In Vitro Evaluation of a Nanoparticle-Based mRNA Delivery System for Cells in the Joint
Abstract
:1. Introduction
2. Materials and Methods
2.1. Nanoparticle Synthesis
2.2. mRNA Synthesis
2.3. Cell Isolation
2.3.1. Bovine Chondrocytes (bCH) Isolation
2.3.2. Isolation of Human Mesenchymal Stem Cells (hBMSC)
2.3.3. Isolation of Human Synovial-Derived Stem Cells (hSDSC)
2.3.4. Isolation of Rat Tendon Derived Stem and Progenitor Cells (rTDSPC)
2.4. Cell Culture
2.5. 3D Cultures
2.6. Delivery of mRNA Using Nanoparticles
2.7. Fluorescence Imaging and Live/Dead Staining
2.8. Metabolic Activity and Toxicity Screening
2.9. Flow Cytometry
2.10. Statistics
3. Results
3.1. Nanoparticle Characterization
3.2. Nanoparticle-Based mRNA Delivery to Different Cell Populations of Articular Joints
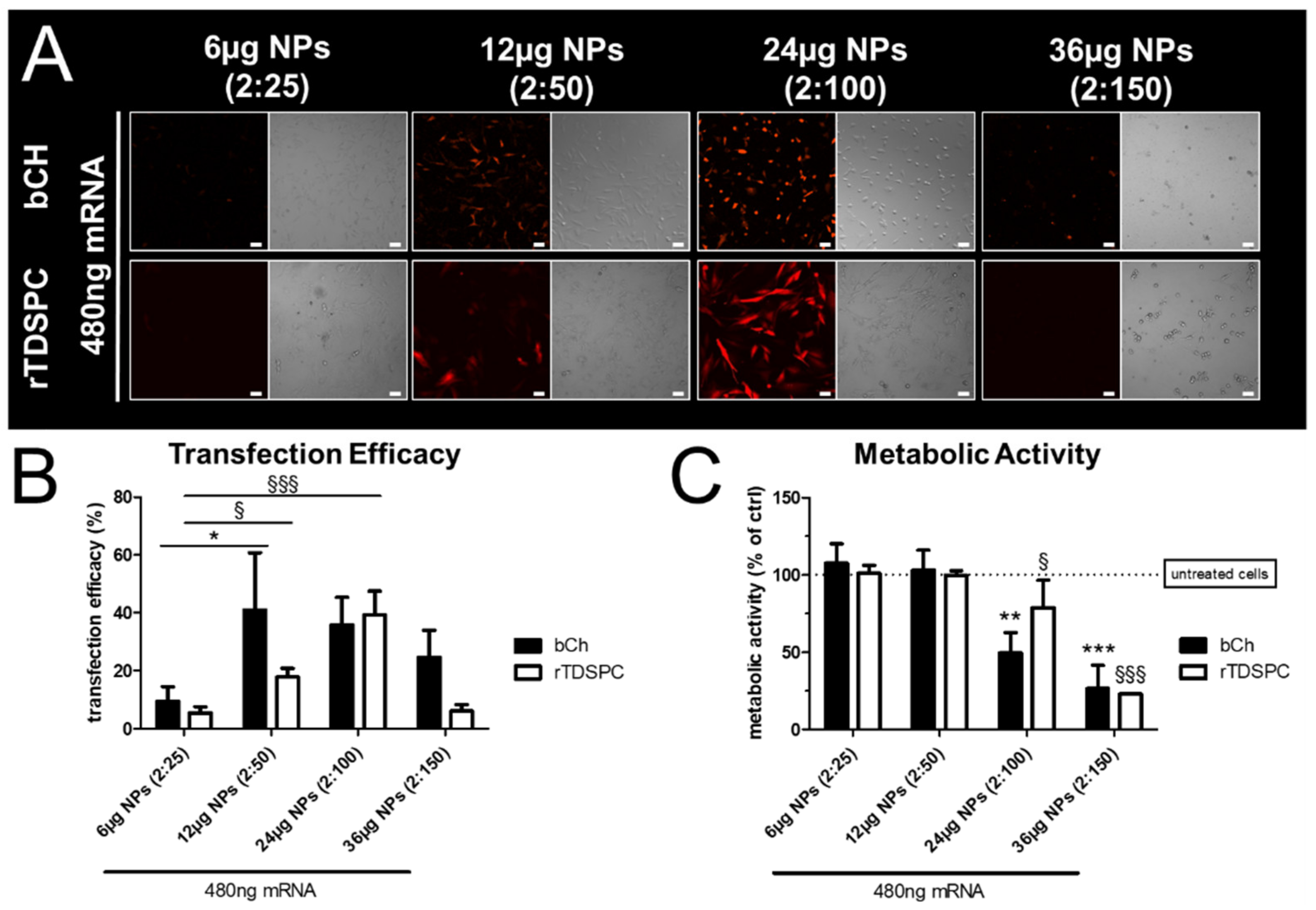
3.3. Effect of Different mRNA:Nanoparticle Ratios on Transfection Efficiency and Cell Viability in bCH and rTDSPC
3.4. Transfection Efficiency and Metabolic Cell Activity after Treatment with Increasing Amounts of mRNA Complexed to 6 or 12 µg of Nanoparticles
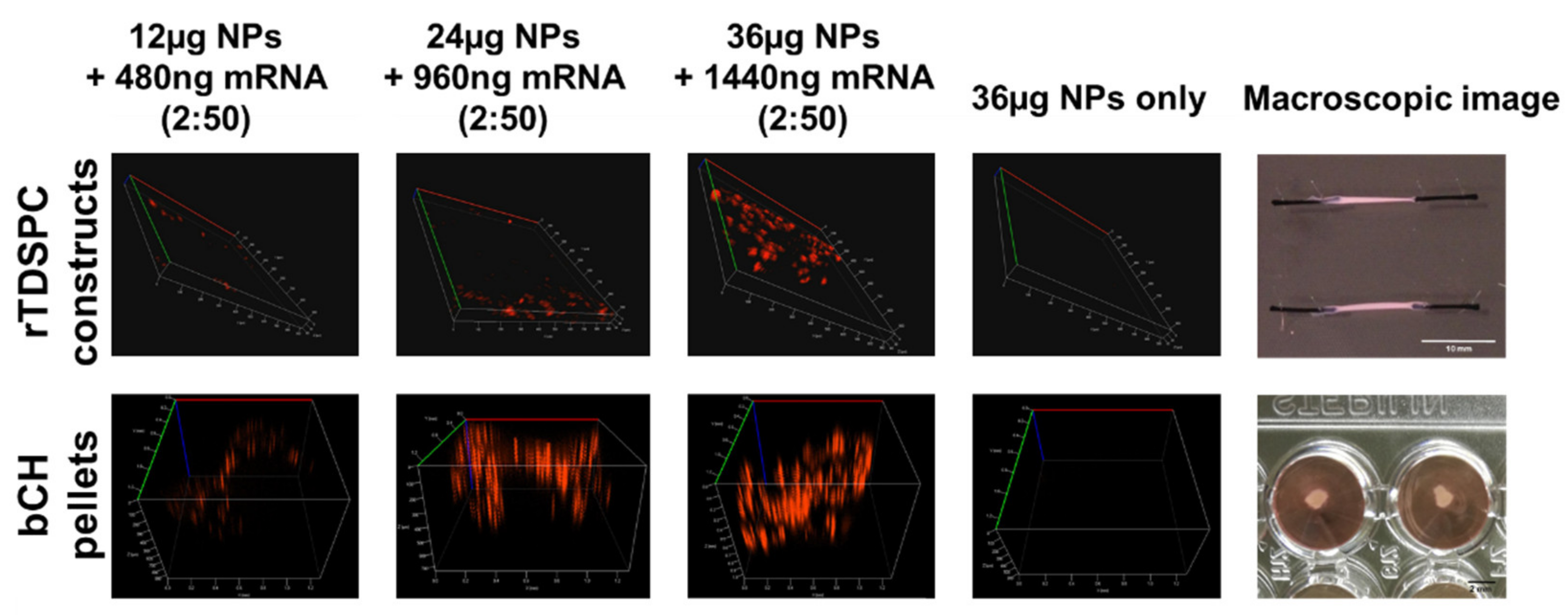
3.5. Treatment of bCH and rTDSPC with the Optimal mRNA:Nanoparticle Loading Ratio of 2:50
4. Discussion
4.1. Strength and Weakness of the Study
4.2. Clinical Outlook
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| bCH | bovine chondrocytes |
| bFGF | basic fibroblast growth factor |
| BMPs | bone morphogenetic proteins |
| FBS | fetal bovine serum |
| hBMSC | human bone marrow stromal cells |
| hSDSC | human synovial derived stem cells |
| IVT | in vitro-transcribed |
| LNP | lipid nanoparticles |
| NPs | nanoparticles |
| NSAID | non-steroidal anti-inflammatory drugs |
| OA | osteoarthritis |
| ORF | open reading frame |
| PBS | phosphate-buffered saline |
| P/S | penicillin/streptomycin |
| RUNX | runt-related transcription factor |
| rTDSPC | rat tendon derived stem/progenitor cells |
| TNFα | tumor necrosis factor-alpha |
| TGFβ | transforming growth factor β |
| UTR | untranslated regions |
References
- Glyn-Jones, S.; Palmer, A.J.R.; Agricola, R.; Price, A.J.; Vincent, T.; Weinans, H.; Carr, A.J. Osteoarthritis. Lancet 2015, 386, 376–387. [Google Scholar] [CrossRef]
- Wallace, S.L.; Robinson, H.; Masi, A.; Decker, J.L.; Mccarty, D.J.; Yü, T.-F. Preliminary criteria for the classification of the acute arthritis of primary gout. Arthritis Rheum. 1977, 20, 895–900. [Google Scholar] [CrossRef] [PubMed]
- McInnes, I.B.; Schett, G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef] [Green Version]
- Gabrielli, A.; Avvedimento, E.V.; Krieg, T. Scleroderma. N. Engl. J. Med. 2009, 360, 1989–2003. [Google Scholar] [CrossRef] [PubMed]
