MicroRNA 320, an Anti-Oncogene Target miRNA for Cancer Therapy
Abstract
:1. Introduction
2. Tumor Suppressive Functions of miR-320
2.1. miR-320 Inhibits EMT and Tumor Metastasis
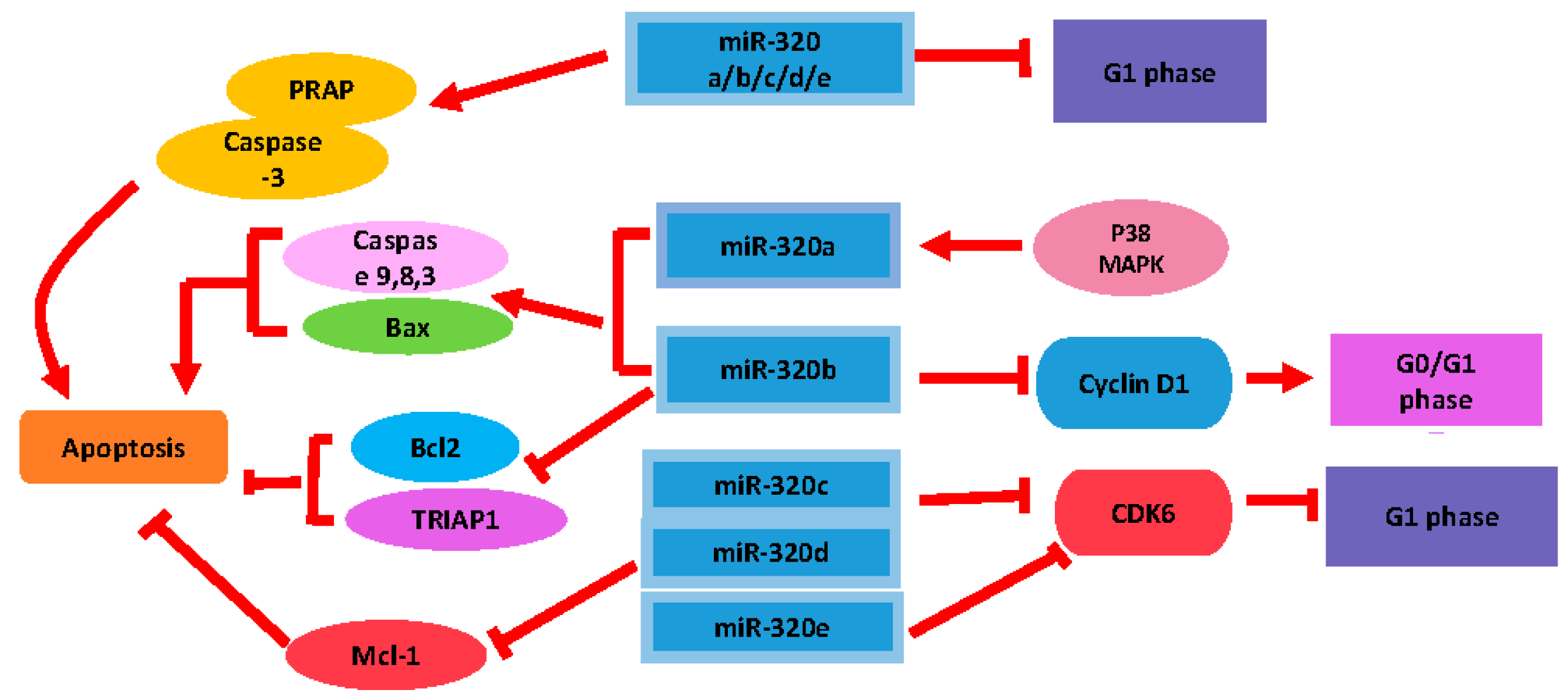
2.2. The Modulatory Role of miR-320 in Cell Proliferation and Apoptosis
2.3. The Modulatory Role of miR-320 in Alleviating Drug Resistance
3. miR-320: A Biomarker for Diagnosis, Prognosis, and Prediction of Cancer
3.1. miR-320 as a Diagnositic Biomarker
3.2. miR-320 as a Prognostic Biomarker
3.3. miR-320 as a Predictive Biomarker
4. Regulation of miR-320 Expression
4.1. Regulation of Transcription Factors
4.2. Epigenetic Modification
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Fabian, M.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [Green Version]
- Nitschke, L.; Tewari, A.; Coffin, S.; Xhako, E.; Pang, K.; Gennarino, V.; Johnson, J.; Blanco, F.; Liu, Z.; Zoghbi, H. miR760 regulates ATXN1 levels via interaction with its 5’ untranslated region. Genes Dev. 2020, 34, 1147–1160. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Rigoutsos, I. MiR-103a-3p targets the 5’ UTR of GPRC5A in pancreatic cells. RNA 2014, 20, 1431–1439. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Wu, X.; Qian, W.; Cai, H.; Sun, X.; Zhang, W.; Tan, S.; Wu, Z.; Qian, P.; Ding, K.; et al. CCAR1 5’ UTR as a natural miRancer of miR-1254 overrides tamoxifen resistance. Cell Res. 2016, 26, 655–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, M.; Reis, R.; Calin, G. MicroRNA history: Discovery, recent applications, and next frontiers. Mutat. Res. 2011, 717, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, T.; Candido, S.; Pilarz, M.; Sicinska, E.; Bronson, R.; Bowden, M.; Lachowicz, I.; Mulry, K.; Fassl, A.; Han, R.; et al. Cell cycle-targeting microRNAs promote differentiation by enforcing cell-cycle exit. Proc. Natl. Acad. Sci. USA 2017, 114, 10660–10665. [Google Scholar] [CrossRef] [Green Version]
- Long, J.; He, Q.; Yin, Y.; Lei, X.; Li, Z.; Zhu, W. The effect of miRNA and autophagy on colorectal cancer. Cell Prolif. 2020, 53, e12900. [Google Scholar] [CrossRef]
- Li, S.; Sun, Y.; Tian, T.; Qin, X.; Lin, S.; Zhang, T.; Zhang, Q.; Zhou, M.; Zhang, X.; Zhou, Y.; et al. MicroRNA-214-3p modified tetrahedral framework nucleic acids target survivin to induce tumour cell apoptosis. Cell Prolif. 2020, 53, e12708. [Google Scholar] [CrossRef] [Green Version]
- Rohan, T.; Wang, T.; Weinmann, S.; Wang, Y.; Lin, J.; Ginsberg, M.; Loudig, O. A miRNA Expression Signature in Breast Tumor Tissue Is Associated with Risk of Distant Metastasis. Cancer Res. 2019, 79, 1705–1713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Zhang, G.; Lv, S.; Wen, X.; Liu, P. miRNA-301b-3p accelerates migration and invasion of high-grade ovarian serous tumor via targeting CPEB3/EGFR axis. J. Cell Biochem. 2019, 120, 12618–12627. [Google Scholar] [CrossRef]
- Nan, Y.; Guo, H.; Guo, L.; Wang, L.; Ren, B.; Yu, K.; Huang, Q.; Zhong, Y. MiRNA-451 Inhibits Glioma Cell Proliferation and Invasion Through the mTOR/HIF-1α/VEGF Signaling Pathway by Targeting CAB39. Hum. Gene Ther. Clin. Dev. 2018, 29, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Mari, L.; Hoefnagel, S.; Zito, D.; van de Meent, M.; van Endert, P.; Calpe, S.; Sancho Serra, M.; Heemskerk, M.; van Laarhoven, H.; Hulshof, M.; et al. microRNA 125a Regulates MHC-I Expression on Esophageal Adenocarcinoma Cells, Associated with Suppression of Antitumor Immune Response and Poor Outcomes of Patients. Gastroenterology 2018, 155, 784–798. [Google Scholar] [CrossRef]
