NRG1 Genetic Variant Influences the Efficacy of Androgen-Deprivation Therapy in Men with Prostate Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Response Evaluation
2.2. SNP Selection and Genotyping
2.3. Bioinformatics Analysis
2.4. Statistical Analysis
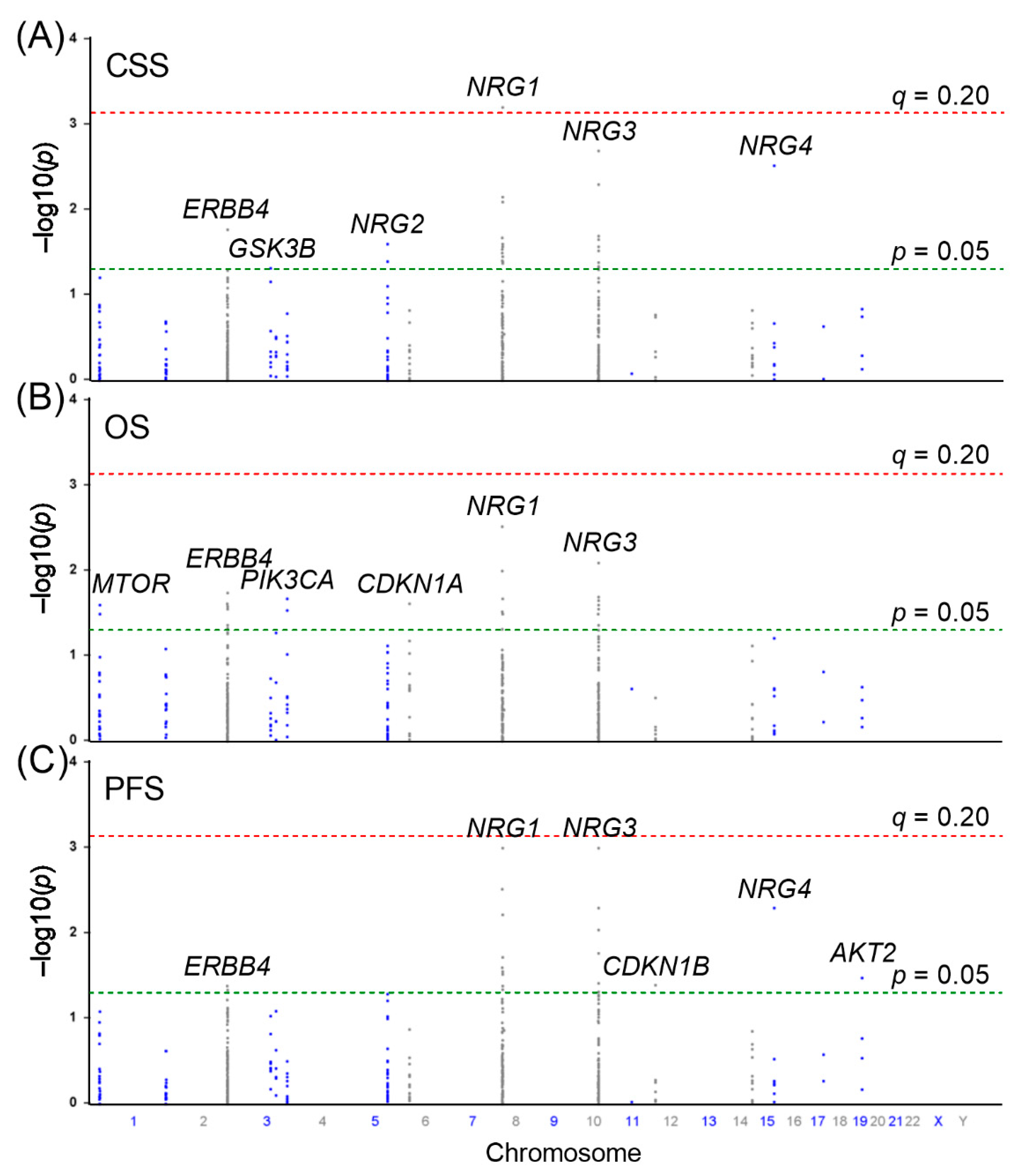
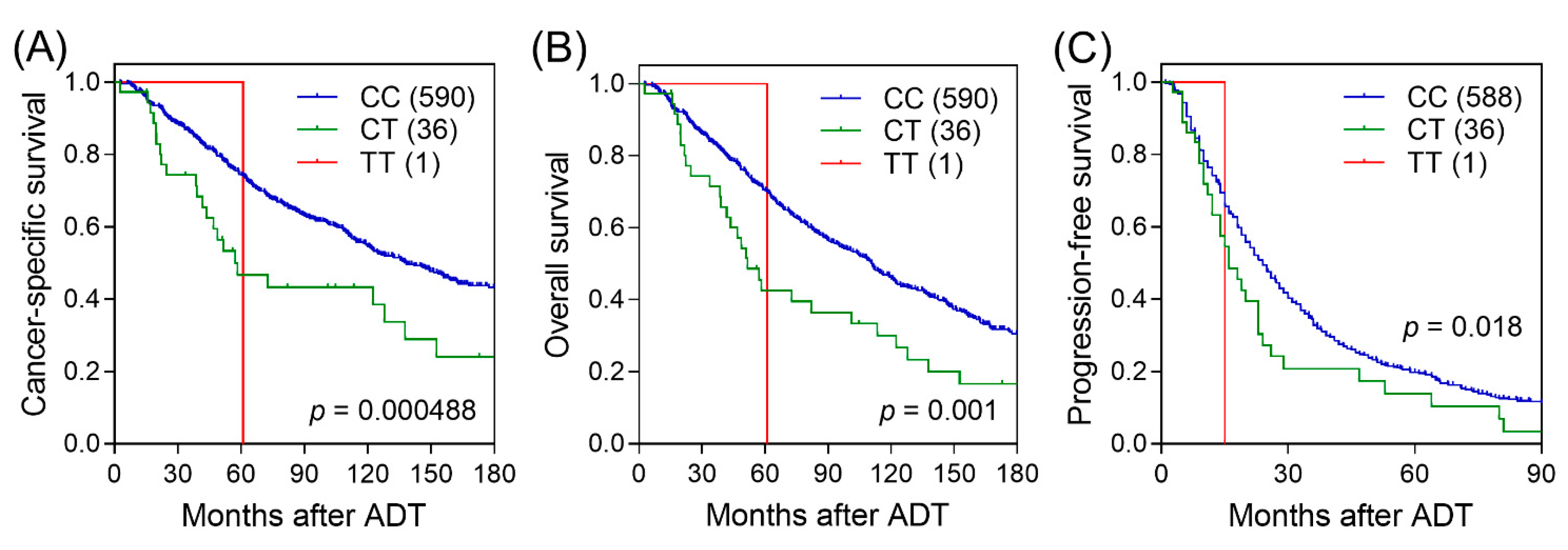
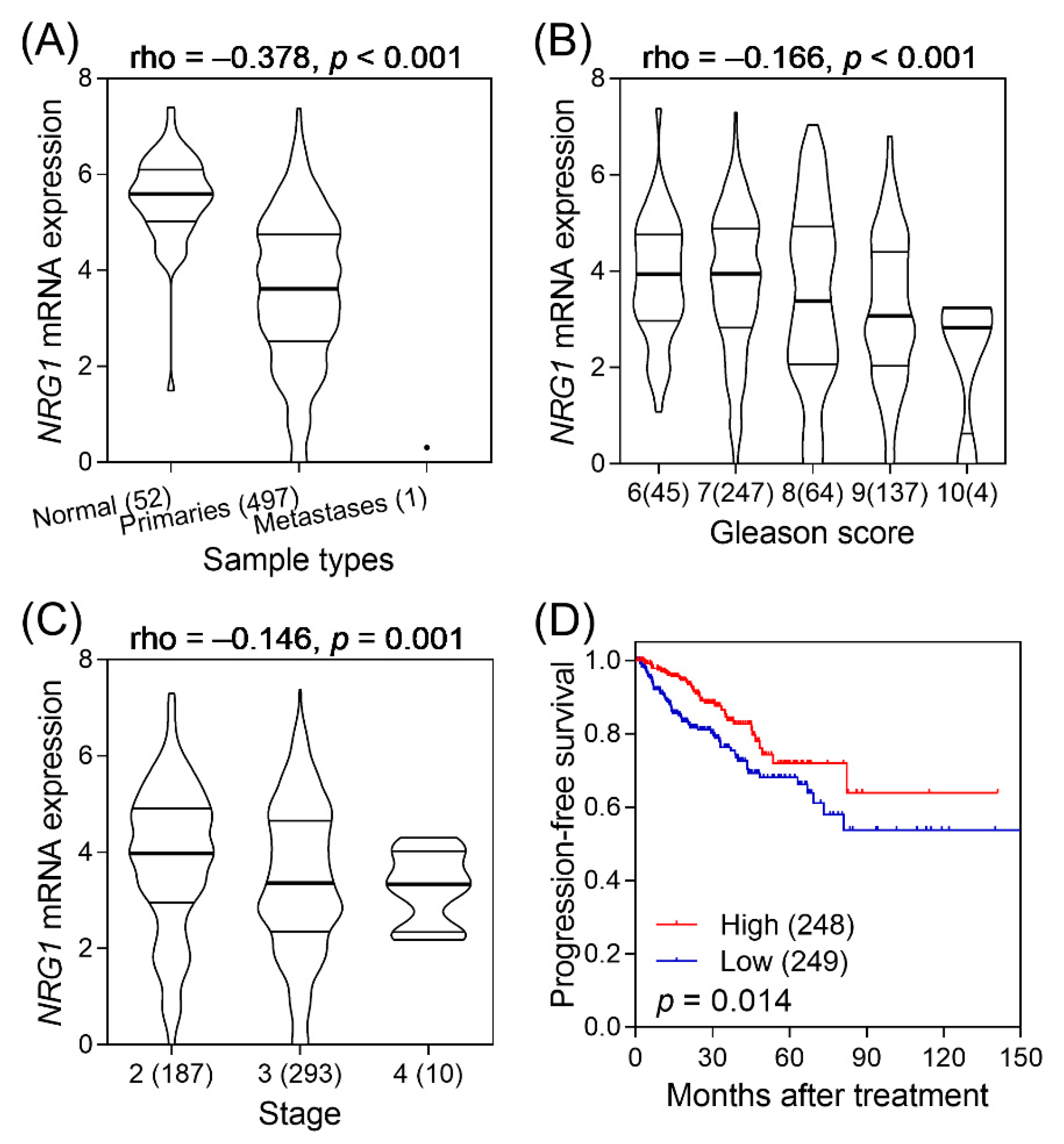
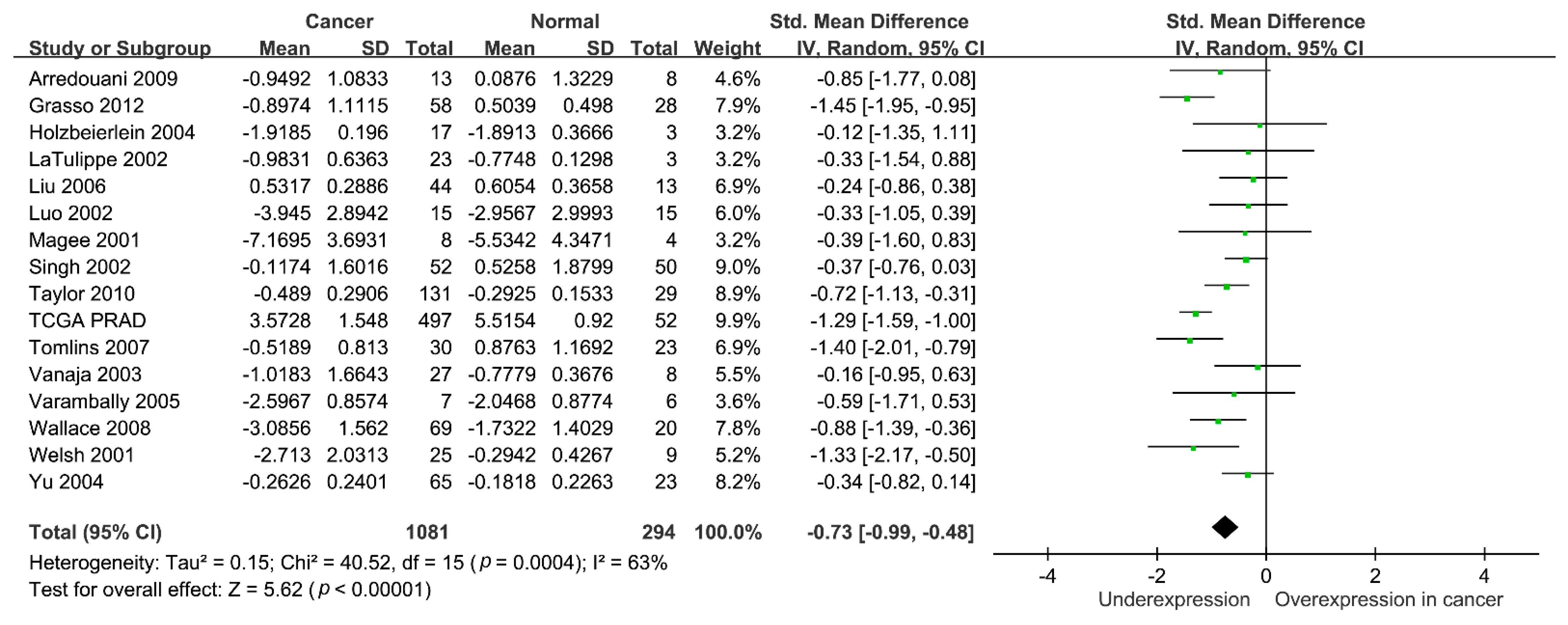
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021. online ahead of print. [Google Scholar] [CrossRef]
- Huggins, C.; Hodges, C.V. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J. Clin. 1972, 22, 232–240. [Google Scholar]
- Kirby, M.; Hirst, C.; Crawford, E.D. Characterising the castration-resistant prostate cancer population: A systematic review. Int. J. Clin. Pract. 2011, 65, 1180–1192. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, P.; Holm, N.V.; Verkasalo, P.K.; Iliadou, A.; Kaprio, J.; Koskenvuo, M.; Pukkala, E.; Skytthe, A.; Hemminki, K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000, 343, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Al Olama, A.A.; Kote-Jarai, Z.; Berndt, S.I.; Conti, D.V.; Schumacher, F.; Han, Y.; Benlloch, S.; Hazelett, D.J.; Wang, Z.; Saunders, E.; et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat. Genet. 2014, 46, 1103–1109. [Google Scholar] [CrossRef] [Green Version]
- Holmes, W.E.; Sliwkowski, M.X.; Akita, R.W.; Henzel, W.J.; Lee, J.; Park, J.W.; Yansura, D.; Abadi, N.; Raab, H.; Lewis, G.D.; et al. Identification of heregulin, a specific activator of p185erbB2. Science 1992, 256, 1205–1210. [Google Scholar] [CrossRef] [PubMed]
- Falls, D.L. Neuregulins: Functions, forms, and signaling strategies. Exp. Cell Res. 2003, 284, 14–30. [Google Scholar] [CrossRef]
