Eps15 Homology Domain-Containing Protein 3 Hypermethylation as a Prognostic and Predictive Marker for Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Specimens
2.2. DNA, ccfDNA, and RNA Extraction
2.3. Assessment of Genome-Wide Methylation Level
2.4. Real-Time Reverse-Transcription Polymerase Chain Reaction
2.5. TaqMan Quantitative Methylation-Specific PCR
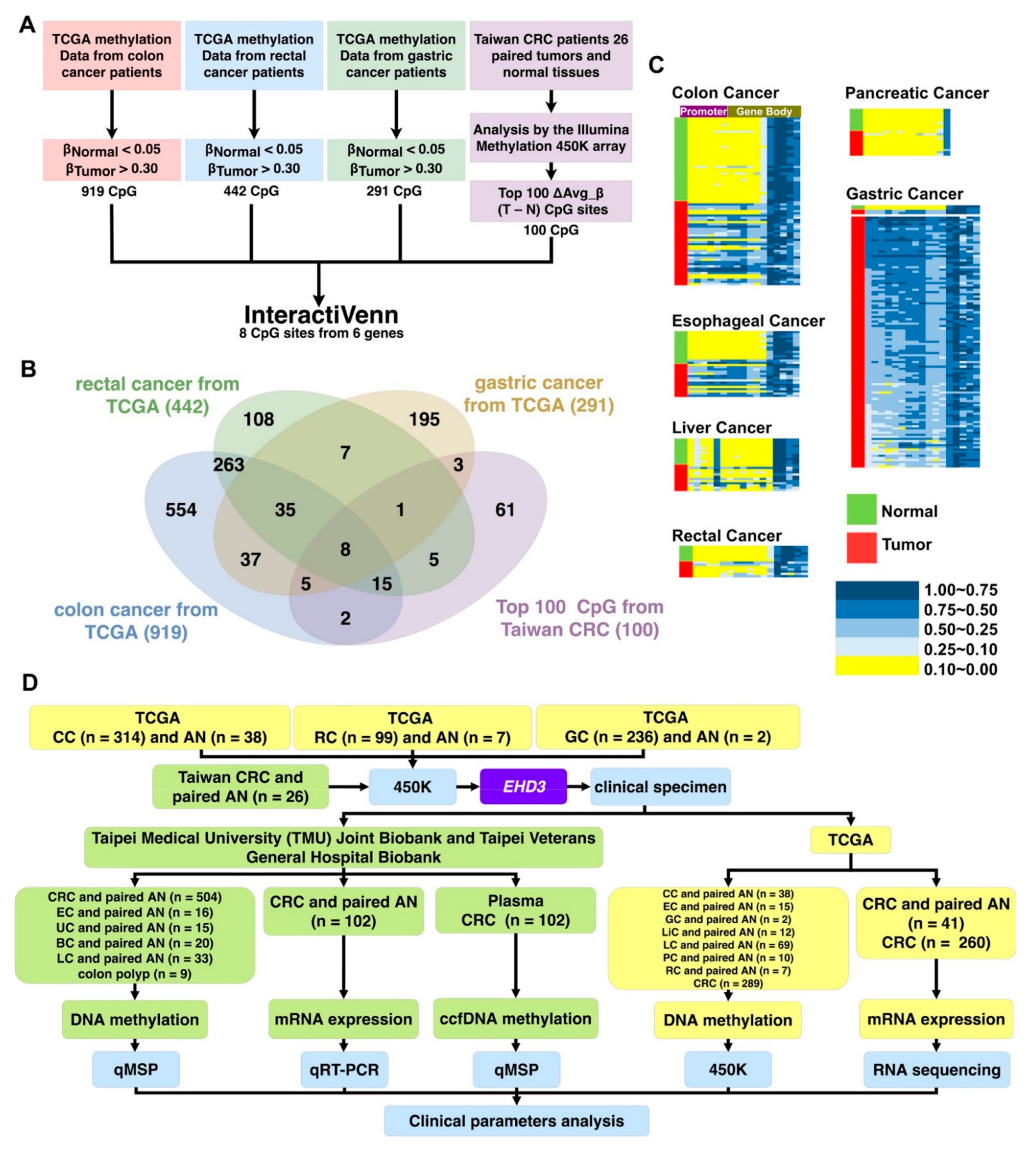
2.6. TCGA Data Analysis and Candidate Gene Selection
2.7. Statistical Analyses
3. Results
3.1. EHD3 Is a Common Target in Alimentary Canal Cancer
3.2. EHD3 Promoter Hypermethylation and Low Expression of mRNA and Protein in Asian Patients with CRC
3.3. Promoter Hypermethylation, Low mRNA, and Protein Expression of EHD3 in Western Patients with CRC and Poor Prognoses
3.4. Promoter Hypermethylation of EHD3 Reduces Drug Sensitivity in Patients with CRC from Western Countries
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Ministry of Health and Welfare. Statistical Results of Deaths of Taiwanese People in the 107th Year of the Republic Era; Ministry of Health and Welfare: Taiwan, China, 2019. [Google Scholar]
- Maida, M.; Macaluso, F.S.; Ianiro, G.; Mangiola, F.; Sinagra, E.; Hold, G.; Maida, C.; Cammarota, G.; Gasbarrini, A.; Scarpulla, G. Screening of colorectal cancer: Present and future. Expert Rev. Anticancer Ther. 2017, 17, 1131–1146. [Google Scholar] [CrossRef]
- Vymetalkova, V.; Vodicka, P.; Vodenkova, S.; Alonso, S.; Schneider-Stock, R. DNA methylation and chromatin modifiers in colorectal cancer. Mol. Asp. Med. 2019, 69, 73–92. [Google Scholar] [CrossRef]
- Muller, M.; Hansmannel, F.; Arnone, D.; Choukour, M.; Ndiaye, N.C.; Kokten, T.; Houlgatte, R.; Peyrin-Biroulet, L. Genomic and molecular alterations in human inflammatory bowel disease-associated colorectal cancer. United Eur. Gastroenterol. J. 2020, 8, 675–684. [Google Scholar] [CrossRef]
- Sakai, E.; Nakajima, A.; Kaneda, A. Accumulation of aberrant DNA methylation during colorectal cancer development. World J. Gastroenterol. 2014, 20, 978–987. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Jung, M.; Pfeifer, G.P. Aging and DNA methylation. BMC Biol. 2015, 13, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wilson, R.; Heiss, J.; Breitling, L.P.; Saum, K.-U.; Schöttker, B.; Holleczek, B.; Waldenberger, M.; Peters, A.; Brenner, H. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat. Commun. 2017, 8, 14617. [Google Scholar] [CrossRef] [PubMed]
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015, 149, 1204–1225.e12. [Google Scholar] [CrossRef] [PubMed]