- Nucera, S.; Giustacchini, A.; Boccalatte, F.; Calabria, A.; Fanciullo, C.; Plati, T.; Ranghetti, A.; Garcia-Manteiga, J.; Cittaro, D.; Benedicenti, F.; et al. miRNA-126 Orchestrates an Oncogenic Program in B Cell Precursor Acute Lymphoblastic Leukemia. Cancer Cell 2016, 29, 905–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juźwik, C.; Drake, S.; Zhang, Y.; Paradis-Isler, N.; Sylvester, A.; Amar-Zifkin, A.; Douglas, C.; Morquette, B.; Moore, C.; Fournier, A. microRNA dysregulation in neurodegenerative diseases: A systematic review. Prog. Neurobiol. 2019, 182, 101664. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Xia, L.; Fan, S.; Zheng, J.; Qin, J.; Fan, X.; Liu, Y.; Tao, J.; Liu, Y.; Li, K.; et al. Circular RNA CircMAP3K5 Acts as a MicroRNA-22-3p Sponge to Promote Resolution of Intimal Hyperplasia Via TET2-Mediated Smooth Muscle Cell Differentiation. Circulation 2021, 143, 354–371. [Google Scholar] [CrossRef] [PubMed]
- Kandettu, A.; Radhakrishnan, R.; Chakrabarty, S.; Sriharikrishnaa, S.; Kabekkodu, S. The emerging role of miRNA clusters in breast cancer progression. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188413. [Google Scholar] [CrossRef]
- McCreight, J.; Schneider, S.; Wilburn, D.; Swanson, W. Evolution of microRNA in primates. PLoS ONE 2017, 12, e0176596. [Google Scholar] [CrossRef]
- Kim, D.; Saetrom, P.; Snøve, O.; Rossi, J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16230–16235. [Google Scholar] [CrossRef] [Green Version]
- Ittmann, M. Cell cycle control of the BN51 cell cycle gene which encodes a subunit of RNA polymerase III. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 1994, 5, 783–788. [Google Scholar]
- Ren, X.; Wu, J.; Wang, X.; Sartor, M.; Jones, K.; Qian, J.; Nicolaou, P.; Pritchard, T.; Fan, G. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation 2009, 119, 2357–2366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, H.; Zhu, H.; Zhao, S.; Wang, K.; Zhang, N.; Tian, Y.; Li, Y.; Wang, Y.; Lv, X.; Wei, T.; et al. The novel circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression. Cell Death Dis. 2019, 10, 950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wu, J.; Lin, Y.; Zhu, Y.; Xu, X.; Xu, X.; Liang, Z.; Li, S.; Hu, Z.; Zheng, X.; et al. MicroRNA-320c inhibits tumorous behaviors of bladder cancer by targeting Cyclin-dependent kinase 6. J. Exp. Clin. Cancer Res. 2014, 33, 69. [Google Scholar] [CrossRef]
- Lv, Q.; Hu, J.; Li, Y.; Xie, N.; Song, D.; Zhao, W.; Yan, Y.; Li, B.; Wang, P.; Xie, S. MiR-320a effectively suppresses lung adenocarcinoma cell proliferation and metastasis by regulating STAT3 signals. Cancer Biol. Ther. 2017, 18, 142–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Q.; Du, H.; Liu, Y.; Huang, Y.; Wang, G.; Zhang, X.; Chen, S.; Zhou, H. Low expression of microRNA-320b correlates with tumorigenesis and unfavorable prognosis in glioma. Oncol. Rep. 2017, 38, 959–966. [Google Scholar] [CrossRef] [Green Version]
- Renard, I.; Joniau, S.; van Cleynenbreugel, B.; Collette, C.; Naômé, C.; Vlassenbroeck, I.; Nicolas, H.; de Leval, J.; Straub, J.; Van Criekinge, W.; et al. Identification and validation of the methylated TWIST1 and NID2 genes through real-time methylation-specific polymerase chain reaction assays for the noninvasive detection of primary bladder cancer in urine samples. Eur. Urol. 2010, 58, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, J.; Vali, M.; McVeigh, M.; Kominsky, S.; Fackler, M.; Lahti-Domenici, J.; Polyak, K.; Sacchi, N.; Garrett-Mayer, E.; Argani, P.; et al. Very high frequency of hypermethylated genes in breast cancer metastasis to the bone, brain, and lung. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2004, 10, 3104–3109. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Hsu, D.; Wang, H.; Wang, H.; Lan, H.; Yang, W.; Huang, C.; Kao, S.; Tzeng, C.; Tai, S.; et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat. Cell Biol. 2010, 12, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Duan, P.; Wang, J.; Lu, X.; Cheng, J. miR-320 inhibited ovarian cancer oncogenicity via targeting TWIST1 expression. Am. J. Transl. Res. 2017, 9, 3705–3713. [Google Scholar]