- Talmage, D.A. Mechanisms of neuregulin action. Novartis Found. Symp. 2008, 289, 74–84. [Google Scholar]
- Gudmundsson, J.; Sulem, P.; Gudbjartsson, D.F.; Jonasson, J.G.; Masson, G.; He, H.; Jonasdottir, A.; Sigurdsson, A.; Stacey, S.N.; Johannsdottir, H.; et al. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat. Genet. 2012, 44, 319–322. [Google Scholar] [CrossRef]
- Slattery, M.L.; John, E.M.; Stern, M.C.; Herrick, J.; Lundgreen, A.; Giuliano, A.R.; Hines, L.; Baumgartner, K.B.; Torres-Mejia, G.; Wolff, R.K. Associations with growth factor genes (FGF1, FGF2, PDGFB, FGFR2, NRG2, EGF, ERBB2) with breast cancer risk and survival: The Breast Cancer Health Disparities Study. Breast Cancer Res. Treat. 2013, 140, 587–601. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.; Liu, H.; Zhao, Y.; Qian, D.; Luo, S.; Patz, E.F., Jr.; Su, L.; Shen, S.; Christian, I.D.; Gao, W.; et al. Genetic variants of BIRC3 and NRG1 in the NLRP3 inflammasome pathway are associated with non-small cell lung cancer survival. Am. J. Cancer Res. 2020, 10, 2582–2595. [Google Scholar]
- Pinto, R.; Assis, J.; Nogueira, A.; Pereira, C.; Coelho, S.; Brandao, M.; Dias, J.; Alves, S.; Pereira, D.; Medeiros, R. Pharmacogenomics in epithelial ovarian cancer first-line treatment outcome: Validation of GWAS-associated NRG3 rs1649942 and BRE rs7572644 variants in an independent cohort. Pharm. J. 2019, 19, 25–32. [Google Scholar] [CrossRef]
- Bao, B.Y.; Pao, J.B.; Huang, C.N.; Pu, Y.S.; Chang, T.Y.; Lan, Y.H.; Lu, T.L.; Lee, H.Z.; Juang, S.H.; Chen, L.M.; et al. Polymorphisms inside microRNAs and microRNA target sites predict clinical outcomes in prostate cancer patients receiving androgen-deprivation therapy. Clin. Cancer Res. 2011, 17, 928–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, J.H.; Lin, V.C.; Yu, C.C.; Huang, C.Y.; Yin, H.L.; Chang, T.Y.; Lu, T.L.; Huang, S.P.; Bao, B.Y. Inherited Variants in Wnt Pathway Genes Influence Outcomes of Prostate Cancer Patients Receiving Androgen Deprivation Therapy. Int. J. Mol. Sci. 2016, 17, 1970. [Google Scholar] [CrossRef] [Green Version]