- Costello, J.F.; Frühwald, M.C.; Smiraglia, D.J.; Rush, L.J.; Robertson, G.P.; Gao, X.; Wright, F.A.; Feramisco, J.D.; Peltomäki, P.; Lang, J.C.; et al. Aberrant CpG-island methylation has non-random and tumour-type–specific patterns. Nat. Genet. 2000, 24, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Vymetalkova, V.; Cervena, K.; Bartu, L.; Vodicka, P. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. Int. J. Mol. Sci. 2018, 19, 3356. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.-C.; Lin, J.-K.; Lin, C.-H.; Lin, H.-H.; Yang, S.-H.; Jiang, J.-K.; Chen, W.-S.; Chou, C.-C.; Tsai, S.-F.; Chang, S.-C. Clinical Relevance of Plasma DNA Methylation in Colorectal Cancer Patients Identified by Using a Genome-Wide High-Resolution Array. Ann. Surg. Oncol. 2014, 22, 1419–1427. [Google Scholar] [CrossRef]
- Zong, L.; Hattori, N.; Yoda, Y.; Yamashita, S.; Takeshima, H.; Takahashi, T.; Maeda, M.; Katai, H.; Nanjo, S.; Ando, T.; et al. Establishment of a DNA methylation marker to evaluate cancer cell fraction in gastric cancer. Gastric Cancer 2016, 19, 361–369. [Google Scholar] [CrossRef]
- Wong, W.T.; Schumacher, C.; Salcini, A.E.; Romano, A.; Castagnino, P.; Pelicci, P.G.; Di Fiore, P. A protein-binding domain, EH, identified in the receptor tyrosine kinase substrate Eps15 and conserved in evolution. Proc. Natl. Acad. Sci. USA 1995, 92, 9530–9534. [Google Scholar] [CrossRef]
- Confalonieri, S.; Di Fiore, P.P. The eps15 homology (eh) domain. FEBS Lett. 2002, 513, 24–29. [Google Scholar] [CrossRef]
- Naslavsky, N.; McKenzie, J.; Altan-Bonnet, N.; Sheff, D.; Caplan, S. EHD3 regulates early-endosome-to-Golgi transport and preserves Golgi morphology. J. Cell Sci. 2009, 122, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Naslavsky, N.; Caplan, S. EHD proteins: Key conductors of endocytic transport. Trends Cell Biol. 2011, 21, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Chukkapalli, S.; Amessou, M.; Dekhil, H.; Dilly, A.K.; Liu, Q.; Bandyopadhyay, S.; Thomas, R.D.; Bejna, A.; Batist, G.; Kandouz, M. Ehd3, a regulator of vesicular trafficking, is silenced in gliomas and functions as a tumor suppressor by controlling cell cycle arrest and apoptosis. Carcinogenesis 2013, 35, 877–885. [Google Scholar] [CrossRef]
- Chen, Y.; Liao, L.-D.; Wu, Z.-Y.; Yang, Q.; Guo, J.-C.; He, J.-Z.; Wang, S.-H.; Xu, X.-E.; Wu, J.-Y.; Pan, F.; et al. Identification of key genes by integrating DNA methylation and next-generation transcriptome sequencing for esophageal squamous cell carcinoma. Aging 2020, 12, 1332–1365. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, G.V.; Da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 1–7. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Yuan, H.; Dong, Q.; Zheng, B.; Hu, X.; Xu, J.-B.; Tu, S. Lymphovascular invasion is a high risk factor for stage I/II colorectal cancer: A systematic review and meta-analysis. Oncotarget 2017, 8, 46565–46579. [Google Scholar] [CrossRef] [PubMed]
