Gender Specific Differences in Disease Susceptibility: The Role of Epigenetics
Abstract
:1. Introduction
2. Diseases with Altered Sex Ratio
2.1. Differential Response to Infections
2.2. Response to Vaccines
2.3. Gender-Specific Reactions to Xenobiotics
3. Epigenetic Processes Involved in Gender Differences
3.1. Genomic Imprinting
3.1.1. Placental-Specific Imprinting
3.1.2. Imprinting Deregulation, Brain Disorders, and Gender Differences
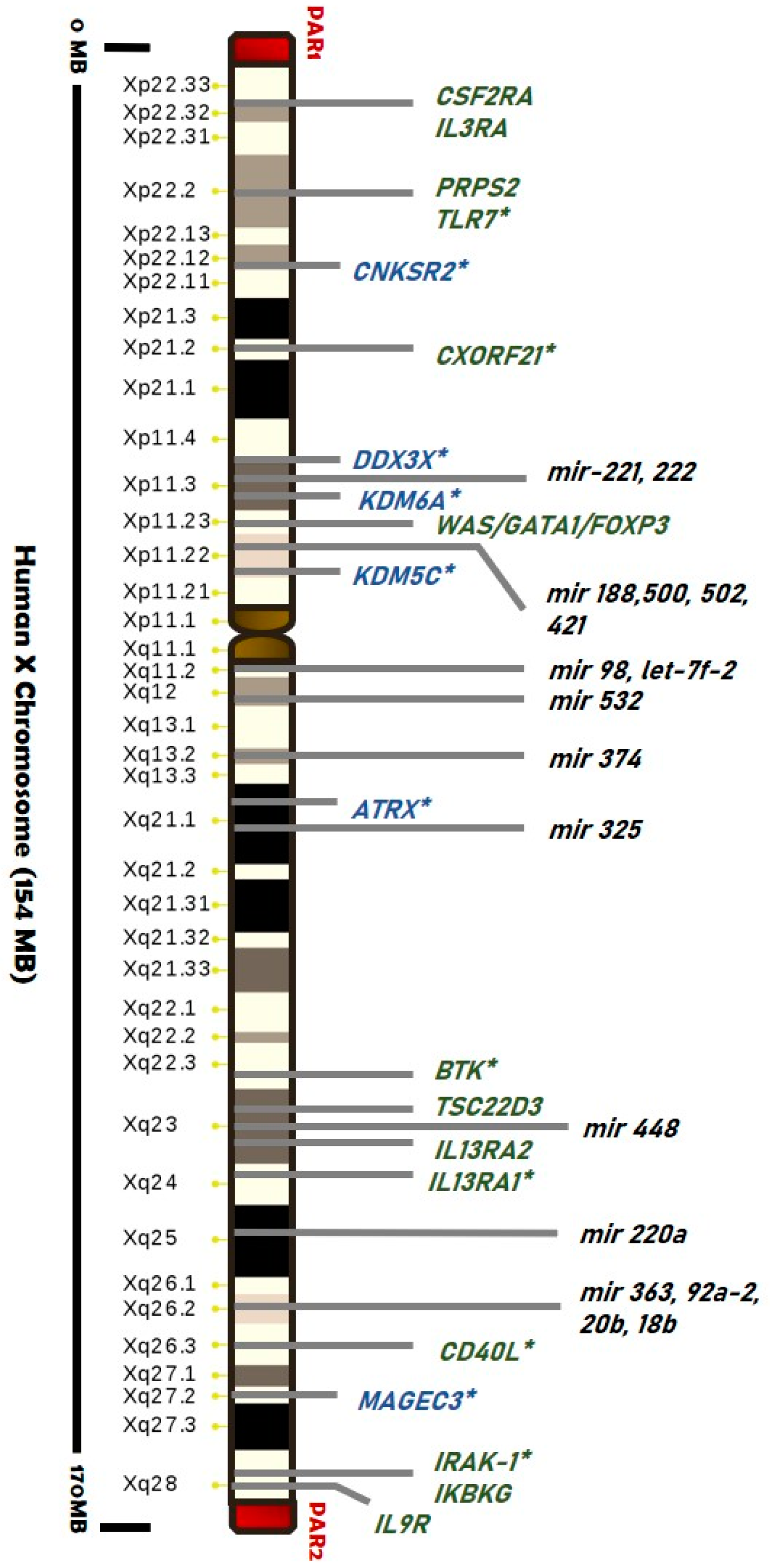
3.2. The X-Chromosome Inactivation in Females
3.2.1. Skewed XCI and Diseases
3.2.2. Is There a Role for Escape Genes in Male-Female Difference?
4. Epigenetics as Unifying Mechanism
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Oertelt-Prigione, S.; Mariman, E. The impact of sex differences on genomic research. Int. J. Biochem. Cell Biol. 2020, 124, 105774. [Google Scholar] [CrossRef]
- Jin, J.-M.; Bai, P.; He, W.; Wu, F.; Liu, X.-F.; Han, D.-M.; Liu, S.; Yang, J.-K. Gender Differences in Patients With COVID-19: Focus on Severity and Mortality. Front. Public Health 2020, 8, 152. [Google Scholar] [CrossRef]
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef]
- Barker, D.; Osmond, C. Infant Mortality, Childhood Nutrition, and Ischaemic Heart Disease in England and wales. Lancet 1986, 327, 1077–1081. [Google Scholar] [CrossRef]
- Angum, F.; Khan, T.; Kaler, J.; Siddiqui, L.; Hussain, A. The Prevalence of Autoimmune Disorders in Women: A Narrative Review. Cureus 2020, 12, e8094. [Google Scholar] [CrossRef] [PubMed]
- Ortona, E.; Pierdominici, M.; Maselli, A.; Veroni, C.; Aloisi, F.; Shoenfeld, Y. Sex-based differences in autoimmune diseases. Ann. Ist. Super. Sanita. 2016, 52, 205–212. [Google Scholar]
- Hanamsagar, R.; Bilbo, S.D. Sex differences in neurodevelopmental and neurodegenerative disorders: Focus on microglial function and neuroinflammation during development. J. Steroid Biochem. Mol. Biol. 2016, 160, 127–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinares-Garcia, P.; Stratikopoulos, M.; Zagato, A.; Loke, H.; Lee, J. Sex: A Significant Risk Factor for Neurodevelopmental and Neurodegenerative Disorders. Brain Sci. 2018, 8, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Miranda, B.R.; Fazzari, M.; Rocha, E.M.; Castro, S.; Greenamyre, J.T. Sex Differences in Rotenone Sensitivity Reflect the Male-to-Female Ratio in Human Parkinson’s Disease Incidence. Toxicol. Sci. 2019, 170, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Shao, X.; Wang, X.; Liu, L.; Liang, H. Sex disparities in cancer. Cancer Lett. 2019, 466, 35–38. [Google Scholar] [CrossRef]
- Yan, H.-X.; Pang, P.; Wang, F.-L.; Tian, W.; Luo, Y.-K.; Huang, W.; Yang, G.-Q.; Jin, N.; Zang, L.; Du, J.; et al. Dynamic profile of differentiated thyroid cancer in male and female patients with thyroidectomy during 2000–2013 in China: A retrospective study. Sci. Rep. 2017, 7, 15832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altamimi, A.; Abu-Saris, R.; El-Metwally, A.; Alaifan, T.; Alamri, A. Demographic Variations of MERS-CoV Infection among Suspected and Confirmed Cases: An Epidemiological Analysis of Laboratory-Based Data from Riyadh Regional Laboratory. BioMed Res. Int. 2020, 2020, 9629747. [Google Scholar] [CrossRef] [PubMed]
- Bunders, M.J.; Altfeld, M. Implications of Sex Differences in Immunity for SARS-CoV-2 Pathogenesis and Design of Therapeutic Interventions. Immunity 2020, 53, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Baig, S. Gender disparity in infections of Hepatitis B virus. J. Coll. Physicians Surg. Pak. 2009, 19, 598–600. [Google Scholar]
- Chamekh, M.; Deny, M.; Romano, M.; Lefèvre, N.; Corazza, F.; Duchateau, J.; Casimir, G. Differential Susceptibility to Infectious Respiratory Diseases between Males and Females Linked to Sex-Specific Innate Immune Inflammatory Response. Front. Immunol. 2017, 8, 1806. [Google Scholar] [CrossRef]
- Billi, A.C.; Kahlenberg, J.M.; Gudjonsson, J.E. Sex bias in autoimmunity. Curr. Opin. Rheumatol. 2019, 31, 53–61. [Google Scholar] [CrossRef]
- Laffont, S.; Guéry, J.-C. Deconstructing the sex bias in allergy and autoimmunity: From sex hormones and beyond. Adv. Immunol. 2019, 142, 35–64. [Google Scholar] [CrossRef]
- Costa, A.R.; De Oliveira, M.L.; Cruz, I.; Gonçalves, I.; Cascalheira, J.F.; Santos, C.R.A. The Sex Bias of Cancer. Trends Endocrinol. Metab. 2020, 31, 785–799. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [Green Version]
- Mauvais-Jarvis, F.; Merz, N.B.; Barnes, P.J.; Brinton, R.D.; Carrero, J.-J.; DeMeo, D.L.; De Vries, G.J.; Epperson, C.N.; Govindan, R.; Klein, S.L.; et al. Sex and gender: Modifiers of health, disease, and medicine. Lancet 2020, 396, 565–582. [Google Scholar] [CrossRef]
- Loke, H.; Harley, V.; Lee, J. Biological factors underlying sex differences in neurological disorders. Int. J. Biochem. Cell Biol. 2015, 65, 139–150. [Google Scholar] [CrossRef]
- Thomason, M.E. Development of Brain Networks In Utero: Relevance for Common Neural Disorders. Biol. Psychiatry 2020, 88, 40–50. [Google Scholar] [CrossRef]
- Van Den Eeden, S.K.; Tanner, C.M.; Bernstein, A.L.; Fross, R.D.; Leimpeter, A.; Bloch, D.A.; Nelson, L.M. Incidence of Parkinson’s Disease: Variation by Age, Gender, and Race/Ethnicity. Am. J. Epidemiol. 2003, 157, 1015–1022. [Google Scholar] [CrossRef]
- Ullah, M.F.; Ahmad, A.; Bhat, S.H.; Abu-Duhier, F.M.; Barreto, G.E.; Ashraf, G.M. Impact of sex differences and gender specificity on behavioral characteristics and pathophysiology of neurodegenerative disorders. Neurosci. Biobehav. Rev. 2019, 102, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tang, L.; Wu, Y.; Fan, C.; Zhang, S.; Xiang, B.; Zhou, M.; Li, X.; Li, Y.; Li, G.; et al. Abnormal X chromosome inactivation and tumor development. Cell. Mol. Life Sci. 2020, 77, 2949–2958. [Google Scholar] [CrossRef] [PubMed]
- Dunford, A.; Weinstock, D.M.; Savova, V.; Schumacher, S.E.; Cleary, J.P.; Yoda, A.; Sullivan, T.J.; Hess, J.M.; Gimelbrant, A.A.A.; Beroukhim, R.; et al. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet. 2017, 49, 10–16. [Google Scholar] [CrossRef] [Green Version]
- Lotze, M.; Domin, M.; Gerlach, F.H.; Gaser, C.; Lueders, E.; Schmidt, C.O.; Neumann, N. Novel findings from 2,838 Adult Brains on Sex Differences in Gray Matter Brain Volume. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef]
- Balaton, B.P.; Dixon-McDougall, T.; Peeters, S.B.; Brown, C.J. The eXceptional nature of the X chromosome. Hum. Mol. Genet. 2018, 27, R242–R249. [Google Scholar] [CrossRef] [Green Version]