- Genomes Project, C.; Abecasis, G.R.; Auton, A.; Brooks, L.D.; DePristo, M.A.; Durbin, R.M.; Handsaker, R.E.; Kang, H.M.; Marth, G.T.; McVean, G.A. An integrated map of genetic variation from 1,092 human genomes. Nature 2012, 491, 56–65. [Google Scholar]
- Bao, B.Y.; Pao, J.B.; Huang, C.N.; Pu, Y.S.; Chang, T.Y.; Lan, Y.H.; Lu, T.L.; Lee, H.Z.; Chen, L.M.; Ting, W.C.; et al. Significant associations of prostate cancer susceptibility variants with survival in patients treated with androgen-deprivation therapy. Int. J. Cancer 2012, 130, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef]
- Frazer, K.A.; Ballinger, D.G.; Cox, D.R.; Hinds, D.A.; Stuve, L.L.; Gibbs, R.A.; Belmont, J.W.; Boudreau, A.; Hardenbol, P.; Leal, S.M.; et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007, 449, 851–861. [Google Scholar]
- Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Li, L.H.; Qiao, H.M.; Yang, X.; Chen, L.; Luo, X.H. Regulation of the Antioxidant Response by MyoD Transcriptional Coactivator in Castration-resistant Prostate Cancer Cells. Urology 2019, 123, 296.e9–296.e18. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.C.; Yuste, L.; Diaz-Rodriguez, E.; Esparis-Ogando, A.; Pandiella, A. Differential shedding of transmembrane neuregulin isoforms by the tumor necrosis factor-alpha-converting enzyme. Mol. Cell Neurosci. 2000, 16, 631–648. [Google Scholar] [CrossRef] [PubMed]
- Muraoka-Cook, R.S.; Feng, S.M.; Strunk, K.E.; Earp, H.S., 3rd. ErbB4/HER4: Role in mammary gland development, differentiation and growth inhibition. J. Mammary Gland Biol. Neoplasia 2008, 13, 235–246. [Google Scholar] [PubMed] [Green Version]