- Knijn, N.; van Exsel, U.E.M.; de Noo, M.E.; Nagtegaal, I.D. The value of intramural vascular invasion in colorectal cancer—A systematic review and meta-analysis. Histopathology 2017, 72, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shi, C.; Zhang, K.; Xu, Q. The gender-specific association of EHD3 polymorphisms with major depressive disorder. Neurosci. Lett. 2014, 567, 11–14. [Google Scholar] [CrossRef]
- Chang, S.-C.; Liew, P.-L.; Ansar, M.; Lin, S.-Y.; Wang, S.-C.; Hung, C.-S.; Chen, J.-Y.; Jain, S.; Lin, R.-K. Hypermethylation and decreased expression of TMEM240 are potential early-onset biomarkers for colorectal cancer detection, poor prognosis, and early recurrence prediction. Clin. Epigenetics 2020, 12, 1–17. [Google Scholar] [CrossRef]
- Cai, B.; Giridharan, S.S.P.; Zhang, J.; Saxena, S.; Bahl, K.; Schmidt, J.A.; Sorgen, P.L.; Guo, W.; Naslavsky, N.; Caplan, S. Differential Roles of C-terminal Eps15 Homology Domain Proteins as Vesiculators and Tubulators of Recycling Endosomes. J. Biol. Chem. 2013, 288, 30172–30180. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Insinna, C.; Ott, C.; Stauffer, J.; Pintado, P.A.; Rahajeng, J.; Baxa, U.; Walia, V.; Cuenca, A.; Hwang, Y.-S.; et al. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat. Cell Biol. 2015, 17, 228–240. [Google Scholar] [CrossRef]



| Characteristics | Total n 2 | EHD3 Methylation 3 | Total n | EHD3 mRNA 4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Low n (%) | High n (%) | Low n (%) | High n (%) | |||||||
| CRC | 504 | 225 | (44.6) | 279 | (55.4) | 102 | 72 | (70.6) | 30 | (29.4) |
| Age | ||||||||||
| <65 | 226 | 104 | (46.0) | 122 | (54.0)0.479 | 27 | 12 | (44.4) | 15 | (55.6)0.210 |
| >65 | 273 | 117 | (42.9) | 156 | (57.9) | 40 | 16 | (40.0) | 24 | (60.0) |
| Sex | ||||||||||
| Male | 300 | 133 | (44.3) | 167 | (55.7)0.960 | 39 | 21 | (53.8) | 18 | (46.2)0.960 |
| Female | 195 | 86 | (44.1) | 109 | (55.9) | 28 | 15 | (53.6) | 13 | (46.4) |
| Tumor Type | ||||||||||
| Adeno | 461 | 203 | (44.0) | 258 | (56.0)0.779 | 65 | 35 | (53.8) | 30 | (46.2)0.439 |
| Others | 30 | 14 | (46.7) | 16 | (53.3) | 1 | 1 | (0.00) | 0 | (100.0) |
| Tumor Stage | ||||||||||
| I | 42 | 18 | (42.9) | 24 | (57.1)0.826 | 5 | 3 | (60.0) | 2 | (40.0)0.388 |
| II | 187 | 90 | (48.1) | 97 | (51.9) | 28 | 14 | (50.0) | 14 | (50.0) |
