2017, 65. [Google Scholar] [CrossRef]
- Jalan, R.; De Chiara, F.; Balasubramaniyan, V.; Andreola, F.; Khetan, V.; Malago, M.; Pinzani, M.; Mookerjee, R.P.; Rombouts, K. Ammonia produces pathological changes in human hepatic stellate cells and is a target for therapy of portal hypertension. J. Hepatol. 2016, 64. [Google Scholar] [CrossRef]
- Rosato, V.; Masarone, M.; Dallio, M.; Federico, A.; Aglitti, A.; Persico, M. NAFLD and extra-hepatic comorbidities: Current evidence on a multi-organ metabolic syndrome. Int. J. Environ. Res. Public Health 2019, 16, 3415. [Google Scholar] [CrossRef]
- Manco, R.; Itzkovitz, S. Liver zonation. J. Hepatol. 2021, 74, 466–468. [Google Scholar] [CrossRef]
- Dunn, J.C.Y.; Tompkins, R.G.; Yarmush, M.L. Long-Term in Vitro Function of Adult Hepatocytes in a Collagen Sandwich Configuration. Biotechnol. Prog. 1991, 7, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Suurmond, C.-A.E.; Lasli, S.; van den Dolder, F.W.; Ung, A.; Kim, H.-J.; Bandaru, P.; Lee, K.; Cho, H.-J.; Ahadian, S.; Ashammakhi, N.; et al. In Vitro Human Liver Model of Nonalcoholic Steatohepatitis by Coculturing Hepatocytes, Endothelial Cells, and Kupffer Cells. Adv. Healthc. Mater. 2019, 8, 1901379. [Google Scholar] [CrossRef]
- Wei, G.; Wang, J.; Lv, Q.; Liu, M.; Xu, H.; Zhang, H.; Jin, L.; Yu, J.; Wang, X. Three-dimensional coculture of primary hepatocytes and stellate cells in silk scaffold improves hepatic morphology and functionality in vitro. J. Biomed. Mater. Res. Part A 2018, 106, 2171–2180. [Google Scholar] [CrossRef] [PubMed]
- Baze, A.; Parmentier, C.; Hendriks, D.F.G.; Hurrell, T.; Heyd, B.; Bachellier, P.; Schuster, C.; Ingelman-Sundberg, M.; Richert, L. Three-Dimensional Spheroid Primary Human Hepatocytes in Monoculture and Coculture with Nonparenchymal Cells. Tissue Eng. Part C Methods 2018, 24, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Abu-Absi, S.F.; Hansen, L.K.; Hu, W.-S. Three-dimensional co-culture of hepatocytes and stellate cells. Cytotechnology 2004, 45, 125–140. [Google Scholar] [CrossRef]
- Kim, Y.; Kang, K.; Jeong, J.; Paik, S.S.; Kim, J.S.; Park, S.A.; Kim, W.D.; Park, J.; Choi, D. Three-dimensional (3D) printing of mouse primary hepatocytes to generate 3D hepatic structure. Ann. Surg. Treat. Res. 2017, 92, 67–72. [Google Scholar] [CrossRef]
- Lewis, P.L.; Green, R.M.; Shah, R.N. 3D-printed gelatin scaffolds of differing pore geometry modulate hepatocyte function and gene expression. Acta Biomater. 2018, 69, 63–70. [Google Scholar] [CrossRef]
- Krüger, M.; Oosterhoff, L.A.; van Wolferen, M.E.; Schiele, S.A.; Walther, A.; Geijsen, N.; De Laporte, L.; van der Laan, L.J.W.; Kock, L.M.; Spee, B. Cellulose Nanofibril Hydrogel Promotes Hepatic Differentiation of Human Liver Organoids. Adv. Healthc. Mater. 2020. [Google Scholar] [CrossRef]
- Török, E.; Lutgehetmann, M.; Bierwolf, J.; Melbeck, S.; Düllmann, J.; Nashan, B.; Ma, P.X.; Pollok, J.M. Primary human hepatocytes on biodegradable poly(l-lactic acid) matrices: A promising model for improving transplantation efficiency with tissue engineering. Liver Transpl. 2011. [Google Scholar] [CrossRef]
- Li, J.; Li, L.; Yu, H.; Cao, H.; Gao, C.; Gong, Y. Growth and metabolism of human hepatocytes on biomodified collagen poly(lactic-co-glycolic acid) three-dimensional scaffold. Asaio J. 2006. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhou, M.; Zhang, M.; Liu, W.; Zhou, Y.; Lang, M. Hepatocyte culture on 3D porous scaffolds of PCL/PMCL. Colloids Surf. B Biointerfaces 2019, 173, 185–193. [Google Scholar] [CrossRef]