- Bacus, S.S.; Huberman, E.; Chin, D.; Kiguchi, K.; Simpson, S.; Lippman, M.; Lupu, R. A ligand for the erbB-2 oncogene product (gp30) induces differentiation of human breast cancer cells. Cell Growth Differ. 1992, 3, 401–411. [Google Scholar]
- Jonna, S.; Feldman, R.A.; Swensen, J.; Gatalica, Z.; Korn, W.M.; Borghaei, H.; Ma, P.C.; Nieva, J.J.; Spira, A.I.; Vanderwalde, A.M.; et al. Detection of NRG1 Gene Fusions in Solid Tumors. Clin. Cancer Res. 2019, 25, 4966–4972. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Cuesta, L.; Thomas, R.K. Molecular Pathways: Targeting NRG1 Fusions in Lung Cancer. Clin. Cancer Res. 2015, 21, 1989–1994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gay, N.D.; Wang, Y.; Beadling, C.; Warrick, A.; Neff, T.; Corless, C.L.; Tolba, K. Durable Response to Afatinib in Lung Adenocarcinoma Harboring NRG1 Gene Fusions. J. Thorac. Oncol. 2017, 12, e107–e110. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.R.; Lim, H.; Shen, Y.; Pleasance, E.; Ch’ng, C.; Reisle, C.; Leelakumari, S.; Zhao, C.; Yip, S.; Ho, J.; et al. Successful targeting of the NRG1 pathway indicates novel treatment strategy for metastatic cancer. Ann. Oncol. 2017, 28, 3092–3097. [Google Scholar] [CrossRef]
- Jones, M.R.; Williamson, L.M.; Topham, J.T.; Lee, M.K.C.; Goytain, A.; Ho, J.; Denroche, R.E.; Jang, G.; Pleasance, E.; Shen, Y.; et al. NRG1 Gene Fusions Are Recurrent, Clinically Actionable Gene Rearrangements in KRAS Wild-Type Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2019, 25, 4674–4681. [Google Scholar] [CrossRef] [Green Version]