- Mok, C.; Lau, C. Pathogenesis of systemic lupus erythematosus. J. Clin. Pathol. 2003, 56, 481–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Siegler, S.; Allard, P.; Kirtley, C.; Leardini, A.; Rosenbaum, D.; Whittle, M.; D’Lima, D.D.; Cristofolini, L.; Witte, H.; et al. ISB recommendation on definitions of joint coordinate system of various joints for the reporting of human joint motion—Part I: Ankle, hip, and spine. J. Biomech. 2002, 35, 543–548. [Google Scholar] [CrossRef]
- Savvidou, O.; Milonaki, M.; Goumenos, S.; Flevas, D.; Papagelopoulos, P.; Moutsatsou, P. Glucocorticoid signaling and osteoarthritis. Mol. Cell. Endocrinol. 2019, 480, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Ding, C. Do NSAIDs affect the progression of osteoarthritis? Inflammation 2002, 26, 139–142. [Google Scholar] [CrossRef]
- Lee, C.M.; Maibach, H.I. Deep Percutaneous Penetration into Muscles and Joints. J. Pharm. Sci. 2006, 95, 1405–1413. [Google Scholar] [CrossRef]
- Dedrick, R.L.; Flessner, M.F. Pharmacokinetic Problems in Peritoneal Drug Administration: Tissue Penetration and Surface Exposure. J. Natl. Cancer Inst. 1997, 89, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Bessa, P.C.; Casal, M.; Reis, R.L. Bone morphogenetic proteins in tissue engineering: The road from the laboratory to the clinic, part I (basic concepts). J. Tissue Eng. Regen. Med. 2008, 2, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Bajpayee, A.G.; Grodzinsky, A.J. Cartilage-targeting drug delivery: Can electrostatic interactions help? Nat. Rev. Rheumatol. 2017, 13, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.F.; Pham, C.T. Intra-articular drug delivery systems for joint diseases. Curr. Opin. Pharmacol. 2018, 40, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Kou, L.; Xiao, S.; Sun, R.; Bao, S.; Yao, Q.; Chen, R. Biomaterial-engineered intra-articular drug delivery systems for osteoarthritis therapy. Drug Deliv. 2019, 26, 870–885. [Google Scholar] [CrossRef]
- Larsen, C.; Østergaard, J.; Larsen, S.; Jensen, H.; Jacobsen, S.; Lindegaard, C.; Andersen, P.H. Intra-articular depot formulation principles: Role in the management of postoperative pain and arthritic disorders. J. Pharm. Sci. 2008, 97, 4622–4654. [Google Scholar] [CrossRef]
- Scallon, B.J. Chimeric anti-TNF-α monoclonal antibody cA2 binds recombinant transmembrane TNF-α and activates immune effector functions. Cytokine 1995, 7, 251–259. [Google Scholar] [CrossRef]
- Strohl, W. Chimeric Genes, Proteins. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar] [CrossRef]