| III | 140 | 60 | (42.9) | 80 | (57.1) | 12 | 9 | (75.0) | 3 | (25.0) |
| IV | 96 | 40 | (41.7) | 56 | (58.3) | 22 | 10 | (40.0) | 12 | (60.0) |
| Tumor Size | ||||||||||
| T0-T1 | 34 | 17 | (50.0) | 17 | (50.0)0.494 | 5 | 2 | (40.0) | 3 | (60.0)0.770 |
| T2-T4 | 455 | 200 | (44.0) | 255 | (56.0) | 62 | 29 | (46.8) | 33 | (53.2) |
| Regional lymph nodes metastasis | ||||||||||
| N = 0 | 255 | 120 | (47.1) | 135 | (52.9)0.208 | 37 | 15 | (40.5) | 22 | (59.5)0.016 * |
| N > 1 | 232 | 96 | (41.4) | 136 | (58.6) | 30 | 21 | (70.0) | 9 | (30.0) |
| Distant metastasis | ||||||||||
| M = 0 | 372 | 171 | (46.0) | 201 | (54.0)0.404 | 41 | 23 | (56.1) | 18 | (43.9)0.332 |
| M > 1 | 97 | 40 | (41.2) | 57 | (58.8) | 23 | 10 | (43.5) | 13 | (56.5) |
| Differentiation grade | ||||||||||
| Well/Moderate | 450 | 201 | (44.7) | 249 | (55.3)0.627 | 63 | 28 | (44.4) | 35 | (55.6)0.876 |
| Poor | 37 | 15 | (40.5) | 22 | (59.5) | 2 | 1 | (50.0) | 1 | (50.0) |
| Location | ||||||||||
| Ascending Colon | 121 | 61 | (50.4) | 60 | (49.6)0.337 | 15 | 10 | (66.7) | 5 | (33.3)0.242 |
| Transverse Colon | 37 | 15 | (40.5) | 22 | (59.5) | 8 | 2 | (25.0) | 6 | (75.0) |
| Descending Colon | 44 | 15 | (34.1) | 29 | (65.9) | 7 | 3 | (42.9) | 4 | (57.1) |
| Sigmoid Colon | 152 | 66 | (43.4) | 86 | (56.6) | 18 | 7 | (38.9) | 11 | (61.1) |
| Rectal | 121 | 58 | (47.9) | 63 | (52.1) | 13 | 7 | (53.8) | 6 | (46.2) |
| Vascular invasion | ||||||||||
| No invasion | 333 | 162 | (48.6) | 171 | (51.4)0.005 ** | 4 | 2 | (50.0) | 2 | (50.0)0.919 |
| invasion | 144 | 50 | (34.7) | 94 | (65.3) | 57 | 30 | (52.6) | 27 | (47.4) |
| MSI | ||||||||||
| MSS/MSI-L MSI-H | 53 | 20 | (37.7) | 33 | (62.3)0.047 * | 32 | 16 | (50.0) | 16 | (50.0)0.677 |
| 7 | 0 | (0.0) | 7 | (100.0) | 5 | 2 | (40.0) | 3 | (60.0) | |
| Variable | Multivariate Analysis 1 | ||
|---|---|---|---|
| HR | 95% CI | p-Value | |
| Overall survival | |||
| Age | 5.003 | 0.065–385.851 | 0.468 |
| Differentiation | 1.157 | 0.020–66.973 | 0.944 |
| Tumor stage | 445.234 | 4.525–43,808.867 | 0.009 ** |
| Location | 2.538 | 0.271–23.731 | 0.414 |
| Female EHD3 DNA methylation 2 | 40.350 | 1.799–904.977 | 0.020 * |
| EHD3 RNA expression | 0.21 | 0.001–0.454 | 0.014 * |
| Recurrence-free survival | |||
| Age | 7.074 | 0.253–197.642 | 0.250 |
| Differentiation | 1.589 | 0.171–14.755 | 0.684 |
| Tumor stage | 174.281 | 5.824–5214.941 | 0.003 ** |
| Location | 1.774 | 0.307–10.250 | 0.522 |
| Female EHD3 DNA methylation | 21.966 | 1.630–296.011 | 0.020 * |
| EHD3 RNA expression | 0.17 | 0.01–3.09 | 0.006 ** |
| Recurrence-free survival | |||
| Age | 1.457 | 0.371–5.717 | 0.589 |