- Honarpisheh, P.; McCullough, L.D. Sex as a biological variable in the pathology and pharmacology of neurodegenerative and neurovascular diseases. Br. J. Pharmacol. 2019, 176, 4173–4192. [Google Scholar] [CrossRef]
- Dearden, L.; Bouret, S.G.; Ozanne, S.E. Sex and gender differences in developmental programming of metabolism. Mol. Metab. 2018, 15, 8–19. [Google Scholar] [CrossRef]
- Scofield, R.H.; Bruner, G.R.; Namjou, B.; Kimberly, R.P.; Ramsey-Goldman, R.; Petri, M.; Reveille, J.D.; Alarcón, G.S.; Vilá, L.M.; Reid, J.; et al. Klinefelter’s syndrome (47,XXY) in male systemic lupus erythematosus patients: Support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008, 58, 2511–2517. [Google Scholar] [CrossRef]
- Cooney, C.M.; Bruner, G.R.; Aberle, T.; Namjou-Khales, B.; Myers, L.K.; Feo, L.; Li, S.; D’Souza, A.; Ramirez, A.; Harley, J.B.; et al. 46,X,del(X)(q13) Turner’s syndrome women with systemic lupus erythematosus in a pedigree multiplex for SLE. Genes Immun. 2009, 10, 478–481. [Google Scholar] [CrossRef] [Green Version]
- Harris, V.M.; Sharma, R.; Cavett, J.; Kurien, B.T.; Liu, K.; Koelsch, K.A.; Rasmussen, A.; Radfar, L.; Lewis, D.; Stone, D.U.; et al. Klinefelter’s syndrome (47,XXY) is in excess among men with Sjögren’s syndrome. Clin. Immunol. 2016, 168, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naftolin, F.; Friedenthal, J.; Nachtigall, R.; Nachtigall, L. Cardiovascular health and the menopausal woman: The role of estrogen and when to begin and end hormone treatment. F1000Research 2019, 8, 1576. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xu, A.; Jia, S.; Huang, J. Recent advances in the molecular mechanism of sex disparity in hepatocellular carcinoma. Oncol. Lett. 2019, 17, 4222–4228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frye, C.A.; Koonce, C.J.; Edinger, K.L.; Osborne, D.M.; Walf, A.A. Androgens with activity at estrogen receptor beta have anxiolytic and cognitive-enhancing effects in male rats and mice. Horm. Behav. 2008, 54, 726–734. [Google Scholar] [CrossRef] [Green Version]
- McDermott, C.M.; Liu, D.; Schrader, L.A. Role of gonadal hormones in anxiety and fear memory formation and inhibition in male mice. Physiol. Behav. 2012, 105, 1168–1174. [Google Scholar] [CrossRef] [PubMed]
- Leckman, J.F.; Scahill, L. Possible Exacerbation of Tics by Androgenic Steroids. N. Engl. J. Med. 1990, 322, 1674. [Google Scholar] [CrossRef] [PubMed]
- Gillies, G.E.; Pienaar, I.S.; Vohra, S.; Qamhawi, Z. Sex differences in Parkinson’s disease. Front. Neuroendocr. 2014, 35, 370–384. [Google Scholar] [CrossRef] [Green Version]
- Flanagan, K.L.; Fink, A.L.; Plebanski, M.; Klein, S.L. Sex and Gender Differences in the Outcomes of Vaccination over the Life Course. Annu. Rev. Cell Dev. Biol. 2017, 33, 577–599. [Google Scholar] [CrossRef]
- Vom Steeg, L.; Klein, S.L. SeXX Matters in Infectious Disease Pathogenesis. PLoS Pathog. 2016, 12, e1005374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giefing-Kröll, C.; Berger, P.; Lepperdinger, G.; Grubeck-Loebenstein, B. How sex and age affect immune responses, susceptibility to infections, and response to vaccination. Aging Cell 2015, 14, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Klein, R.S. Sex Drives Dimorphic Immune Responses to Viral Infections. J. Immunol. 2017, 198, 1782–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef]
- Grindstaff, J.L.; Brodie, E.D.; Ketterson, E.D. Immune function across generations: Integrating mechanism and evolutionary process in maternal antibody transmission. Proc. Biol. Sci. 2003, 270, 2309–2319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fink, A.L.; Klein, S.L. The evolution of greater humoral immunity in females than males: Implications for vaccine efficacy. Curr. Opin. Physiol. 2018, 6, 16–20. [Google Scholar] [CrossRef]
- Libert, C.; Dejager, L.; Pinheiro, I. The X chromosome in immune functions: When a chromosome makes the difference. Nat. Rev. Immunol. 2010, 10, 594–604. [Google Scholar] [CrossRef]
- Jaillon, S.; Berthenet, K.; Garlanda, C. Sexual Dimorphism in Innate Immunity. Clin. Rev. Allergy Immunol. 2019, 56, 308–321. [Google Scholar] [CrossRef]
- Schmiedel, B.J.; Singh, D.; Madrigal, A.; Valdovino-Gonzalez, A.G.; White, B.M.; Zapardiel-Gonzalo, J.; Ha, B.; Altay, G.; Greenbaum, J.A.; McVicker, G.; et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell 2018, 175, 1701–1715.e16. [Google Scholar] [CrossRef] [Green Version]
- Klein, S.L.; Hodgson, A.; Robinson, D.P. Mechanisms of sex disparities in influenza pathogenesis. J. Leukoc. Biol. 2012, 92, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Kloc, M.; Ghobrial, R.M.; Kubiak, J.Z. The Role of Genetic Sex and Mitochondria in Response to COVID-19 Infection. Int. Arch. Allergy Immunol. 2020, 181, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Fett, C.; Mack, M.; Eyck, P.P.T.; Meyerholz, D.; Perlman, S. Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection. J. Immunol. 2017, 198, 4046–4053. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.; Groot, R.J.D.; Drosten, C.; Gulyaeva, A.A.; Penzar, D. Severe acute respiratory syndrome-related coronavirus: The species and its viruses–A statement of the Coronavirus Study Group. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, C.; Regitz-Zagrosek, V.; Neuhauser, H.K.; Morgan, R.; Klein, S.L. Impact of sex and gender on COVID-19 outcomes in Europe. Biol. Sex. Differ. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Acheampong, D.O.; Barffour, I.K.; Boye, A.; Aninagyei, E.; Ocansey, S.; Morna, M.T. Male predisposition to severe COVID-19: Review of evidence and potential therapeutic prospects. Biomed. Pharmacother. 2020, 131, 110748. [Google Scholar] [CrossRef] [PubMed]
- Scully, E.P.; Haverfield, J.; Ursin, R.L.; Tannenbaum, C.; Klein, S.L. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020, 20, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Gargaglioni, L.H.; Marques, D.A. Let’s talk about sex in the context of COVID-19. J. Appl. Physiol. 2020, 128, 1533–1538. [Google Scholar] [CrossRef] [PubMed]
- Márquez, E.J.; Trowbridge, J.; Kuchel, G.A.; Banchereau, J.; Ucar, D. The lethal sex gap: COVID-19. Immun. Ageing 2020, 17, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Al-Lami, R.A.; Urban, R.J.; Volpi, E.; Algburi, A.M.; Baillargeon, J. Sex Hormones and Novel Corona Virus Infectious Disease (COVID-19). Mayo Clin. Proc. 2020, 95, 1710–1714. [Google Scholar] [CrossRef]
- Bwire, G.M. Coronavirus: Why Men are More Vulnerable to Covid-19 Than Women? SN Compr. Clin. Med. 2020, 2, 874–876. [Google Scholar] [CrossRef]