- Knight, D.M.; Trinh, H.; Le, J.; Siegel, S.; Shealy, D.; McDonough, M.; Scallon, B.; Moore, M.A.; Vilcek, J.; Daddona, P.; et al. Construction and initial characterization of a mouse-human chimeric anti-TNF antibody. Mol. Immunol. 1993, 30, 1443–1453. [Google Scholar] [CrossRef]
- Van Dullemen, H.M.; van Deventer, S.J.; Hommes, D.W.; Bijl, H.A.; Jansen, J.; Tytgat, G.N.; Woody, J. Treatment of Crohn’s disease with anti-tumor necrosis factor chimeric monoclonal antibody (cA2). Gastroenterology 1995, 109, 129–135. [Google Scholar] [CrossRef]
- Elliott, M.; Maini, R.; Feldmann, M.; Kalden, J.; Antoni, C.; Smolen, J.; Leeb, B.; Breedveld, F.; Macfarlane, J.; Bijl, J.; et al. Randomised double-blind comparison of chimeric monoclonal antibody to tumour necrosis factor α (cA2) versus placebo in rheumatoid arthritis. Lancet 1994, 344, 1105–1110. [Google Scholar] [CrossRef]
- Tratschin, J.D.; Miller, I.L.; Smith, M.G.; Carter, B.J. Adeno-associated virus vector for high-frequency integration, expression, and rescue of genes in mammalian cells. Mol. Cell. Biol. 1985, 5. [Google Scholar] [CrossRef] [Green Version]
- Büning, H.; Perabo, L.; Coutelle, O.; Quadt-Humme, S.; Hallek, M. Recent developments in adeno-associated virus vector technology. J. Gene Med. 2008, 10, 717–733. [Google Scholar] [CrossRef]
- Knott, G.J.; Doudna, J.A. CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fellmann, C.; Gowen, B.G.; Lin, P.-C.; Doudna, J.A.; Corn, J.E. Cornerstones of CRISPR–Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 2017, 16, 89–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahin, U.; Karikó, K.; Türeci, Ö. mRNA-based therapeutics—Developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef] [PubMed]
- Dowdy, S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017, 35, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, J.C.; Kowalski, P.; Anderson, D.G. Advances in the delivery of RNA therapeutics: From concept to clinical reality. Genome Med. 2017, 9, 1–16. [Google Scholar] [CrossRef] [Green Version]
- U.S. National Library of Medicine. Available online: https://www.clinicaltrials.gov/ct2/results?cond=&term=mRNA&cntry=&state=&city=&dist= (accessed on 26 April 2021).
- Weng, Y.; Li, C.; Yang, T.; Hu, B.; Zhang, M.; Guo, S.; Xiao, H.; Liang, X.-J.; Huang, Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020, 40, 107534. [Google Scholar] [CrossRef]
- Evans, C.H.; Ghivizzani, S.C.; Robbins, P.D. Gene Delivery to Joints by Intra-Articular Injection. Hum. Gene Ther. 2018, 29, 2–14. [Google Scholar] [CrossRef]
- Gao, X.; Huang, L. Cationic liposome-mediated gene transfer. Gene Ther. 1995, 2, 710–722. [Google Scholar]