- Qi, X.; Li, J.; Zhou, C.; Lv, C.; Tian, M. MicroRNA-320a inhibits cell proliferation, migration and invasion by targeting BMI-1 in nasopharyngeal carcinoma. FEBS Lett. 2014, 588, 3732–3738. [Google Scholar] [CrossRef] [Green Version]
- Sugano, T.; Masuda, M.; Takeshita, F.; Motoi, N.; Hirozane, T.; Goto, N.; Kashimoto, S.; Uno, Y.; Moriyama, H.; Sawa, M.; et al. Pharmacological blockage of transforming growth factor-β signalling by a Traf2- and Nck-interacting kinase inhibitor, NCB-0846. Br. J. Cancer 2021, 124, 228–236. [Google Scholar] [CrossRef]
- Zhang, H.; Li, R.; Li, Y.; Yu, X.; Sun, Q.; Li, A.; Kong, Y. eIF4E-related miR-320a and miR-340-5p inhibit endometrial carcinoma cell metastatic capability by preventing TGF-β1-induced epithelial-mesenchymal transition. Oncol. Rep. 2020, 43, 447–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Li, X.; Sun, W.; Yue, S.; Yang, J.; Li, J.; Ma, B.; Wang, J.; Yang, X.; Pu, M.; et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017, 397, 33–42. [Google Scholar] [CrossRef]
- Kahn, M. Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 2014, 13, 513–532. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.; Deng, J.; Xiang, X.; Zhang, L.; Yu, F.; Chen, J.; Sun, Z.; Feng, M.; Xiong, J. miR-320 enhances the sensitivity of human colon cancer cells to chemoradiotherapy in vitro by targeting FOXM1. Biochem. Biophys. Res. Commun. 2015, 457, 125–132. [Google Scholar] [CrossRef]
- Hsieh, I.; Chang, K.; Tsai, Y.; Ke, J.; Lu, P.; Lee, K.; Yeh, S.; Hong, T.; Chen, Y. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis 2013, 34, 530–538. [Google Scholar] [CrossRef] [Green Version]
- Aljagthmi, A.; Hill, N.; Cooke, M.; Kazanietz, M.; Abba, M.; Long, W.; Kadakia, M. ΔNp63α suppresses cells invasion by downregulating PKCγ/Rac1 signaling through miR-320a. Cell Death Dis. 2019, 10, 680. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Sun, Q.; Yu, Z.; Mao, S.; Jin, Y.; Li, J.; Jiang, Z.; Zhang, Y.; Chen, M.; Chen, P.; et al. MiR-320a-3p/ELF3 axis regulates cell metastasis and invasion in non-small cell lung cancer via PI3K/Akt pathway. Gene 2018, 670, 31–37. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; Li, J.; Geng, H.; Zhou, B.; Zhang, B.; Chen, H. miR-320/ELF3 axis inhibits the progression of breast cancer via the PI3K/AKT pathway. Oncol. Lett. 2020, 19, 3239–3248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, T.; Feng, Y.; Liu, Q.; Yang, X.; Jiang, T.; Chen, Y.; Zhang, Q. MicroRNA-320a suppresses in GBM patients and modulates glioma cell functions by targeting IGF-1R. Tumor Biol. 2014, 35, 11269–11275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yin, P.; Zhang, Z.; Xu, B.; Che, D.; Dai, Z.; Dong, C.; Jiang, P.; Hong, H.; Yang, Z.; et al. Deficiency of pigment epithelium-derived factor in nasopharyngeal carcinoma cells triggers the epithelial-mesenchymal transition and metastasis. Cell Death Dis. 2017, 8, e2838. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Sun, Q.; Yan, W.; Han, Q.; Wang, G.; Hu, Y. The role of miR-320a and its target gene GMEB1 in epithelial-mesenchymal transition and invasion of colorectal cancer. J. Gene Med. 2021, e3327. [Google Scholar] [CrossRef]
- Qin, H.; Liu, J.; Du, Z.; Hu, R.; Yu, Y.; Wang, Q. Circular RNA hsa_circ_0012673 facilitates lung cancer cell proliferation and invasion via miR-320a/LIMK18521 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1841–1852. [Google Scholar] [PubMed]