- Klein, S.L.; Jedlicka, A.; Pekosz, A. The Xs and Y of immune responses to viral vaccines. Lancet Infect. Dis. 2010, 10, 338–349. [Google Scholar] [CrossRef]
- Fish, E.N. The X-files in immunity: Sex-based differences predispose immune responses. Nat. Rev. Immunol. 2008, 8, 737–744. [Google Scholar] [CrossRef]
- Klein, S.L.; Pekosz, A. Sex-based Biology and the Rational Design of Influenza Vaccination Strategies. J. Infect. Dis. 2014, 209, S114–S119. [Google Scholar] [CrossRef] [Green Version]
- Burger, J.; Fossi, C.; McClellan-Green, P.; Orlando, E.F. Methodologies, bioindicators, and biomarkers for assessing gender-related differences in wildlife exposed to environmental chemicals. Environ. Res. 2007, 104, 135–152. [Google Scholar] [CrossRef] [PubMed]
- Díez-Villanueva, P.; Vicent, L.; Alfonso, F. Gender disparities in treatment response in octogenarians with acute coronary syndrome. J. Thorac. Dis. 2020, 12, 1277–1279. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Singh, V.; Sobolewski, M.; Cory-Slechta, D.A.; Schneider, J.S. Sex-Dependent Effects of Developmental Lead Exposure on the Brain. Front. Genet. 2018, 9, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranzani, O.T.; Milà, C.; Sanchez, M.; Bhogadi, S.; Kulkarni, B.; Balakrishnan, K.; Sambandam, S.; Sunyer, J.; Marshall, J.D.; Kinra, S.; et al. Personal exposure to particulate air pollution and vascular damage in peri-urban South India. Environ. Int. 2020, 139, 105734. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.B.-Z.; Paré, P.D.; Man, S.F.P.; Sin, D.D. The Growing Burden of Chronic Obstructive Pulmonary Disease and Lung Cancer in Women: Examining Sex Differences in Cigarette Smoke Metabolism. Am. J. Respir. Crit. Care Med. 2007, 176, 113–120. [Google Scholar] [CrossRef]
- Jee, H.J.; Lee, S.G.; Bormate, K.J.; Jung, Y.-S. Effect of Caffeine Consumption on the Risk for Neurological and Psychiatric Disorders: Sex Differences in Human. Nutrients 2020, 12, 3080. [Google Scholar] [CrossRef]
- Kautzky-Willer, A.; Kosi, L.; Lin, J.; Mihaljevic, R. Gender-based differences in glycaemic control and hypoglycaemia prevalence in patients with type 2 diabetes: Results from patient-level pooled data of six randomized controlled trials. Diabetes Obes. Metab. 2015, 17, 533–540. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Obeidat, M.; Zhou, G.; Leung, J.M.; Tashkin, D.; Wise, R.; Connett, J.; Joubert, P.; Bossé, Y.; van den Berge, M.; et al. Responsiveness to Ipratropium Bromide in Male and Female Patients with Mild to Moderate Chronic Obstructive Pulmonary Disease. EBioMedicine 2017, 19, 139–145. [Google Scholar] [CrossRef] [Green Version]
- Carè, A.; Bellenghi, M.; Matarrese, P.; Gabriele, L.; Salvioli, S.; Malorni, W. Sex disparity in cancer: Roles of microRNAs and related functional players. Cell Death Differ. 2018, 25, 477–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaidonis, G.; Rao, A.N.; Ouyang, Y.-B.; Stary, C.M. Elucidating sex differences in response to cerebral ischemia: Immunoregulatory mechanisms and the role of microRNAs. Prog. Neurobiol. 2019, 176, 73–85. [Google Scholar] [CrossRef]
- Carcel, C.; Woodward, M.; Wang, X.; Bushnell, C.; Sandset, E.C. Sex matters in stroke: A review of recent evidence on the differences between women and men. Front. Neuroendocr. 2020, 59, 100870. [Google Scholar] [CrossRef] [PubMed]
- Gazerani, P.; Cairns, B.E. Sex-Specific Pharmacotherapy for Migraine: A Narrative Review. Front. Neurosci. 2020, 14, 222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frega, S.; Dal Maso, A.; Ferro, A.; Bonanno, L.; Conte, P.; Pasello, G. Heterogeneous tumor features and treatment outcome between males and females with lung cancer (LC): Do gender and sex matter? Crit. Rev. Oncol. 2019, 138, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Kalisch-Smith, J.I.; Simmons, D.G.; Dickinson, H.; Moritz, K.M. Review: Sexual dimorphism in the formation, function and adaptation of the placenta. Placenta 2017, 54, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Bronson, S.L.; Bale, T.L. Prenatal Stress-Induced Increases in Placental Inflammation and Offspring Hyperactivity Are Male-Specific and Ameliorated by Maternal Antiinflammatory Treatment. Endocrinology 2014, 155, 2635–2646. [Google Scholar] [CrossRef] [Green Version]
- Weinstock, M. Sex-dependent changes induced by prenatal stress in cortical and hippocampal morphology and behaviour in rats: An update. Stress 2011, 14, 604–613. [Google Scholar] [CrossRef]
- Palanza, P.; Paterlini, S.; Brambilla, M.M.; Ramundo, G.; Caviola, G.; Gioiosa, L.; Parmigiani, S.; Vom Saal, F.S.; Ponzi, D. Sex-biased impact of endocrine disrupting chemicals on behavioral development and vulnerability to disease: Of mice and children. Neurosci. Biobehav. Rev. 2021, 121, 29–46. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, L.; Li, L.-X.; Xie, C.-M.; Li, D.; Shi, H.-J.; Zhang, Y.-H. Gender-specific relationship between prenatal exposure to phthalates and intrauterine growth restriction. Pediatr. Res. 2014, 76, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Tucci, V.; Isles, A.R.; Kelsey, G.; Ferguson-Smith, A.C.; Bartolomei, M.S.; Benvenisty, N.; Bourc’his, D.; Charalambous, M.; Dulac, C.; Feil, R.; et al. Genomic Imprinting and Physiological Processes in Mammals. Cell 2019, 176, 952–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassidy, F.C.; Charalambous, M. Genomic imprinting, growth and maternal–fetal interactions. J. Exp. Biol. 2018, 221, jeb164517. [Google Scholar] [CrossRef] [Green Version]
- Ishida, M.; Moore, G.E. The role of imprinted genes in humans. Mol. Asp. Med. 2013, 34, 826–840. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.I.; Ye, B.; Staples, J.; Marcketta, A.; Gao, C.; Center, R.G.; Collaboration, G.R.D.; Shuldiner, A.R.; Hout, C.V.V. Ge-nome-Wide Survey of Parent-of-Origin Specific Associations across Clinical Traits Derived from Electronic Health Records. medRxiv 2020. [Google Scholar] [CrossRef]
- Renfree, M.B.; Suzuki, S.; Kaneko-Ishino, T. The origin and evolution of genomic imprinting and viviparity in mammals. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frost, J.M.; Moore, G.E. The Importance of Imprinting in the Human Placenta. PLoS Genet. 2010, 6, e1001015. [Google Scholar] [CrossRef] [Green Version]
- Court, F.; Tayama, C.; Romanelli, V.; Martin-Trujillo, A.; Iglesias-Platas, I.; Okamura, K.; Sugahara, N.; Simón, C.; Moore, H.; Harness, J.V.; et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014, 24, 554–569. [Google Scholar] [CrossRef] [Green Version]