- Farhood, H.; Serbina, N.; Huang, L. The role of dioleoyl phosphatidylethanolamine in cationic liposome mediated gene transfer. Biochim. Biophys. Acta (BBA) Biomembr. 1995, 1235, 289–295. [Google Scholar] [CrossRef] [Green Version]
- Filipczak, N.; Pan, J.; Yalamarty, S.S.K.; Torchilin, V.P. Recent advancements in liposome technology. Adv. Drug Deliv. Rev. 2020, 156, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Samaridou, E.; Heyes, J.; Lutwyche, P. Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv. Drug Deliv. Rev. 2020, 154–155, 37–63. [Google Scholar] [CrossRef]
- Masotti, A.; Mossa, G.; Cametti, C.; Ortaggi, G.; Bianco, A.; Del Grosso, N.; Malizia, D.; Esposito, C. Comparison of different commercially available cationic liposome–DNA lipoplexes: Parameters influencing toxicity and transfection efficiency. Colloids Surf. B Biointerfaces 2009, 68, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Tabar, M.S.; Hesaraki, M.; Esfandiari, F.; Samani, F.S.; Vakilian, H.; Baharvand, H. Evaluating Electroporation and Lipofectamine Approaches for Transient and Stable Transgene Expressions in Human Fibroblasts and Embryonic Stem Cells. Cell J. 2015, 17, 438–450. [Google Scholar]
- Colella, F.; Garcia, J.P.; Sorbona, M.; Lolli, A.; Antunes, B.; D’Atri, D.; Barré, F.P.; Oieni, J.; Vainieri, M.L.; Zerrillo, L.; et al. Drug delivery in intervertebral disc degeneration and osteoarthritis: Selecting the optimal platform for the delivery of disease-modifying agents. J. Control. Release 2020, 328, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Zhong, Z.; Lok, M.C.; Jiang, X.; Hennink, W.E.; Feijen, A.J.; Engbersen, J.F.J. Novel Bioreducible Poly(amido amine)s for Highly Efficient Gene Delivery. Bioconjug. Chem. 2007, 18, 138–145. [Google Scholar] [CrossRef]
- Lin, C.; Engbersen, J.F. Effect of chemical functionalities in poly(amido amine)s for non-viral gene transfection. J. Control. Release 2008, 132, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Lammens, T.; Zhong, Z.; Gu, H.; Lok, M.; Jiang, X.; Hennink, W.; Feijen, J.; Engbersen, J. Disulfide-containing poly (β-amino ester)s for gene delivery. J. Control. Release 2006, 116, e79–e81. [Google Scholar] [CrossRef]
- Lin, C.; Zhong, Z.; Lok, M.C.; Jiang, X.; Hennink, W.E.; Feijen, J.; Engbersen, J.F. Linear poly(amido amine)s with secondary and tertiary amino groups and variable amounts of disulfide linkages: Synthesis and in vitro gene transfer properties. J. Control. Release 2006, 116, 130–137. [Google Scholar] [CrossRef]