- Chua, Y.L.; Ito, Y.; Pole, J.C.; Newman, S.; Chin, S.F.; Stein, R.C.; Ellis, I.O.; Caldas, C.; O’Hare, M.J.; Murrell, A.; et al. The NRG1 gene is frequently silenced by methylation in breast cancers and is a strong candidate for the 8p tumour suppressor gene. Oncogene 2009, 28, 4041–4052. [Google Scholar] [CrossRef] [Green Version]
- Vermeer, P.D.; Einwalter, L.A.; Moninger, T.O.; Rokhlina, T.; Kern, J.A.; Zabner, J.; Welsh, M.J. Segregation of receptor and ligand regulates activation of epithelial growth factor receptor. Nature 2003, 422, 322–326. [Google Scholar] [CrossRef]
- Birnbaum, D.; Adelaide, J.; Popovici, C.; Charafe-Jauffret, E.; Mozziconacci, M.J.; Chaffanet, M. Chromosome arm 8p and cancer: A fragile hypothesis. Lancet Oncol. 2003, 4, 639–642. [Google Scholar] [CrossRef]
- Pole, J.C.; Courtay-Cahen, C.; Garcia, M.J.; Blood, K.A.; Cooke, S.L.; Alsop, A.E.; Tse, D.M.; Caldas, C.; Edwards, P.A. High-resolution analysis of chromosome rearrangements on 8p in breast, colon and pancreatic cancer reveals a complex pattern of loss, gain and translocation. Oncogene 2006, 25, 5693–5706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinstein, E.J.; Grimm, S.; Leder, P. The oncogene heregulin induces apoptosis in breast epithelial cells and tumors. Oncogene 1998, 17, 2107–2113. [Google Scholar] [CrossRef] [Green Version]
- Lyne, J.C.; Melhem, M.F.; Finley, G.G.; Wen, D.; Liu, N.; Deng, D.H.; Salup, R. Tissue expression of neu differentiation factor/heregulin and its receptor complex in prostate cancer and its biologic effects on prostate cancer cells in vitro. Cancer J. Sci. Am. 1997, 3, 21–30. [Google Scholar]
- Gregory, C.W.; Whang, Y.E.; McCall, W.; Fei, X.; Liu, Y.; Ponguta, L.A.; French, F.S.; Wilson, E.M.; Earp, H.S., 3rd. Heregulin-induced activation of HER2 and HER3 increases androgen receptor transactivation and CWR-R1 human recurrent prostate cancer cell growth. Clin. Cancer Res. 2005, 11, 1704–1712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimsley, S.J.; Shini, S.; Underwood, M.A.; Edwards, J. Heregulin expression and prognosis in prostate adenocarcinoma. Urol. Int. 2011, 87, 363–368. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | n (%) | CSS a | OS a | PFS a | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Events, n | Median, Months | p | Events, n | Median, Months | p | Events, n | Median, Months | p | ||
| Total, n (%) | 630 | 314 | 135 | 413 | 109 | 518 | 23 | |||
| Age at Diagnosis, Years | ||||||||||
| Median (IQR) | 73 (67–79) | |||||||||
| <74 | 344 (54.7) | 168 | 154 | 0.014 | 201 | 128 | <0.001 | 295 | 20 | <0.001 |
| ≥74 | 285 (45.3) | 145 | 120 | 211 | 86 | 222 | 29 | |||
| PSA at ADT Initiation, ng/mL | ||||||||||
| Median (IQR) | 34.5 (11.25–129) | |||||||||
| <35 | 307 (50.6) | 115 | 196 | <0.001 | 167 | 138 | <0.001 | 245 | 26 | 0.028 |
| ≥35 | 300 (49.4) | 187 | 88 | 232 | 72 | 256 | 19 | |||
| PSA Nadir, ng/mL | ||||||||||
| Median (IQR) | 0.14 (0.01–1.16) | |||||||||
| <0.15 | 314 (50.7) | 109 | 202 | <0.001 | 167 | 159 | <0.001 | 243 | 34 | <0.001 |
| ≥0.15 | 305 (49.3) | 200 | 65 | 239 | 59 | 270 | 15 | |||
| Time to PSA Nadir, Months | ||||||||||
| Median (IQR) | 11 (5–20) | |||||||||
| <12 | 323 (52.2) | 177 | 96 | <0.001 | 216 | 77 | <0.001 | 276 | 12 | <0.001 |
| ≥12 | 296 (47.8) | 132 | 162 | 190 | 123 | 237 | 36 | |||
| Clinical Stage at Diagnosis | ||||||||||
| T1/T2 | 187 (29.9) | 70 | NR | <0.001 | 103 | 138 | <0.001 | 144 | 26 | <0.001 |
| T3/T4/N1 | 205 (32.8) | 81 | 196 | 119 | 138 | 162 | 30 | |||
| M1 | 233 (37.3) | 162 | 63 | 189 | 59 | 209 | 16 | |||
| Gleason Score at Diagnosis | ||||||||||
| 2–6 | 188 (30.6) | 81 | 185 | <0.001 | 112 | 133 | <0.001 | 144 | 28 | 0.001 |
| 7 | 194 (31.6) | 84 | 184 | 115 | 121 | 164 | 25 | |||