- Hanna, C.W.; Peñaherrera, M.S.; Saadeh, H.; Andrews, S.; McFadden, D.E.; Kelsey, G.; Robinson, W.P. Pervasive polymorphic imprinted methylation in the human placenta. Genome Res. 2016, 26, 756–767. [Google Scholar] [CrossRef]
- Sanchez-Delgado, M.; Court, F.; Vidal, E.; Medrano, J.; Monteagudo-Sánchez, A.; Martin-Trujillo, A.; Tayama, C.; Iglesias-Platas, I.; Kondova, I.; Bontrop, R.; et al. Human Oocyte-Derived Methylation Differences Persist in the Placenta Revealing Widespread Transient Imprinting. PLoS Genet. 2016, 12, e1006427. [Google Scholar] [CrossRef]
- Thamban, T.; Agarwaal, V.; Khosla, S. Role of genomic imprinting in mammalian development. J. Biosci. 2020, 45, 1–21. [Google Scholar] [CrossRef]
- Lambertini, L.; Marsit, C.J.; Sharma, P.; Maccani, M.; Ma, Y.; Hu, J.; Chen, J. Imprinted gene expression in fetal growth and development. Placenta 2012, 33, 480–486. [Google Scholar] [CrossRef] [Green Version]
- Kappil, M.A.; Green, B.B.; Armstrong, D.A.; Sharp, A.J.; Lambertini, L.; Marsit, C.J.; Chen, J. Placental expression profile of imprinted genes impacts birth weight. Epigenetics 2015, 10, 842–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deyssenroth, M.A.; Marsit, C.J.; Chen, J.; Lambertini, L. In-depth characterization of the placental imprintome reveals novel differentially methylated regions across birth weight categories. Epigenetics 2020, 15, 47–60. [Google Scholar] [CrossRef]
- Green, B.B.; Kappil, M.; Lambertini, L.; Armstrong, D.A.; Guerin, D.J.; Sharp, A.J.; Lester, B.M.; Chen, J.; Marsit, C.J. Expression of imprinted genes in placenta is associated with infant neurobehavioral development. Epigenetics 2015, 10, 834–841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kingsley, S.L.; Deyssenroth, M.A.; Kelsey, K.T.; Awad, Y.A.; Kloog, I.; Schwartz, J.D.; Lambertini, L.; Chen, J.; Marsit, C.J.; Wellenius, G.A. Maternal residential air pollution and placental imprinted gene expression. Environ. Int. 2017, 108, 204–211. [Google Scholar] [CrossRef]
- Carter, R.C.; Chen, J.; Li, Q.; Deyssenroth, M.; Dodge, N.C.; Wainwright, H.C.; Molteno, C.D.; Meintjes, E.M.; Jacobson, J.L.; Jacobson, S.W. Alcohol-Related Alterations in Placental Imprinted Gene Expression in Humans Mediate Effects of Prenatal Alcohol Exposure on Postnatal Growth. Alcohol. Clin. Exp. Res. 2018, 42, 1431–1443. [Google Scholar] [CrossRef]
- Strakovsky, R.S.; Schantz, S.L. Impacts of bisphenol A (BPA) and phthalate exposures on epigenetic outcomes in the human placenta. Environ. Epigenetics 2018, 4, dvy022. [Google Scholar] [CrossRef] [Green Version]
- Al-Qaraghouli, M.; Fang, Y.M.V. Effect of Fetal Sex on Maternal and Obstetric Outcomes. Front. Pediatr. 2017, 5, 144. [Google Scholar] [CrossRef] [Green Version]
- Bale, T.L. The placenta and neurodevelopment: Sex differences in prenatal vulnerability. Dialogues Clin. Neurosci. 2016, 18, 459. [Google Scholar]
- Adam, I.; Salih, M.M.; Mohmmed, A.A.; Rayis, D.A.; Elbashir, M.I. Pregnant women carrying female fetuses are at higher risk of placental malaria infection. PLoS ONE 2017, 12, e0182394. [Google Scholar] [CrossRef]
- Carpenter, T.; Grecian, S.M.; Reynolds, R.M. Sex differences in early-life programming of the hypothalamic–pituitary–adrenal axis in humans suggest increased vulnerability in females: A systematic review. J. Dev. Orig. Health Dis. 2017, 8, 244–255. [Google Scholar] [CrossRef] [Green Version]
- Hebert, J.F.; Myatt, L. Placental mitochondrial dysfunction with metabolic diseases: Therapeutic approaches. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2021, 1867, 165967. [Google Scholar] [CrossRef] [PubMed]
- Nugent, B.M.; McCarthy, M.M. Epigenetic influences on the developing brain: Effects of hormones and nutrition. Adv. Genom. Genet. 2015, 5, 215–225. [Google Scholar] [CrossRef] [Green Version]
- Ruigrok, A.N.V.; Salimi-Khorshidi, G.; Lai, M.-C.; Baron-Cohen, S.; Lombardo, M.V.; Tait, R.J.; Suckling, J. A meta-analysis of sex differences in human brain structure. Neurosci. Biobehav. Rev. 2014, 39, 34–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawson, H.A.; Cheverud, J.M.; Wolf, J.B. Genomic imprinting and parent-of-origin effects on complex traits. Nat. Rev. Genet. 2013, 14, 609–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshney, M.; Nalvarte, I. Genes, Gender, Environment, and Novel Functions of Estrogen Receptor Beta in the Susceptibility to Neurodevelopmental Disorders. Brain Sci. 2017, 7, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez, J.D.; Rubinstein, N.D.; Dulac, C. New Perspectives on Genomic Imprinting, an Essential and Multifaceted Mode of Epigenetic Control in the Developing and Adult Brain. Annu. Rev. Neurosci. 2016, 39, 347–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badcock, C.; Crespi, B. Battle of the sexes may set the brain. Nature 2008, 454, 1054–1055. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, L.S.; Davies, W.; Isles, A.R. Genomic imprinting effects on brain development and function. Nat. Rev. Neurosci. 2007, 8, 832–843. [Google Scholar] [CrossRef]
- Gregg, C.; Zhang, J.; Butler, J.E.; Haig, D.; Dulac, C. Sex-Specific Parent-of-Origin Allelic Expression in the Mouse Brain. Science 2010, 329, 682–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schurz, H.; Salie, M.; Tromp, G.; Hoal, E.G.; Kinnear, C.J.; Möller, M. The X chromosome and sex-specific effects in infectious disease susceptibility. Hum. Genom. 2019, 13, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Veyver, I.B. Skewed X Inactivation in X-Linked Disorders. Semin. Reprod. Med. 2001, 19, 183–192. [Google Scholar] [CrossRef]
- Tukiainen, T.; Villani, A.-C.; Yen, A.; Rivas, M.A.; Marshall, J.L.; Satija, R.; Aguirre, M.; Gauthier, L.; Fleharty, M.; Kirby, A.; et al. Landscape of X chromosome inactivation across human tissues. Nature 2017, 550, 244–248. [Google Scholar] [CrossRef] [Green Version]
- Ciccodicola, A.; D’Esposito, M.; Esposito, T.; Gianfrancesco, F.; Migliaccio, C.; Miano, M.G.; Matarazzo, M.R.; Vacca, M.; Franzè, A.; Cuccurese, M.; et al. Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum. Mol. Genet. 2000, 9, 395–401. [Google Scholar] [CrossRef] [Green Version]
- Balaton, B.P.; Cotton, A.M.; Brown, C.J. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol. Sex. Differ. 2015, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Posynick, B.J.; Brown, C.J. Escape From X-Chromosome Inactivation: An Evolutionary Perspective. Front. Cell Dev. Biol. 2019, 7, 241. [Google Scholar] [CrossRef]