- Fu, G.; Lu, J.; Zheng, Y.; Wang, P.; Shen, Q. MiR-320a inhibits malignant phenotype of melanoma cells via targeting PBX3. JBUON Off. J. Balk. Union Oncol. 2020, 25, 2071–2077. [Google Scholar]
- Chou, L.; Chen, C.; Yang, W.; Chen, C.; Chang, J.; Leu, Y.; Liou, M.; Wang, T. Suppression of Hepatocellular Carcinoma Progression through FOXM1 and EMT Inhibition via Hydroxygenkwanin-Induced miR-320a Expression. Biomolecules 2019, 10, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Q.; Guo, M.; Zheng, J.; Zheng, X.; Ye, Z.; Wei, P. Study on Targeting Relationship Between miR-320b and FGD5-AS1 and Its Effect on Biological Function of Osteosarcoma Cells. Cancer Manag. Res. 2020, 12, 13589–13598. [Google Scholar] [CrossRef]
- Cao, W.; Zhou, G. LncRNA SNHG12 contributes proliferation, invasion and epithelial-mesenchymal transition of pancreatic cancer cells by absorbing miRNA-320b. Biosci. Rep. 2020, 40, BSR20200805. [Google Scholar] [CrossRef]
- Wu, P.; Zhang, X.; Zhu, J.; Guo, X.; Wang, J. Low expression of microRNA-146b-5p and microRNA-320d predicts poor outcome of large B-cell lymphoma treated with cyclophosphamide, doxorubicin, vincristine, and prednisone. Hum. Pathol. 2014, 45, 1664–1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Yu, J.; Wang, L.; Ding, D.; Zhang, L.; Chu, C.; Chen, Q.; Xu, Z.; Zou, Q.; Liu, X. miR-320a is an independent prognostic biomarker for invasive breast cancer. Oncol. Lett. 2014, 8, 1043–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Carbonell, L.; Sinicrope, F.; Alberts, S.; Oberg, A.; Balaguer, F.; Castells, A.; Boland, C.; Goel, A. MiR-320e is a novel prognostic biomarker in colorectal cancer. Br. J. Cancer 2015, 113, 83–90. [Google Scholar] [CrossRef]
- Perez-Carbonell, L.; Sinicrope, F.; Alberts, S.; Oberg, A.; Balaguer, F.; Castells, A.; Boland, C.; Goel, A. Downregulation of MicroRNA-320d predicts poor overall survival and promotes the growth and invasive abilities in glioma. Chem. Biol. Drug Des. 2017, 89, 806–814. [Google Scholar]
- Peng, X.; Yu, R.; Wu, X.; Wu, S.; Pi, C.; Chen, Z.; Zhang, X.; Gao, C.; Shao, Y.; Liu, L.; et al. EGFRCorrelation of plasma exosomal microRNAs with the efficacy of immunotherapy in wild-type advanced non-small cell lung cancer. J. Immunother. Cancer 2020, 8, e000376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Ding, X.; Wang, S.; Xu, L.; Yin, T.; Han, S.; Geng, J.; Sun, W. Downregulation of serum exosomal miR-320d predicts poor prognosis in hepatocellular carcinoma. J. Clin. Lab. Anal. 2020, 34, e23239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Gao, S.; Zhao, Z.; Liang, G.; Kong, J.; Feng, X. MicroRNA-320d regulates tumor growth and invasion by promoting FoxM1 and predicts poor outcome in gastric cardiac adenocarcinoma. Cell Biosci. 2020, 10, 80. [Google Scholar] [CrossRef]
- Yao, J.; Liang, L.; Zhang, Y.; Ding, J.; Tian, Q.; Li, J.; He, X. GNAI1 Suppresses Tumor Cell Migration and Invasion and is Post-Transcriptionally Regulated by Mir-320a/c/d in Hepatocellular Carcinoma. Cancer Biol. Med. 2012, 9, 234–241. [Google Scholar]
- Tang, W.; Wang, D.; Shao, L.; Liu, X.; Zheng, J.; Xue, Y.; Ruan, X.; Yang, C.; Liu, L.; Ma, J.; et al. LINC00680 and TTN-AS1 Stabilized by EIF4A3 Promoted Malignant Biological Behaviors of Glioblastoma Cells. Molecular therapy. Nucleic Acids 2020, 19, 905–921. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, M.; Huang, Y.; Feng, L.; Chen, H.; Hu, Y.; Chen, H.; Zhang, K.; Zheng, L.; Zheng, S. MicroRNA-320b promotes colorectal cancer proliferation and invasion by competing with its homologous microRNA-320a. Cancer Lett. 2015, 356, 669–675. [Google Scholar] [CrossRef] [Green Version]