| 8–10 | 232 (37.8) | 143 | 73 | 177 | 63 | 196 | 18 | |||
| Genotype | Frequency | CSS | HR (95% CI) | p | q | HR (95% CI) a | pa | ||
|---|---|---|---|---|---|---|---|---|---|
| CC/CT/TT | 591/36/1 | 286/25/1 | 1.94 (1.33–2.83) | 0.00062 | 0.168 | 1.60 (1.08–2.35) | 0.018 | ||
| OS | HR (95% CI) | p | HR (95% CI) a | pa | PFS | HR (95% CI) | p | HR (95% CI) a | pa |
| 381/30/1 | 1.78 (1.26–2.52) | 0.001 | 1.57 (1.10–2.24) | 0.014 | 482/43/1 | 1.49 (1.06–2.08) | 0.021 | 1.16 (0.83–1.63) | 0.375 |
| Reference Allele | Alternate Allele | AFR Frequency | AMR Frequency | ASN Frequency | EUR Frequency | Variant Type | Promoter Histone Marks | Enhancer Histone Marks | DNAse | Motifs Changed |
|---|---|---|---|---|---|---|---|---|---|---|
| C | T | 0.00 | 0.00 | 0.02 | 0.00 | intronic | BLD | FAT, BRST, MUS, LNG, VAS, BONE | ESDR, SKIN, SKIN, LNG, MUS, MUS, MUS, SKIN, SKIN | Myf |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.-P.; Chen, Y.-T.; Chen, L.-C.; Lee, C.-H.; Huang, C.-Y.; Yu, C.-C.; Lin, V.C.; Lu, T.-L.; Bao, B.-Y. NRG1 Genetic Variant Influences the Efficacy of Androgen-Deprivation Therapy in Men with Prostate Cancer. Biomedicines 2021, 9, 528. https://doi.org/10.3390/biomedicines9050528
Huang S-P, Chen Y-T, Chen L-C, Lee C-H, Huang C-Y, Yu C-C, Lin VC, Lu T-L, Bao B-Y. NRG1 Genetic Variant Influences the Efficacy of Androgen-Deprivation Therapy in Men with Prostate Cancer. Biomedicines. 2021; 9(5):528. https://doi.org/10.3390/biomedicines9050528
Chicago/Turabian StyleHuang, Shu-Pin, Yei-Tsung Chen, Lih-Chyang Chen, Cheng-Hsueh Lee, Chao-Yuan Huang, Chia-Cheng Yu, Victor C. Lin, Te-Ling Lu, and Bo-Ying Bao. 2021. "NRG1 Genetic Variant Influences the Efficacy of Androgen-Deprivation Therapy in Men with Prostate Cancer" Biomedicines 9, no. 5: 528. https://doi.org/10.3390/biomedicines9050528