- Pinheiro, I.; Dejager, L.; Libert, C. X-chromosome-located microRNAs in immunity: Might they explain male/female differences? The X Chromosome-Genomic Context May Affect X-Located MiRNAs and Downstream Signaling, Thereby Contributing to the Enhanced Immune Response of Females. Bioessays 2011, 33, 791–802. [Google Scholar] [CrossRef]
- Youness, A.; Miquel, C.-H.; Guéry, J.-C. Escape from X Chromosome Inactivation and the Female Predominance in Autoimmune Diseases. Int. J. Mol. Sci. 2021, 22, 1114. [Google Scholar] [CrossRef]
- Balaton, B.P.; Brown, C.J. Escape Artists of the X Chromosome. Trends Genet. 2016, 32, 348–359. [Google Scholar] [CrossRef]
- Shvetsova, E.; Sofronova, A.; Monajemi, R.; Gagalova, K.; Draisma, H.H.; White, S.J.; Santen, G.W.E.; Chuva de Sousa Lopes, S.M.C.; Heijmans, B.T.; van Meurs, J.; et al. Skewed X-inactivation is common in the general female population. Eur. J. Hum. Genet. 2019, 27, 455–465. [Google Scholar] [CrossRef] [Green Version]
- Zito, A.; Davies, M.N.; Tsai, P.-C.; Roberts, S.; Andres-Ejarque, R.; Nardone, S.; Bell, J.T.; Wong, C.C.Y.; Small, K.S. Heritability of skewed X-inactivation in female twins is tissue-specific and associated with age. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Mengel-From, J.; Lindahl-Jacobsen, R.; Nygaard, M.; Soerensen, M.; Ørstavik, K.H.; Hertz, J.M.; Andersen-Ranberg, K.; Tan, Q.; Christensen, K. Skewness of X-chromosome inactivation increases with age and varies across birth cohorts in elderly Danish women. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Ørstavik, K.H. X chromosome inactivation in clinical practice. Hum. Genet. 2009, 126, 363–373. [Google Scholar] [CrossRef]
- Ishido, N.; Inoue, N.; Watanabe, M.; Hidaka, Y.; Iwatani, Y. The Relationship between Skewed X Chromosome Inactivation and the Prognosis of Graves’ and Hashimoto’s Diseases. Thyroid 2015, 25, 256–261. [Google Scholar] [CrossRef] [Green Version]
- Chabchoub, G.; Uz, E.; Maalej, A.; Mustafa, C.A.; Rebai, A.; Mnif, M.; Bahloul, Z.; Farid, N.R.; Ozcelik, T.; Ayadi, H. Analysis of skewed X-chromosome inactivation in females with rheumatoid arthritis and autoimmune thyroid diseases. Arthritis Res. Ther. 2009, 11, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santiwatana, S.; Mahachoklertwattana, P.; Limwongse, C.; Khlairit, P.; Pongratanakul, S.; Roothumnong, E.; Prangphan, K.; Choubtum, L.; Songdej, D.; Poomthavorn, P. Skewed X chromosome inactivation in girls and female adolescents with autoimmune thyroid disease. Clin. Endocrinol. 2018, 89, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, M.J.; Kavvoura, F.K.; Brand, O.J.; Newby, P.R.; Jackson, L.E.; Hargreaves, C.E.; Franklyn, J.A.; Gough, S.C.L. Skewed X Chromosome Inactivation and Female Preponderance in Autoimmune Thyroid Disease: An Association Study and Meta-Analysis. J. Clin. Endocrinol. Metab. 2014, 99, E127–E131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Li, X.; Gibson, A.; Edberg, J.; Kimberly, R.P.; Absher, D.M. Skewed allelic expression on X chromosome associated with aberrant expression of XIST on systemic lupus erythematosus lymphocytes. Hum. Mol. Genet. 2020, 29, 2523–2534. [Google Scholar] [CrossRef]
- Özbalkan, Z.; Baǧışlar, S.; Kiraz, S.; Akyerli, C.B.; Özer, H.T.E.; Yavuz, Ş.; Birlik, A.M.; Çalgüneri, M.; Özçelik, T. Skewed X chromosome inactivation in blood cells of women with scleroderma. Arthritis Rheum. 2005, 52, 1564–1570. [Google Scholar] [CrossRef]
- Kanaan, S.B.; Onat, O.E.; Balandraud, N.; Martin, G.V.; Nelson, J.L.; Azzouz, D.F.; Auger, I.; Arnoux, F.; Martin, M.; Roudier, J.; et al. Evaluation of X Chromosome Inactivation with Respect to HLA Genetic Susceptibility in Rheumatoid Arthritis and Systemic Sclerosis. PLoS ONE 2016, 11, e0158550. [Google Scholar] [CrossRef]
- Pessach, I.M.; Notarangelo, L.D. X-linked primary immunodeficiencies as a bridge to better understandingX-chromosome related autoimmunity. J. Autoimmun. 2009, 33, 17–24. [Google Scholar] [CrossRef]
- Bajic, V.; Mandusic, V.; Stefanova, E.; Bozovic, A.; Davidović, R.; Zivkovic, L.; Cabarkapa, A.; Spremo-Potparevic, B. Skewed X-Chromosome Inactivation in Women Affected by Alzheimer’s Disease. J. Alzheimer’s Dis. 2015, 43, 1251–1259. [Google Scholar] [CrossRef]
- Taherian, M.; Maghsoudi, H.; Bidaki, K.; Taherian, R. The Relationship BetweenSkewed X-Chromosome Inactivation and Neurological Disorders Development: A Review. Int. Clin. Neurosci. J. 2016, 3, 81–91. [Google Scholar] [CrossRef]
- Bajic, V.P.; Essack, M.; Zivkovic, L.; Stewart, A.; Zafirovic, S.; Bajic, V.B.; Gojobori, T.; Isenovic, E.; Spremo-Potparevic, B. The X Files: “The Mystery of X Chromosome Instability in Alzheimer’s Disease”. Front. Genet. 2020, 10, 1368. [Google Scholar] [CrossRef]
- Villard, L.; Levy, N.; Xiang, F.; Kpebe, A.; Labelle, V.; Chevillard, C.; Zhang, Z.; Schwartz, C.E.; Tardieu, M.; Chelly, J.; et al. Segregation of a totally skewed pattern of X chromosome inactivation in four familial cases of Rett syndrome without MECP2 mutation: Implications for the disease. J. Med. Genet. 2001, 38, 435–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fieremans, N.; Van Esch, H.; Holvoet, M.; Van Goethem, G.; Devriendt, K.; Roselló, M.; Mayo, S.; Martinez, F.; Jhangiani, S.; Muzny, D.M.D.M.; et al. Identification of Intellectual Disability Genes in Female Patients with a Skewed X-Inactivation Pattern. Hum. Mutat. 2016, 37, 804–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talebizadeh, Z.; Bittel, D.C.; Veatch, O.J.; Kibiryeva, N.; Butler, M.G. Brief Report: Non-Random X Chromosome Inactivation in Females with Autism. J. Autism Dev. Disord. 2005, 35, 675–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Li, Y.; Ma, L.; Zhang, G.; Liu, M.; Wang, C.; Zheng, Y.; Li, R. A new sex-specific underlying mechanism for female schizophrenia: Accelerated skewed X chromosome inactivation. Biol. Sex. Differ. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Medema, R.H.; Burgering, B.M.T. The X Factor: Skewing X Inactivation towards Cancer. Cell 2007, 129, 1253–1254. [Google Scholar] [CrossRef] [Green Version]
- Carrel, L.; Willard, H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 2005, 434, 400–404. [Google Scholar] [CrossRef]
- Mousavi, M.J.; Mahmoudi, M.; Ghotloo, S. Escape from X chromosome inactivation and female bias of autoimmune diseases. Mol. Med. 2020, 26, 1–20. [Google Scholar] [CrossRef]