- Loh, Q.L.; Choong, C. Three-dimensional scaffolds for tissue engineering applications: Role of porosity and pore size. Tissue Eng. Part B Rev. 2013, 19, 485–502. [Google Scholar] [CrossRef]
- Krieghoff, J.; Picke, A.-K.; Salbach-Hirsch, J.; Rother, S.; Heinemann, C.; Bernhardt, R.; Kascholke, C.; Möller, S.; Rauner, M.; Schnabelrauch, M.; et al. Increased pore size of scaffolds improves coating efficiency with sulfated hyaluronan and mineralization capacity of osteoblasts. Biomater. Res. 2019, 23, 26. [Google Scholar] [CrossRef] [PubMed]
- Tytgat, L.; Kollert, M.R.; Van Damme, L.; Thienpont, H.; Ottevaere, H.; Duda, G.N.; Geissler, S.; Dubruel, P.; Van Vlierberghe, S.; Qazi, T.H. Evaluation of 3D Printed Gelatin-Based Scaffolds with Varying Pore Size for MSC-Based Adipose Tissue Engineering. Macromol. Biosci. 2020, 20, 1900364. [Google Scholar] [CrossRef] [PubMed]
- Roulot, D.; Czernichow, S.; Le Clésiau, H.; Costes, J.L.; Vergnaud, A.C.; Beaugrand, M. Liver stiffness values in apparently healthy subjects: Influence of gender and metabolic syndrome. J. Hepatol. 2008, 48, 606–613. [Google Scholar] [CrossRef]
- Ruoß, M.; Rebholz, S.; Weimer, M.; Grom-Baumgarten, C.; Athanasopulu, K.; Kemkemer, R.; Käß, H.; Ehnert, S.; Nussler, A.K. Development of Scaffolds with Adjusted Stiffness for Mimicking Disease-Related Alterations of Liver Rigidity. J. Funct. Biomater. 2020, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, V.; Maroufi, N.F.; Saghati, S.; Asadi, N.; Darabi, M.; Ahmad, S.N.S.; Hosseinkhani, H.; Rahbarghazi, R. Current progress in hepatic tissue regeneration by tissue engineering. J. Transl. Med. 2019, 17, 383. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Ostuni, E.; LeDuc, P.; Naruse, K.; Ingber, D.E.; Whitesides, G.M. Subcellular positioning of small molecules. Nature 2001. [Google Scholar] [CrossRef]
- Li Jeon, N.; Baskaran, H.; Dertinger, S.K.W.; Whitesides, G.M.; Van De Water, L.; Toner, M. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nat. Biotechnol. 2002. [Google Scholar] [CrossRef]
- Prentice-Mott, H.V.; Chang, C.H.; Mahadevan, L.; Mitchison, T.J.; Irimia, D.; Shah, J.V. Biased migration of confined neutrophil-like cells in asymmetric hydraulic environments. Proc. Natl. Acad. Sci. USA 2013. [Google Scholar] [CrossRef]
- Radisic, M.; Deen, W.; Langer, R.; Vunjak-Novakovic, G. Mathematical model of oxygen distribution in engineered cardiac tissue with parallel channel array perfused with culture medium containing oxygen carriers. Am. J. Physiol. Hear. Circ. Physiol. 2005. [Google Scholar] [CrossRef]
- Xiao, R.R.; Zeng, W.J.; Li, Y.T.; Zou, W.; Wang, L.; Pei, X.F.; Xie, M.; Huang, W.H. Simultaneous generation of gradients with gradually changed slope in a microfluidic device for quantifying axon response. Anal. Chem. 2013. [Google Scholar] [CrossRef]
- Peng, C.C.; Liao, W.H.; Chen, Y.H.; Wu, C.Y.; Tung, Y.C. A microfluidic cell culture array with various oxygen tensions. Lab Chip 2013. [Google Scholar] [CrossRef]
- Cimetta, E.; Cannizzaro, C.; James, R.; Biechele, T.; Moon, R.T.; Elvassore, N.; Vunjak-Novakovic, G. Microfluidic device generating stable concentration gradients for long term cell culture: Application to Wnt3a regulation of β-catenin signaling. Lab Chip 2010. [Google Scholar] [CrossRef] [PubMed]
- Seidi, A.; Kaji, H.; Annabi, N.; Ostrovidov, S.; Ramalingam, M.; Khademhosseini, A. A microfluidic-based neurotoxin concentration gradient for the generation of an in vitro model of Parkinson’s disease. Biomicrofluidics 2011. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lee, L.P. Non-invasive microfluidic gap junction assay. Integr. Biol. 2010. [Google Scholar] [CrossRef]