| Sex | 0.821 | 0.222–3.037 | 0.767 |
| Differentiation | 1.355 | 0.234–7.833 | 0.734 |
| Tumor stage | 11.622 | 3.505–38.543 | <0.000 *** |
| Location | 1.482 | 0.443–4.957 | 0.523 |
| EHD3 DNA methylation | 1.878 | 0.567–6.217 | 0.302 |
| EHD3 RNA expression | 0.283 | 0.091–0.881 | 0.029 * |
| Characteristics | Total | Complete Response n (%) | Progressive Disease n (%) | p Value 1 |
|---|---|---|---|---|
| Chemotherapy 2 | 25 | 13 (52.0) | 12 (48.0) | |
| Low methylation | 13 | 10 (76.9) | 3 (23.1) | 0.017 * |
| High methylation | 12 | 3 (25.0) | 9 (75.0) | |
| Antimetabolites | ||||
| Low methylation | 12 | 9 (75.0) | 3 (25.0) | 0.039 * |
| High methylation | 12 | 3 (25.0) | 9 (75.0) | |
| DNA Alkylating drugs | ||||
| Low methylation | 6 | 5 (83.3) | 1 (16.7) | 0.545 |
| High methylation | 6 | 3 (50.0) | 3 (50.0) | |
| Topoisomerase inhibitors | ||||
| Low methylation | 4 | 2 (50.0) | 2 (50.0) | 0.491 |
| High methylation | 7 | 1 (14.3) | 6 (85.7) | |
| Targeted Molecular therapy 3 | ||||
| Low methylation | 4 | 3 (75.0) | 1 (25.0) | 0.524 |
| High methylation | 6 | 2 (33.3) | 4 (66.7) | |
| Antimetabolites | ||||
| 5-fluorouracil | ||||
| Low methylation | 8 | 5 (62.5) | 3 (37.5) | 0.074 |
| High methylation | 11 | 2 (18.2) | 9 (81.8) | |
| Capecitabine | ||||
| Low methylation | 5 | 5 (100.0) | 0 (0.00) | 0.375 |
| High methylation | 3 | 2 (66.7) | 1(33.3) | |
| Folinic acid | ||||
| Low methylation | 6 | 3 (50.0) | 3 (50.0) | 0.600 |
| High methylation | 11 | 3 (27.3) | 8 (72.7) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-H.; Chang, S.-C.; Ansar, M.; Hung, C.-S.; Lin, R.-K. Eps15 Homology Domain-Containing Protein 3 Hypermethylation as a Prognostic and Predictive Marker for Colorectal Cancer. Biomedicines 2021, 9, 453. https://doi.org/10.3390/biomedicines9050453
Wang Y-H, Chang S-C, Ansar M, Hung C-S, Lin R-K. Eps15 Homology Domain-Containing Protein 3 Hypermethylation as a Prognostic and Predictive Marker for Colorectal Cancer. Biomedicines. 2021; 9(5):453. https://doi.org/10.3390/biomedicines9050453
Chicago/Turabian StyleWang, Yu-Han, Shih-Ching Chang, Muhamad Ansar, Chin-Sheng Hung, and Ruo-Kai Lin. 2021. "Eps15 Homology Domain-Containing Protein 3 Hypermethylation as a Prognostic and Predictive Marker for Colorectal Cancer" Biomedicines 9, no. 5: 453. https://doi.org/10.3390/biomedicines9050453
APA StyleWang, Y.-H., Chang, S.-C., Ansar, M., Hung, C.-S., & Lin, R.-K. (2021). Eps15 Homology Domain-Containing Protein 3 Hypermethylation as a Prognostic and Predictive Marker for Colorectal Cancer. Biomedicines, 9(5), 453. https://doi.org/10.3390/biomedicines9050453