- Kormann, M.S.D.; Hasenpusch, G.; Aneja, M.K.; Nica, G.; Flemmer, A.W.; Herber-Jonat, S.; Huppmann, M.; E Mays, L.; Illenyi, M.; Schams, A.; et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011, 29, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Rothweiler, R.; Basoli, V.; Duttenhoefer, F.; Kubosch, D.; Schmelzeisen, R.; Johnstone, B.; Alini, M.; Stoddart, M.J. Predicting and Promoting Human Bone Marrow MSC Chondrogenesis by Way of TGFβ Receptor Profiles: Toward Personalized Medicine. Front. Bioeng. Biotechnol. 2020, 8, 618. [Google Scholar] [CrossRef]
- Kovermann, N.J.; Basoli, V.; Della Bella, E.; Alini, M.; Lischer, C.; Schmal, H.; Kubosch, E.J.; Stoddart, M.J. BMP2 and TGF-β Cooperate Differently during Synovial-Derived Stem-Cell Chondrogenesis in a Dexamethasone-Dependent Manner. Cells 2019, 8, 636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehwolf, R.; Spitzer, G.; Wagner, A.; Lehner, C.; Weissenbacher, N.; Tempfer, H.; Traweger, A. 3D-Embedded Cell Cultures to Study Tendon Biology. Methods Mol. Biol. 2019, 2045, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Leonhardt, H.; Cardoso, M.C. DNA labeling in living cells. Cytom. Part A 2005, 67A, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.J.; Patterson, J.; Oreilly, R.; Gibson, M. Glutathione-triggered disassembly of isothermally responsive polymer nanoparticles obtained by nanoprecipitation of hydrophilic polymers. Polym. Chem. 2014, 5, 126–131. [Google Scholar] [CrossRef]
- Dessau, W.; Vertel, B.M.; Von Der Mark, H. Extracellular matrix formation by chondrocytes in monolayer culture. J. Cell Biol. 1981, 90, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Brodkin, K.; García, A.; Levenston, M. Chondrocyte phenotypes on different extracellular matrix monolayers. Biomaterials 2004, 25, 5929–5938. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Tanzil, G.; Mobasheri, A.; Clegg, P.D.; Sendzik, J.; John, T.; Shakibaei, M. Cultivation of human tenocytes in high-density culture. Histochem. Cell Biol. 2004, 122, 219–228. [Google Scholar] [CrossRef]
- Fröhlich, E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012, 7, 5577–5591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Lilly, G.D.; Doty, R.C.; Podsiadlo, P.; Kotov, N.A. In vitro Toxicity Testing of Nanoparticles in 3D Cell Culture. Small 2009, 5, 1213–1221. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.; Jackson, S.; Haycock, J.; MacNeil, S. Culture of skin cells in 3D rather than 2D improves their ability to survive exposure to cytotoxic agents. J. Biotechnol. 2006, 122, 372–381. [Google Scholar] [CrossRef]