- Carraro, A.; Hsu, W.M.; Kulig, K.M.; Cheung, W.S.; Miller, M.L.; Weinberg, E.J.; Swart, E.F.; Kaazempur-Mofrad, M.; Borenstein, J.T.; Vacanti, J.P.; et al. In vitro analysis of a hepatic device with intrinsic microvascular-based channels. Biomed. Microdev. 2008. [Google Scholar] [CrossRef]
- Griep, L.M.; Wolbers, F.; de Wagenaar, B.; ter Braak, P.M.; Weksler, B.B.; Romero, I.A.; Couraud, P.O.; Vermes, I.; van der Meer, A.D.; van den Berg, A. BBB ON CHIP: Microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed. Microdev. 2013, 15, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.J.; Hung, P.J.; Lee, L.P. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol. Bioeng. 2007. [Google Scholar] [CrossRef]
- Ortega, M.A.; Fernández-Garibay, X.; Castaño, A.G.; De Chiara, F.; Hernández-Albors, A.; Balaguer-Trias, J.; Ramón-Azcón, J. Muscle-on-a-chip with an on-site multiplexed biosensing system for: In situ monitoring of secreted IL-6 and TNF-α. Lab Chip 2019, 19. [Google Scholar] [CrossRef]
- Lopez-Muñoz, G.A.; Ortega, M.A.; Ferret-Miñana, A.; De Chiara, F.; Ramón-Azcón, J. Direct and Label-Free Monitoring of Albumin in 2D Fatty Liver Disease Model Using Plasmonic Nanogratings. Nanomaterials 2020, 10, 2520. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, M.P.; Lamon-Fava, S.; Fielding, R.A. Skeletal muscle lipid deposition and insulin resistance: Effect of dietary fatty acids and exercise. Am. J. Clin. Nutr. 2007, 85, 662–677. [Google Scholar]
- Osaki, T.; Sivathanu, V.; Kamm, R.D. Crosstalk between developing vasculature and optogenetically engineered skeletal muscle improves muscle contraction and angiogenesis. Biomaterials 2018. [Google Scholar] [CrossRef] [PubMed]
- Theberge, A.B.; Yu, J.; Young, E.W.K.; Ricke, W.A.; Bushman, W.; Beebe, D.J. Microfluidic Multiculture Assay to Analyze Biomolecular Signaling in Angiogenesis. Anal. Chem. 2015. [Google Scholar] [CrossRef] [PubMed]
- Uzel, S.G.M.; Platt, R.J.; Subramanian, V.; Pearl, T.M.; Rowlands, C.J.; Chan, V.; Boyer, L.A.; So, P.T.C.; Kamm, R.D. Microfluidic device for the formation of optically excitable, three-dimensional, compartmentalized motor units. Sci. Adv. 2016. [Google Scholar] [CrossRef]
- Wevers, N.R.; Van Vught, R.; Wilschut, K.J.; Nicolas, A.; Chiang, C.; Lanz, H.L.; Trietsch, S.J.; Joore, J.; Vulto, P. High-throughput compound evaluation on 3D networks of neurons and glia in a microfluidic platform. Sci. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Ryu, H.; Tahk, D.; Ko, J.; Chung, Y.; Lee, H.K.; Lee, T.R.; Jeon, N.L. “open-top” microfluidic device for in vitro three-dimensional capillary beds. Lab Chip 2017. [Google Scholar] [CrossRef]
- Jang, K.J.; Suh, K.Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 2010. [Google Scholar] [CrossRef]
- Suzuki, H.; Hirakawa, T.; Watanabe, I.; Kikuchi, Y. Determination of blood pO2 using a micromachined Clark-type oxygen electrode. Anal. Chim. Acta 2001. [Google Scholar] [CrossRef]
- Wang, L.; Acosta, M.A.; Leach, J.B.; Carrier, R.L. Spatially monitoring oxygen level in 3D microfabricated cell culture systems using optical oxygen sensing beads. Lab Chip 2013. [Google Scholar] [CrossRef]
- Bellin, D.L.; Sakhtah, H.; Rosenstein, J.K.; Levine, P.M.; Thimot, J.; Emmett, K.; Dietrich, L.E.P.; Shepard, K.L. Integrated circuit-based electrochemical sensor for spatially resolved detection of redox-active metabolites in biofilms. Nat. Commun. 2014. [Google Scholar] [CrossRef]