- Costa, C.; Indovina, P.; Mattioli, E.; Forte, I.; Iannuzzi, C.; Luzzi, L.; Bellan, C.; De Summa, S.; Bucci, E.; Di Marzo, D.; et al. P53-regulated miR-320a targets PDL1 and is downregulated in malignant mesothelioma. Cell Death Dis. 2020, 11, 748. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wang, Z.; Ren, H.; Bao, X.; Zhang, Y.; Wang, B.; Ruan, D. Long Non-Coding RNA GAS5 Suppresses Tumor Progression and Enhances the Radiosensitivity of Prostate Cancer Through the miR-320a/RAB21 Axis. Cancer Manag. Res. 2020, 12, 8833–8845. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, L.; Yang, H.; Ding, D.; Zhang, L.; Wang, J.; Chen, Q.; Zou, Q.; Jin, Y.; Liu, X. Rab14 Suppression Mediated by MiR-320a Inhibits Cell Proliferation, Migration and Invasion in Breast Cancer. J. Cancer 2016, 7, 2317–2326. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Yang, Z.; Wang, H.; Cao, Z.; Zhao, Y.; Gong, C.; Ma, L.; Wang, X.; Hu, X.; Chen, S. MicroRNA-320a inhibits proliferation and invasion of breast cancer cells by targeting RAB11A. Am. J. Cancer Res. 2015, 5, 2719–2729. [Google Scholar] [PubMed]
- Liu, J.; Song, Z.; Feng, C.; Lu, Y.; Zhou, Y.; Lin, Y.; Dong, C. The long non-coding RNA SUMO1P3 facilitates breast cancer progression by negatively regulating miR-320a. Am. J. Transl. Res. 2017, 9, 5594–5602. [Google Scholar]
- Shang, C.; Zhang, H.; Guo, Y.; Hong, Y.; Liu, Y.; Xue, Y. MiR-320a down-regulation mediates bladder carcinoma invasion by targeting ITGB3. Mol. Biol. Rep. 2014, 41, 2521–2527. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yan, H.; Yang, W.; Hu, L.; Yu, L.; Liu, Q.; Li, L.; Huang, D.; Ding, J.; Shen, F.; et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J. Hepatol. 2009, 50, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhai, B.; He, C.; Li, Z.; Gao, H.; Niu, Z.; Jiang, X.; Lu, J.; Sun, X. LncRNA TTN-AS1 promotes the progression of cholangiocarcinoma via the miR-320a/neuropilin-1 axis. Cell Death Dis. 2020, 11, 637. [Google Scholar] [CrossRef]
- Sun, J.; Huang, Y.; Li, J.; Zhang, X.; Wang, L.; Meng, Y.; Yan, B.; Bian, Y.; Zhao, J.; Wang, W.; et al. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochem. Biophys. Res. Commun. 2012, 420, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, X.; Liu, Y.; Ye, Y.; Zhang, H.; He, P.; Zhang, Q.; Dong, L.; Liu, Y.; Dong, J. microRNA-320a inhibits tumor invasion by targeting neuropilin 1 and is associated with liver metastasis in colorectal cancer. Oncol. Rep. 2012, 27, 685–694. [Google Scholar]
- Gattolliat, C.; Uguen, A.; Pesson, M.; Trillet, K.; Simon, B.; Doucet, L.; Robaszkiewicz, M.; Corcos, L. MicroRNA and targeted mRNA expression profiling analysis in human colorectal adenomas and adenocarcinomas. Eur. J. Cancer. 2015, 51, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, J.; Pan, J.; Geng, X.; Li, L.; Wu, J.; Song, P.; Wang, Y.; Liu, J.; Wang, L. MiR-320a inhibits gastric carcinoma by targeting activity in the FoxM1-P27KIP1 axis. Oncotarget 2016, 7, 29275–29286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, X.; Cui, H.; Zhou, Y.; Yin, D.; Feng, Y.; Xin, Q.; Xu, X.; Liu, W.; Liu, S.; Zhang, Q. miR-320a modulates cell growth and chemosensitivity via regulating ADAM10 in gastric cancer. Mol. Med. Rep. 2017, 16, 9664–9670. [Google Scholar] [CrossRef]