- Katsir, K.W.; Linial, M. Human genes escaping X-inactivation revealed by single cell expression data. BMC Genom. 2019, 20, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Berletch, J.B.; Yang, F.; Xu, J.; Carrel, L.; Disteche, C.M. Genes that escape from X inactivation. Hum. Genet. 2011, 130, 237–245. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Kain, M.; Wang, L. Inactivation of X-linked tumor suppressor genes in human cancer. Future Oncol. 2012, 8, 463–481. [Google Scholar] [CrossRef] [Green Version]
- Wijchers, P.J.; Yandim, C.; Panousopoulou, E.; Ahmad, M.; Harker, N.; Saveliev, A.; Burgoyne, P.S.; Festenstein, R. Sexual dimorphism in mammalian autosomal gene regulation is determined not only by Sry but by sex chromosome complement as well. Dev. Cell 2010, 19, 477–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neri, G.; Schwartz, C.E.; Lubs, H.A.; Stevenson, R.E. X-linked intellectual disability update 2017. Am. J. Med. Genet. Part. A 2018, 176, 1375–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos-Rebouças, C.B.; Boy, R.; Vianna, E.Q.; Gonçalves, A.P.; Piergiorge, R.M.; Abdala, B.B.; Dos Santos, J.M.; Calassara, V.; Machado, F.B.; Medina-Acosta, E.; et al. Skewed X-Chromosome Inactivation and Compensatory Upregulation of Escape Genes Precludes Major Clinical Symptoms in a Female With a Large Xq Deletion. Front. Genet. 2020, 11, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umesono, K.; Evans, R.M. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell 1989, 57, 1139–1146. [Google Scholar] [CrossRef]
- Ikeda, K.; Horie-Inoue, K.; Inoue, S. Identification of estrogen-responsive genes based on the DNA binding properties of estrogen receptors using high-throughput sequencing technology. Acta Pharmacol. Sin. 2015, 36, 24–31. [Google Scholar] [CrossRef] [Green Version]
- García-Carpizo, V.; Ruiz Llorente, L.; Fernández Fraga, M.; Aranda, A. The growing role of gene methylation on endocrine function. J. Mol. Endocrinol. 2011, 47, R75–R89. [Google Scholar] [CrossRef]
- Kouzmenko, A.; Ohtake, F.; Fujiki, R.; Kato, S. Hormonal gene regulation through DNA methylation and demethylation. Epigenomics 2010, 2, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Hegde, M.; Joshi, M.B. Comprehensive analysis of regulation of DNA methyltransferase isoforms in human breast tumors. J. Cancer Res. Clin. Oncol. 2021, 147, 937–971. [Google Scholar] [CrossRef]
- Mamrut, S.; Avidan, N.; Staun-Ram, E.; Ginzburg, E.; Truffault, F.; Berrih-Aknin, S.; Miller, A. Integrative analysis of methylome and transcriptome in human blood identifies extensive sex- and immune cell-specific differentially methylated regions. Epigenetics 2015, 10, 943–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Dongen, J.; Nivard, M.G.; Willemsen, G.; Hottenga, J.-J.; Helmer, Q.; Dolan, C.V.; Ehli, E.A.; Davies, G.E.; Van Iterson, M.; Breeze, C.E.; et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat. Commun. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Calzón, S.; Perfilyev, A.; De Mello, V.D.; Pihlajamäki, J.; Ling, C. Sex Differences in the Methylome and Transcriptome of the Human Liver and Circulating HDL-Cholesterol Levels. J. Clin. Endocrinol. Metab. 2018, 103, 4395–4408. [Google Scholar] [CrossRef]
- Boks, M.P.; Derks, E.M.; Weisenberger, D.J.; Strengman, E.; Janson, E.; Sommer, I.E.; Kahn, R.S.; Ophoff, R.A. The Relationship of DNA Methylation with Age, Gender and Genotype in Twins and Healthy Controls. PLoS ONE 2009, 4, e6767. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Morgan, M.; Hutchison, K.; Calhoun, V.D. A Study of the Influence of Sex on Genome Wide Methylation. PLoS ONE 2010, 5, e10028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, Q.K.-W.; Galvez, J.H.; Xiao, Q.; AlOgayil, N.; Hyacinthe, J.; Taketo, T.; Bourque, G.; Naumova, A.K. Sex Chromosomes and Sex Phenotype Contribute to Biased DNA Methylation in Mouse Liver. Cells 2020, 9, 1436. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Huang, B.; Sinha, N.; Wang, J.; Sen, A. Androgens regulate ovarian gene expression by balancing Ezh2-Jmjd3 mediated H3K27me3 dynamics. PLoS Genet. 2021, 17, e1009483. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Newcomb, D.C. Sex Bias in Asthma Prevalence and Pathogenesis. Front. Immunol. 2018, 9, 2997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potaczek, D.P.; Harb, H.; Michel, S.; Alhamwe, B.A.; Renz, H.; Tost, J. Epigenetics and allergy: From basic mechanisms to clinical applications. Epigenomics 2017, 9, 539–571. [Google Scholar] [CrossRef] [PubMed]
- Naumova, A.K.; Al Tuwaijri, A.; Morin, A.; Vaillancout, V.T.; Madore, A.-M.; Berlivet, S.; Kohan-Ghadr, H.-R.; Moussette, S.; Laprise, C. Sex- and age-dependent DNA methylation at the 17q12-q21 locus associated with childhood asthma. Hum. Genet. 2013, 132, 811–822. [Google Scholar] [CrossRef]
- Csaba, G. Phylogeny and Ontogeny of Hormone Receptors: The Selection Theory of Receptor Formation and Hormonal Imprinting. Biol. Rev. 1980, 55, 47–63. [Google Scholar] [CrossRef]
- Csaba, G. Hormonal Imprinting: The First Cellular-level Evidence of Epigenetic Inheritance and its Present State. Curr. Genom. 2019, 20, 409–418. [Google Scholar] [CrossRef]
- Titus, L.; Hatch, E.E.; Drake, K.M.; Parker, S.E.; Hyer, M.; Palmer, J.R.; Strohsnitter, W.C.; Adam, E.; Herbst, A.L.; Huo, D.; et al. Reproductive and hormone-related outcomes in women whose mothers were exposed in utero to diethylstilbestrol (DES): A report from the US National Cancer Institute DES Third Generation Study. Reprod. Toxicol. 2019, 84, 32–38. [Google Scholar] [CrossRef]
- Yousefi, P.; Huen, K.; Davé, V.; Barcellos, L.; Eskenazi, B.; Holland, N. Sex differences in DNA methylation assessed by 450 K BeadChip in newborns. BMC Genom. 2015, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, M.; Honda, C.; The Osaka Twin Research Group; Iwatani, Y.; Yorifuji, S.; Iso, H.; Kamide, K.; Hatazawa, J.; Kihara, S.; Sakai, N.; et al. Within-pair differences of DNA methylation levels between monozygotic twins are different between male and female pairs. BMC Med. Genom. 2016, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Bjornsson, H.T. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015, 25, 1473–1481. [Google Scholar] [CrossRef] [Green Version]
- McCabe, C.; Anderson, O.S.; Montrose, L.; Neier, K.; Dolinoy, D.C. Sexually Dimorphic Effects of Early-Life Exposures to Endocrine Disruptors: Sex-Specific Epigenetic Reprogramming as a Potential Mechanism. Curr. Environ. Health Rep. 2017, 4, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Montrose, L.; Faulk, C.; Francis, J.; Dolinoy, D.C. Perinatal lead (Pb) exposure results in sex and tissue-dependent adult DNA methylation alterations in murine IAP transposons. Environ. Mol. Mutagen. 2017, 58, 540–550. [Google Scholar] [CrossRef] [Green Version]