- Eklund, S.E.; Cliffel, D.E.; Kozlov, E.; Prokop, A.; Wikswo, J.; Baudenbacher, F. Modification of the CytosensorTM microphysiometer to simultaneously measure extracellular acidification and oxygen consumption rates. Anal. Chim. Acta 2003. [Google Scholar] [CrossRef]
- Wu, M.H.; Lin, J.L.; Wang, J.; Cui, Z.; Cui, Z. Development of high throughput optical sensor array for on-line pH monitoring in micro-scale cell culture environment. Biomed. Microdev. 2009. [Google Scholar] [CrossRef]
- Obregón, R.; Ahadian, S.; Ramón-Azcón, J.; Chen, L.; Fujita, T.; Shiku, H.; Chen, M.; Matsue, T. Non-invasive measurement of glucose uptake of skeletal muscle tissue models using a glucose nanobiosensor. Biosens. Bioelectron. 2013. [Google Scholar] [CrossRef]
- Hernández-Albors, A.; Castaño, A.G.; Fernández-Garibay, X.; Ortega, M.A.; Balaguer, J.; Ramón-Azcón, J. Microphysiological sensing platform for an in-situ detection of tissue-secreted cytokines. Biosens. Bioelectron. X 2019. [Google Scholar] [CrossRef]
- Schwartz, W.B. Medicine and the Computer. New Engl. J. Med. 1970, 283, 1257–1264. [Google Scholar] [CrossRef]
- Andrade, M.A.; Bork, P. Automated extraction of information in molecular biology. FEBS Lett. 2000, 476, 12–17. [Google Scholar] [CrossRef]
- Ghassemi, M.; Naumann, T.; Schulam, P.; Beam, A.L.; Chen, I.Y.; Ranganath, R. Practical guidance on artificial intelligence for health-care data. Lancet Digit. Heal. 2019, 1, e157–e159. [Google Scholar] [CrossRef]
- Stein, H.S.; Gregoire, J.M. Progress and prospects for accelerating materials science with automated and autonomous workflows. Chem. Sci. 2019. [Google Scholar] [CrossRef] [PubMed]
- Ambinder, E.P. Electronic health records. J. Oncol. Pr. 2005, 1, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Abràmoff, M.D.; Lavin, P.T.; Birch, M.; Shah, N.; Folk, J.C. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digit. Med. 2018. [Google Scholar] [CrossRef]
- Raghu, A.; Komorowski, M.; Singh, S. Model-based reinforcement learning for sepsis treatment. arXiv 2018, arXiv:1811.09602. Available online: https://arxiv.org/abs/1811.09602 (accessed on 23 November 2018).
- Yom-Tov, E.; Feraru, G.; Kozdoba, M.; Mannor, S.; Tennenholtz, M.; Hochberg, I. Encouraging Physical Activity in Patients With Diabetes: Intervention Using a Reinforcement Learning System. J. Med. Internet Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Xu, C.; Shen, Z.; Yu, C.; Li, Y. Application of Machine Learning Techniques for Clinical Predictive Modeling: A Cross-Sectional Study on Nonalcoholic Fatty Liver Disease in China. Biomed. Res. Int. 2018, 2018, 4304376. [Google Scholar] [CrossRef]
- Heinemann, F.; Birk, G.; Stierstorfer, B. Deep learning enables pathologist-like scoring of NASH models. Sci. Rep. 2019, 9, 18454. [Google Scholar] [CrossRef] [PubMed]
- Gerke, S.; Minssen, T.; Cohen, G. Ethical and legal challenges of artificial intelligence-driven healthcare. Artif. Intell. Healthc. 2020, 295–336. [Google Scholar] [CrossRef]
- Guo, B.; Lei, C.; Kobayashi, H.; Ito, T.; Yalikun, Y.; Jiang, Y.; Tanaka, Y.; Ozeki, Y.; Goda, K. High-throughput, label-free, single-cell, microalgal lipid screening by machine-learning-equipped optofluidic time-stretch quantitative phase microscopy. Cytom. Part A 2017, 91, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Mahjoubfar, A.; Tai, L.-C.; Blaby, I.K.; Huang, A.; Niazi, K.R.; Jalali, B. Deep Learning in Label-free Cell Classification. Sci. Rep. 2016, 6, 21471. [Google Scholar] [CrossRef] [PubMed]
- Kozyra, M.; Johansson, I.; Nordling, Å.; Ullah, S.; Lauschke, V.M.; Ingelman-Sundberg, M. Human hepatic 3D spheroids as a model for steatosis and insulin resistance. Sci. Rep. 2018, 8, 14297. [Google Scholar] [CrossRef]