- Feng, L.; Rao, M.; Zhou, Y.; Zhang, Y.; Zhu, Y. Long noncoding RNA 00460 (LINC00460) promotes glioma progression by negatively regulating miR-320a. J. Cell. Biochem. 2019, 120, 9556–9563. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Yu, L.; Liu, J.; Bian, X.; Shi, C.; Sun, C.; Zhou, X.; Wen, Y.; Hua, D.; Zhao, S.; et al. miR-320a functions as a suppressor for gliomas by targeting SND1 and β-catenin, and predicts the prognosis of patients. Oncotarget 2017, 8, 19723–19737. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Huang, D.; Huang, J.; Nie, K.; Li, X.; Yang, X. lncRNA TMPO-AS1 Exerts Oncogenic Roles in HCC Through Regulating miR-320a/SERBP1 Axis. Oncotargets Ther. 2020, 13, 6539–6551. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Xiu, C.; Song, K.; Pei, R.; Miao, S.; Mao, X.; Sun, J.; Jia, S. Long non-coding RNA AFAP1-AS1/miR-320a/RBPJ axis regulates laryngeal carcinoma cell stemness and chemoresistance. J. Cell. Mol. Med. 2018, 22, 4253–4262. [Google Scholar] [CrossRef]
- Yang, X.; Lin, F.; Gao, F. Up-regulated long non-coding RNA ILF3-AS1 indicates poor prognosis of nasopharyngeal carcinoma and promoted cell metastasis. Int. J. Biol. Markers 2020, 35, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhu, X.; Ke, Y.; Xiao, D.; Liang, C.; Chen, J.; Chang, Y. LncRNA FTX inhibition restrains osteosarcoma proliferation and migration via modulating miR-320a/TXNRD1. Cancer Biol. Ther. 2020, 21, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Katsushima, K.; Shinjo, K.; Hatanaka, A.; Ohka, F.; Suzuki, S.; Naiki-Ito, A.; Soga, N.; Takahashi, S.; Kondo, Y. Histone Deacetylase Inhibition in Prostate Cancer Triggers miR-320-Mediated Suppression of the Androgen Receptor. Cancer Res. 2016, 76, 4192–4204. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.; Kim, Y.; Lee, S.; Kang, H.; Kim, D.; Park, J.; Chung, D.; Kong, H.; Yoo, K.; Kim, Y.; et al. Inhibition of Chk1 by miR-320c increases oxaliplatin responsiveness in triple-negative breast cancer. Oncogenesis 2020, 9, 91. [Google Scholar] [CrossRef]
- Xie, N.; Wang, C.; Zhuang, Z.; Hou, J.; Liu, X.; Wu, Y.; Liu, H.; Huang, H. Decreased miR-320a promotes invasion and metastasis of tumor budding cells in tongue squamous cell carcinoma. Oncotarget 2016, 7, 65744–65757. [Google Scholar] [CrossRef] [Green Version]
- Tadano, T.; Kakuta, Y.; Hamada, S.; Shimodaira, Y.; Kuroha, M.; Kawakami, Y.; Kimura, T.; Shiga, H.; Endo, K.; Masamune, A.; et al. MicroRNA-320 family is downregulated in colorectal adenoma and affects tumor proliferation by targeting CDK6. World J. Gastrointest. Oncol. 2016, 8, 532–542. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Li, W.; Zhang, M.; Zhu, S.; Zhang, D.; Wang, X. Inhibitory roles of miR-320 in osteosarcoma via regulating E2F1. J. Cancer Res. Ther. 2016, 12, 68–71. [Google Scholar]
- Lu, Y.; Wu, D.; Wang, J.; Li, Y.; Chai, X.; Kang, Q. miR-320a regulates cell proliferation and apoptosis in multiple myeloma by targeting pre-B-cell leukemia transcription factor 3. Biochem. Biophys. Res. Commun. 2016, 473, 1315–1320. [Google Scholar] [CrossRef]
- Lin, C.; Chen, C.; Lin, C.; Cheng, C.; Lee, C.; Hsieh, Y. Norcantharidin induces mitochondrial-dependent apoptosis through Mcl-1 inhibition in human prostate cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1867–1876. [Google Scholar] [CrossRef]
- Li, Y.; Tang, X.; He, Q.; Yang, X.; Ren, X.; Wen, X.; Zhang, J.; Wang, Y.; Liu, N.; Ma, J. Overexpression of Mitochondria Mediator Gene TRIAP1 by miR-320b Loss Is Associated with Progression in Nasopharyngeal Carcinoma. PLoS Genet. 2016, 12, e1006183. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Tie, Y.; Lv, G.; Zhu, J.; Fu, H.; Zheng, X. Transcriptional activation of miR-320a by ATF2, ELK1 and YY1 induces cancer cell apoptosis under ionizing radiation conditions. Int. J. Oncol. 2018, 53, 1691–1702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, W.; Zhang, H.; Guo, Z.; Yang, L.; Shao, Y.; Liu, X.; Zhao, Y.; Wang, Z.; Zhang, M.; Guo, C.; et al. Aquaporin 1 promotes sensitivity of anthracycline chemotherapy in breast cancer by inhibiting β-catenin degradation to enhance TopoIIα activity. Cell Death Differ. 2021, 28, 382–400. [Google Scholar] [CrossRef]