- Sowa, M.B.; Chrisler, W.B.; Zens, K.; Ashjian, E.J.; Opresko, L.K. Three-dimensional culture conditions lead to decreased radiation induced cytotoxicity in human mammary epithelial cells. Mutat. Res. Mol. Mech. Mutagen. 2010, 687, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Voigt, N.; Henrich-Noack, P.; Kockentiedt, S.; Hintz, W.; Tomas, J.; Sabel, B.A. Toxicity of polymeric nanoparticles in vivo and in vitro. J. Nanopart. Res. 2014, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bajpayee, A.G.; Scheu, M.; Grodzinsky, A.J.; Porter, R.M. Electrostatic interactions enable rapid penetration, enhanced uptake and retention of intra-articular injected avidin in rat knee joints. J. Orthop. Res. 2014, 32, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Morgen, M.; Tung, D.; Boras, B.; Miller, W.; Malfait, A.-M.; Tortorella, M. Nanoparticles for Improved Local Retention after Intra-Articular Injection into the Knee Joint. Pharm. Res. 2012, 30, 257–268. [Google Scholar] [CrossRef] [Green Version]
- Anselmo, A.C.; Mitragotri, S. Nanoparticles in the clinic: An update. Bioeng. Transl. Med. 2019, 4, e10143. [Google Scholar] [CrossRef] [Green Version]
- Aini, H.; Itaka, K.; Fujisawa, A.; Uchida, H.; Uchida, S.; Fukushima, S.; Kataoka, K.; Saito, T.; Chung, U.-I.; Ohba, S. Messenger RNA delivery of a cartilage-anabolic transcription factor as a disease-modifying strategy for osteoarthritis treatment. Sci. Rep. 2016, 6, 18743. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sturm, L.; Schwemberger, B.; Menzel, U.; Häckel, S.; Albers, C.E.; Plank, C.; Rip, J.; Alini, M.; Traweger, A.; Grad, S.; et al. In Vitro Evaluation of a Nanoparticle-Based mRNA Delivery System for Cells in the Joint. Biomedicines 2021, 9, 794. https://doi.org/10.3390/biomedicines9070794
Sturm L, Schwemberger B, Menzel U, Häckel S, Albers CE, Plank C, Rip J, Alini M, Traweger A, Grad S, et al. In Vitro Evaluation of a Nanoparticle-Based mRNA Delivery System for Cells in the Joint. Biomedicines. 2021; 9(7):794. https://doi.org/10.3390/biomedicines9070794
Chicago/Turabian StyleSturm, Lisa, Bettina Schwemberger, Ursula Menzel, Sonja Häckel, Christoph E. Albers, Christian Plank, Jaap Rip, Mauro Alini, Andreas Traweger, Sibylle Grad, and et al. 2021. "In Vitro Evaluation of a Nanoparticle-Based mRNA Delivery System for Cells in the Joint" Biomedicines 9, no. 7: 794. https://doi.org/10.3390/biomedicines9070794
APA StyleSturm, L., Schwemberger, B., Menzel, U., Häckel, S., Albers, C. E., Plank, C., Rip, J., Alini, M., Traweger, A., Grad, S., & Basoli, V. (2021). In Vitro Evaluation of a Nanoparticle-Based mRNA Delivery System for Cells in the Joint. Biomedicines, 9(7), 794. https://doi.org/10.3390/biomedicines9070794