- Blasi, T.; Hennig, H.; Summers, H.D.; Theis, F.J.; Cerveira, J.; Patterson, J.O.; Davies, D.; Filby, A.; Carpenter, A.E.; Rees, P. Label-free cell cycle analysis for high-throughput imaging flow cytometry. Nat. Commun. 2016, 7, 10256. [Google Scholar] [CrossRef] [PubMed]
- Caldez, M.J.; Bjorklund, M.; Kaldis, P. Cell cycle regulation in NAFLD: When imbalanced metabolism limits cell division. Hepatol. Int. 2020, 14, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Chu, A.; Nguyen, D.; Talathi, S.S.; Wilson, A.C.; Ye, C.; Smith, W.L.; Kaplan, A.D.; Duoss, E.B.; Stolaroff, J.K.; Giera, B. Automated detection and sorting of microencapsulation via machine learning. Lab Chip 2019, 19, 1808–1817. [Google Scholar] [CrossRef]
- Das, D.K.; Ghosh, M.; Pal, M.; Maiti, A.K.; Chakraborty, C. Machine learning approach for automated screening of malaria parasite using light microscopic images. Micron 2013, 45, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Hetherington, A.M.; Sawyez, C.G.; Zilberman, E.; Stoianov, A.M.; Robson, D.L.; Borradaile, N.M. Differential Lipotoxic Effects of Palmitate and Oleate in Activated Human Hepatic Stellate Cells and Epithelial Hepatoma Cells. Cell. Physiol. Biochem. 2016, 39, 1648–1662. [Google Scholar] [CrossRef]
- Mirsky, S.K.; Barnea, I.; Levi, M.; Greenspan, H.; Shaked, N.T. Automated analysis of individual sperm cells using stain-free interferometric phase microscopy and machine learning. Cytom. Part A 2017. [Google Scholar] [CrossRef]
- Feldstein, A.E.; Werneburg, N.W.; Canbay, A.; Guicciardi, M.E.; Bronk, S.F.; Rydzewski, R.; Burgart, L.J.; Gores, G.J. Free fatty acids promote hepatic lipotoxicity by stimulating TNF-α expression via a lysosomal pathway. Hepatology 2004, 40, 185–194. [Google Scholar] [CrossRef]
- Ko, J.; Bhagwat, N.; Yee, S.S.; Ortiz, N.; Sahmoud, A.; Black, T.; Aiello, N.M.; McKenzie, L.; O’Hara, M.; Redlinger, C.; et al. Combining Machine Learning and Nanofluidic Technology to Diagnose Pancreatic Cancer Using Exosomes. ACS Nano 2017. [Google Scholar] [CrossRef]
- Lee, S.Y.; Sung, J.H. Gut–liver on a chip toward an in vitro model of hepatic steatosis. Biotechnol. Bioeng. 2018, 115, 2817–2827. [Google Scholar] [CrossRef]
- Ahluwalia, A.; Misto, A.; Vozzi, F.; Magliaro, C.; Mattei, G.; Marescotti, M.C.; Avogaro, A.; Iori, E. Systemic and vascular inflammation in an in-vitro model of central obesity. PLoS ONE 2018, 13, e0192824. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Chiara, F.; Ferret-Miñana, A.; Ramón-Azcón, J. The Synergy between Organ-on-a-Chip and Artificial Intelligence for the Study of NAFLD: From Basic Science to Clinical Research. Biomedicines 2021, 9, 248. https://doi.org/10.3390/biomedicines9030248
De Chiara F, Ferret-Miñana A, Ramón-Azcón J. The Synergy between Organ-on-a-Chip and Artificial Intelligence for the Study of NAFLD: From Basic Science to Clinical Research. Biomedicines. 2021; 9(3):248. https://doi.org/10.3390/biomedicines9030248
Chicago/Turabian StyleDe Chiara, Francesco, Ainhoa Ferret-Miñana, and Javier Ramón-Azcón. 2021. "The Synergy between Organ-on-a-Chip and Artificial Intelligence for the Study of NAFLD: From Basic Science to Clinical Research" Biomedicines 9, no. 3: 248. https://doi.org/10.3390/biomedicines9030248
APA StyleDe Chiara, F., Ferret-Miñana, A., & Ramón-Azcón, J. (2021). The Synergy between Organ-on-a-Chip and Artificial Intelligence for the Study of NAFLD: From Basic Science to Clinical Research. Biomedicines, 9(3), 248. https://doi.org/10.3390/biomedicines9030248