- Iwagami, Y.; Eguchi, H.; Nagano, H.; Akita, H.; Hama, N.; Wada, H.; Kawamoto, K.; Kobayashi, S.; Tomokuni, A.; Tomimaru, Y.; et al. miR-320c regulates gemcitabine-resistance in pancreatic cancer via SMARCC1. Br. J. Cancer 2013, 109, 502–511. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhao, H.; Yu, J.; Xu, X.; Jing, H.; Li, N.; Tang, Y.; Wang, S.; Li, Y.; Cai, J.; et al. MiR-320b/RAD21 axis affects hepatocellular carcinoma radiosensitivity to ionizing radiation treatment through DNA damage repair signaling. Cancer Sci. 2021, 112, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Lü, M.; Ding, K.; Zhang, G.; Yin, M.; Yao, G.; Tian, H.; Lian, J.; Liu, L.; Liang, M.; Zhu, T.; et al. MicroRNA-320a sensitizes tamoxifen-resistant breast cancer cells to tamoxifen by targeting ARPP-19 and ERRγ. Sci. Rep. 2015, 5, 8735. [Google Scholar] [CrossRef] [Green Version]
- He, D.; Gu, X.; Jiang, L.; Jin, J.; Ma, X. A methylation-based regulatory network for microRNA 320a in chemoresistant breast cancer. Mol. Pharmacol. 2014, 86, 536–547.9. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Li, L.; Li, Y.; Sun, H.; Zeng, C. Long noncoding RNA SNHG12 mediates doxorubicin resistance of osteosarcoma via miR-320a/MCL1 axis. Biomed. Pharmacother. 2018, 106, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Lieb, V.; Weigelt, K.; Scheinost, L.; Fischer, K.; Greither, T.; Marcou, M.; Theil, G.; Klocker, H.; Holzhausen, H.; Lai, X.; et al. Serum levels of miR-320 family members are associated with clinical parameters and diagnosis in prostate cancer patients. Oncotarget 2018, 9, 10402–10416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Z.; Tang, J.; Bai, Y.; Lin, H.; You, H.; Jin, H.; Lin, L.; You, P.; Li, J.; Dai, Z.; et al. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar]
- Ozawa, P.; Vieira, E.; Lemos, D.; Souza, I.; Zanata, S.; Pankievicz, V.; Tuleski, T.; Souza, E.; Wowk, P.; Urban, C.; et al. Identification of miRNAs Enriched in Extracellular Vesicles Derived from Serum Samples of Breast Cancer Patients. Biomolecules 2020, 10, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Wang, Y.; Sun, Y.; Miao, L.; Wang, J.; Li, Y.; Liu, H.; Liu, Q. Plasma microRNA-320, microRNA-let-7e and microRNA-21 as novel potential biomarkers for the detection of retinoblastoma. Biomed. Rep. 2014, 2, 424–428. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Liu, B.; Lin, Z.; Yao, Y.; Chen, Y.; Li, Y.; Chen, J.; Yu, D.; Tang, Z.; Wang, B.; et al. MiR-320a acts as a prognostic factor and Inhibits metastasis of salivary adenoid cystic carcinoma by targeting ITGB3. Mol. Cancer 2015, 14, 96. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.; Hu, C.; Wu, F.; Shu, L.; Pan, Y.; Liu, X.; Liu, P.; Ma, F.; Deng, C.; Huang, M. MiR-320a is associated with cisplatin resistance in lung adenocarcinoma and its clinical value in non-small cell lung cancer: A comprehensive analysis based on microarray data. Lung Cancer. 2020, 147, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Yuan, Y.; Xie, L.; Ran, P.; Xiang, X.; Huang, Q.; Qi, G.; Guo, X.; Xiao, C.; Zheng, S. miRNA-320a inhibits tumor proliferation and invasion by targeting c-Myc in human hepatocellular carcinoma. Oncotargets Ther. 2017, 10, 885–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasmyth, K.; Haering, C. Cohesin: Its roles and mechanisms. Annu. Rev. Genet. 2009, 43, 525–558. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Zhang, L.; Wan, H.; Liu, M.; Li, X.; Tang, H. CREB1-driven expression of miR-320a promotes mitophagy by down-regulating VDAC1 expression during serum starvation in cervical cancer cells. Oncotarget 2015, 6, 34924–34940. [Google Scholar] [CrossRef] [Green Version]