- Sobolewski, M.; Varma, G.; Adams, B.; Anderson, D.W.; Schneider, J.S.; Cory-Slechta, D.A. Developmental Lead Exposure and Prenatal Stress Result in Sex-Specific Reprograming of Adult Stress Physiology and Epigenetic Profiles in Brain. Toxicol. Sci. 2018, 163, 478–489. [Google Scholar] [CrossRef]
- Sen, A.; Heredia, N.; Senut, M.-C.; Hess, M.; Land, S.; Qu, W.; Hollacher, K.; Dereski, M.O.; Ruden, D.M. Early life lead exposure causes gender-specific changes in the DNA methylation profile of DNA extracted from dried blood spots. Epigenomics 2015, 7, 379–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, A.; Cingolani, P.; Senut, M.-C.; Land, S.; Mercado-Garcia, A.; Tellez-Rojo, M.M.; Baccarelli, A.A.; Wright, R.O.; Ruden, D.M. Lead exposure induces changes in 5-hydroxymethylcytosine clusters in CpG islands in human embryonic stem cells and umbilical cord blood. Epigenetics 2015, 10, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xie, C.; Murphy, S.K.; Skaar, D.; Nye, M.; Vidal, A.C.; Cecil, K.M.; Dietrich, K.N.; Puga, A.; Jirtle, R.L.; et al. Lead Exposure during Early Human Development and DNA Methylation of Imprinted Gene Regulatory Elements in Adulthood. Environ. Health Perspect. 2016, 124, 666–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Martín, F.J.; Lindquist, D.M.; Landero-Figueroa, J.; Zhang, X.; Chen, J.; Cecil, K.M.; Medvedovic, M.; Puga, A. Sex- and tissue-specific methylome changes in brains of mice perinatally exposed to lead. NeuroToxicology 2015, 46, 92–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montrose, L.; Padmanabhan, V.; Goodrich, J.; Domino, S.E.; Treadwell, M.C.; Meeker, J.; Watkins, D.J.; Dolinoy, D.C. Maternal levels of endocrine disrupting chemicals in the first trimester of pregnancy are associated with infant cord blood DNA methylation. Epigenetics 2018, 13, 301–309. [Google Scholar] [CrossRef] [Green Version]
- Goodrich, J.M.; Dolinoy, D.C.; Sánchez, B.N.; Zhang, Z.; Meeker, J.D.; Mercado-Garcia, A.; Solano-González, M.; Hu, H.; Téllez-Rojo, M.M.; Peterson, K.E. Adolescent epigenetic profiles and environmental exposures from early life through peri-adolescence. Environ. Epigenetics 2016, 2, dvw018. [Google Scholar] [CrossRef] [Green Version]
- Hewagama, A. Role of X-Chromosome Encoded MiRNAs in Autoimmunity: Suppressing the Suppressor and Female Predis-position. Rheumatol. Curr. Res. 2013, 3, 118. [Google Scholar] [CrossRef] [Green Version]
- Otaegui, D.; Baranzini, S.E.; Armañanzas, R.; Calvo, B.; Muñoz-Culla, M.; Khankhanian, P.; Inza, I.; Lozano, J.A.; Castillo-Triviño, T.; Asensio, A.; et al. Differential Micro RNA Expression in PBMC from Multiple Sclerosis Patients. PLoS ONE 2009, 4, e6309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egeland, N.G.; Jonsdottir, K.; Aure, M.R.; Sahlberg, K.; Kristensen, V.N.; Cronin-Fenton, D.; Skaland, I.; Gudlaugsson, E.; Baak, J.P.A.; Janssen, E.A.M. MiR-18a and miR-18b are expressed in the stroma of oestrogen receptor alpha negative breast cancers. BMC Cancer 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Dar, A.A.; Majid, S.; Rittsteuer, C.; De Semir, D.; Bezrookove, V.; Tong, S.; Nosrati, M.; Sagebiel, R.; Miller, J.R., III; Kashani-Sabet, M. The Role of miR-18b in MDM2-p53 Pathway Signaling and Melanoma Progression. JNCI J. Natl. Cancer Inst. 2013, 105, 433–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, Y.; Tamori, A.; Itami, S.; Tanahashi, T.; Toyoda, H.; Tanaka, M.; Wu, W.; Brojigin, N.; Kaneoka, Y.; Maeda, A.; et al. The expression level of miR-18b in hepatocellular carcinoma is associated with the grade of malignancy and prognosis. BMC Cancer 2013, 13, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medzikovic, L.; Aryan, L.; Eghbali, M. Connecting sex differences, estrogen signaling, and microRNAs in cardiac fibrosis. J. Mol. Med. 2019, 56, 1385–1398. [Google Scholar] [CrossRef] [PubMed]
- Cattalini, M.; Soliani, M.; Caparello, M.C.; Cimaz, R. Sex Differences in Pediatric Rheumatology. Clin. Rev. Allergy Immunol. 2019, 56, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Hewagama, A.; Gorelik, G.; Patel, D.; Liyanarachchi, P.; McCune, W.J.; Somers, E.; Gonzalez-Rivera, T.; Cohort, T.M.L.; Strickland, F.; Richardson, B. Overexpression of X-Linked genes in T cells from women with lupus. J. Autoimmun. 2013, 41, 60–71. [Google Scholar] [CrossRef] [Green Version]
- Al Emran, A.; Gallagher, S.J.; Tiffen, J.C.; Hersey, P. Sex bias of females in survival from cancer and infections. Is X the answer? Br. J. Cancer 2021, 124, 1184–1186. [Google Scholar] [CrossRef]
- Marcos-Villar, L.; Díaz-Colunga, J.; Sandoval, J.; Zamarreño, N.; Landeras-Bueno, S.; Esteller, M.; Falcón, A.; Nieto, A. Epigenetic control of influenza virus: Role of H3K79 methylation in interferon-induced antiviral response. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Alhamwe, B.A.; Miethe, S.; von Strandmann, E.P.; Potaczek, D.P.; Garn, H. Epigenetic Regulation of Airway Epithelium Immune Functions in Asthma. Front. Immunol. 2020, 11, 1747. [Google Scholar] [CrossRef]
- Menachery, V.D.; Schäfer, A.; Burnum-Johnson, K.E.; Mitchell, H.D.; Eisfeld, A.J.; Walters, K.B.; Nicora, C.D.; Purvine, S.O.; Casey, C.P.; Monroe, M.E.; et al. MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc. Natl. Acad. Sci. USA 2018, 115, E1012–E1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chlamydas, S.; Papavassiliou, A.G.; Piperi, C. Epigenetic mechanisms regulating COVID-19 infection. Epigenetics 2021, 16, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Pinto, B.G.G.; Oliveira, A.E.R.; Singh, Y.; Jimenez, L.; Gonçalves, A.N.A.; Ogava, R.L.T.; Creighton, R.; Peron, J.P.S.; I Nakaya, H.I. ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19. J. Infect. Dis. 2020, 222, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.T.; Grafham, D.V.; Coffey, A.J.; Scherer, S.; McLay, K.; Muzny, D.; Platzer, M.; Howell, G.R.; Burrows, C.; Bird, C.P.; et al. The DNA sequence of the human X chromosome. Nature 2005, 434, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Sawalha, A.H.; Zhao, M.; Coit, P.; Lu, Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin. Immunol. 2020, 215, 108410. [Google Scholar] [CrossRef] [PubMed]
- PINHO, A.C. EMA Recommends COVID-19 Vaccine AstraZeneca for Authorisation in the EU. Available online: https://www.ema.europa.eu/en/news/ema-recommends-covid-19-vaccine-astrazeneca-authorisation-eu (accessed on 30 April 2021).