- Huang, A.; Zhao, H.; Quan, Y.; Jin, R.; Feng, B.; Zheng, M. E2A predicts prognosis of colorectal cancer patients and regulates cancer cell growth by targeting miR-320a. PLoS ONE 2014, 9, e85201. [Google Scholar] [CrossRef]
- Saito, Y.; Jones, P. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006, 5, 2220–2222. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zou, Y.; Dai, D. MicroRNA-320a suppresses tumor progression by targeting PBX3 in gastric cancer and is downregulated by DNA methylation. World J. Gastrointest. Oncol. 2019, 11, 842–856. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Chatterjee, A.; Das, D.; Ray, A.; Singh, R.; Chattopadhyay, E.; Sarkar, N.; Eccles, M.; Pal, M.; Maitra, A.; et al. Genome-wide miRNA methylome analysis in oral cancer: Possible biomarkers associated with patient survival. Epigenomics 2019, 11, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Alzrigat, M.; Jernberg-Wiklund, H. The miR-125a and miR-320c are potential tumor suppressor microRNAs epigenetically silenced by the polycomb repressive complex 2 in multiple myeloma. RNA Dis 2017, 4, e1529. [Google Scholar] [PubMed] [Green Version]
- Sun, Y.; Zhang, Y.; Zhang, D.; Xu, C.; Chen, S.; Zhang, J.; Ruan, Y.; Chen, F.; Zhang, R.; Qian, Y.; et al. XCI-escaping gene KDM5C contributes to ovarian development via downregulating miR-320a. Hum. Genet. 2017, 136, 227–239. [Google Scholar] [CrossRef] [PubMed]


| Tumor Types | miR-320 Family Member | Number of Tumor Types | Expression Level Compared with Normal Tissues | Ref. |
|---|---|---|---|---|
| BC | miR-320a | 19 | Down | [59] |
| BC | miR-320a | 30 | Down | [60] |
| BC | miR-320a | 36 | Down | [61] |
| Bladder cancer | miR-320a | 65 | Down | [62] |
| Bladder cancer | miR-320c | 13 | Down | [22] |
| CCA | miR-320a | 27 | Down | [63] |
| CCA | miR-320a | 39 | Down | [64] |
| Cervical cancer | miR-320a | 48 | Down | [21] |
| Colon cancer | miR-320a | 40 | Down | [65] |
| CRC | miR-320a | 62 | Down | [66] |
| CRC | miR-320b | 26 | Down | [67] |
| Gastric cancer | miR-320a | 22 | Down | [68] |
| Gastric cancer | miR-320a | 40 | Down | [69] |
| GCA | miR-320d | 60 | Down | [53] |
| Glioma | miR-320a | 42 | Down | [70] |
| Glioma | miR-320a | 120 | Down | [71] |
| HCC | miR-320a/c/d | 50 | Up | [54] |
| HCC | miR-320a | 42 | Down | [72] |
| LAC | miR-320a | 18 | Down | [23] |
| MPM | miR-320a | 14 | Up | [57] |
| NPC | miR-320a | 24 | Down | [73] |
| NPC | miR-320a | 68 | Down | [74] |
| NSCLC | miR-320a-3p | 80 | Down | [37] |
| OSC | miR-320a | 25 | Down | [75] |
| PCa | miR-320a | 51 | Up | [58] |
| PCa | miR-320a | 10 | Down | [76] |
| TNBC | miR-320c | 97 | Down | [77] |
| TSCC | miR-320a | 5 | Down | [78] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Y.; Li, S.; Tang, L. MicroRNA 320, an Anti-Oncogene Target miRNA for Cancer Therapy. Biomedicines 2021, 9, 591. https://doi.org/10.3390/biomedicines9060591
Liang Y, Li S, Tang L. MicroRNA 320, an Anti-Oncogene Target miRNA for Cancer Therapy. Biomedicines. 2021; 9(6):591. https://doi.org/10.3390/biomedicines9060591
Chicago/Turabian StyleLiang, Yuanyuan, Shun Li, and Liling Tang. 2021. "MicroRNA 320, an Anti-Oncogene Target miRNA for Cancer Therapy" Biomedicines 9, no. 6: 591. https://doi.org/10.3390/biomedicines9060591
APA StyleLiang, Y., Li, S., & Tang, L. (2021). MicroRNA 320, an Anti-Oncogene Target miRNA for Cancer Therapy. Biomedicines, 9(6), 591. https://doi.org/10.3390/biomedicines9060591