- Aleem, A.; Nadeem, A.J. Coronavirus (COVID-19) Vaccine-Induced Immune Thrombotic Thrombocytopenia (VITT). In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar] [PubMed]
- Angeli, F.; Spanevello, A.; Reboldi, G.; Visca, D.; Verdecchia, P. SARS-CoV-2 vaccines: Lights and shadows. Eur. J. Intern. Med. 2021, 88, 1–8. [Google Scholar] [CrossRef] [PubMed]
- McGonagle, D.; De Marco, G.; Bridgewood, C. Mechanisms of Immunothrombosis in Vaccine-Induced Thrombotic Thrombocytopenia (VITT) Compared to Natural SARS-CoV-2 Infection. J. Autoimmun. 2021, 121, 102662. [Google Scholar] [CrossRef]
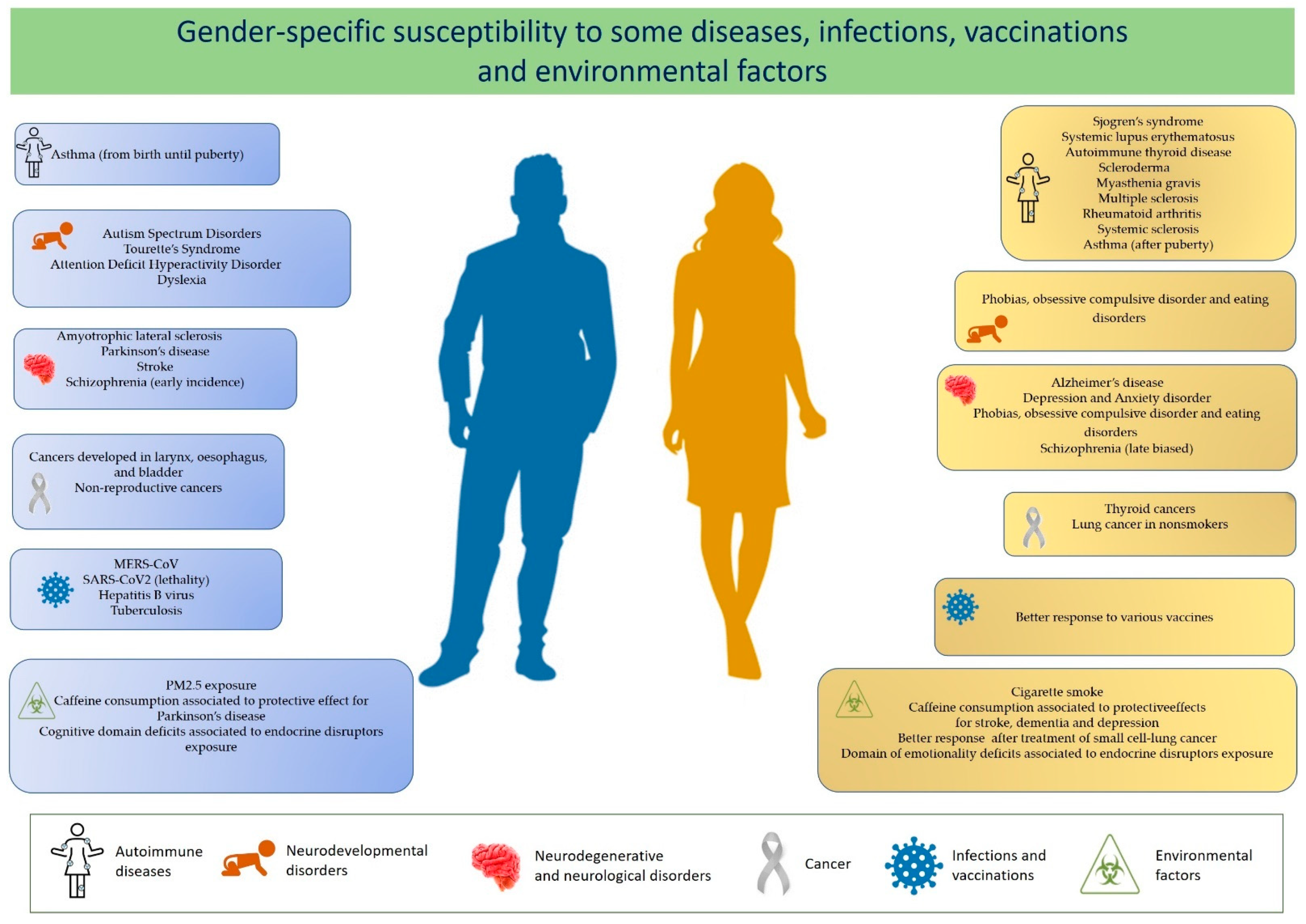

| Female to Male Ratio | References | |
|---|---|---|
| Autoimmune disorders | ||
| Sjogren’s syndrome (SS) | 9:1 | [5] |
| Systemic lupus erythematosus (SLE) | 7:1 | [5] |
| Autoimmune thyroid disease (AITD) | 7:1–10:1 | [6] |
| Scleroderma | 7:1 | [6] |
| Myasthenia gravis (MG) | 2:1–3:1 | [6] |
| Rheumatoid arthritis (RA) | 2–3:1 | [5,6] |
| Multiple sclerosis | 2–3:1 | [7] |
| Systemic sclerosis | 3:1 | [6] |
| Neurodevelopmental and Neurodegenerative diseases | ||
| Autism Spectrum Disorders (ASD) | 1:3–4 1:9 (High-functioning patients) | [7,8] |
| Attention Deficit Hyperactivity Disorder (ADHD) | 1:3 | [8] |
| Tourette’s Syndrome | 1:4 | [8] |
| Depression and Anxiety disorder | 2:1 | [7,8] |
| Amyotrophic lateral sclerosis | 1:1.6 | [7] |
| Schizophrenia | Early incidence male-biased Late incidence female-biased | [7] |
| Stroke | 1:2 | [7] |
| Parkinson’s disease | 1:1.5 | [8,9] |
| Alzheimer’s disease | 1.6–3:1 | [7] |
| Cancer | ||
| Cancers developed in larynx, esophagus, and bladder | 1:4 | [10] |
| Non-reproductive cancers | 1:2 | [10] |
| Thyroid cancers | 3:1 | [11] |
| Infectious diseases | ||
| MERS-CoV | 1:2 | [12] |
| SARS-CoV2 (lethality) | 1:1.5 | [13] |
| Hepatitis B virus | 1:3.8 | [14] |
| Tuberculosis | 1:1.6 | [15] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Migliore, L.; Nicolì, V.; Stoccoro, A. Gender Specific Differences in Disease Susceptibility: The Role of Epigenetics. Biomedicines 2021, 9, 652. https://doi.org/10.3390/biomedicines9060652
Migliore L, Nicolì V, Stoccoro A. Gender Specific Differences in Disease Susceptibility: The Role of Epigenetics. Biomedicines. 2021; 9(6):652. https://doi.org/10.3390/biomedicines9060652
Chicago/Turabian StyleMigliore, Lucia, Vanessa Nicolì, and Andrea Stoccoro. 2021. "Gender Specific Differences in Disease Susceptibility: The Role of Epigenetics" Biomedicines 9, no. 6: 652. https://doi.org/10.3390/biomedicines9060652
APA StyleMigliore, L., Nicolì, V., & Stoccoro, A. (2021). Gender Specific Differences in Disease Susceptibility: The Role of Epigenetics. Biomedicines, 9(6), 652. https://doi.org/10.3390/biomedicines9060652






