Validation of Cell-Free RNA and Circulating Tumor Cells for Molecular Marker Analysis in Metastatic Prostate Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients’ Characteristics
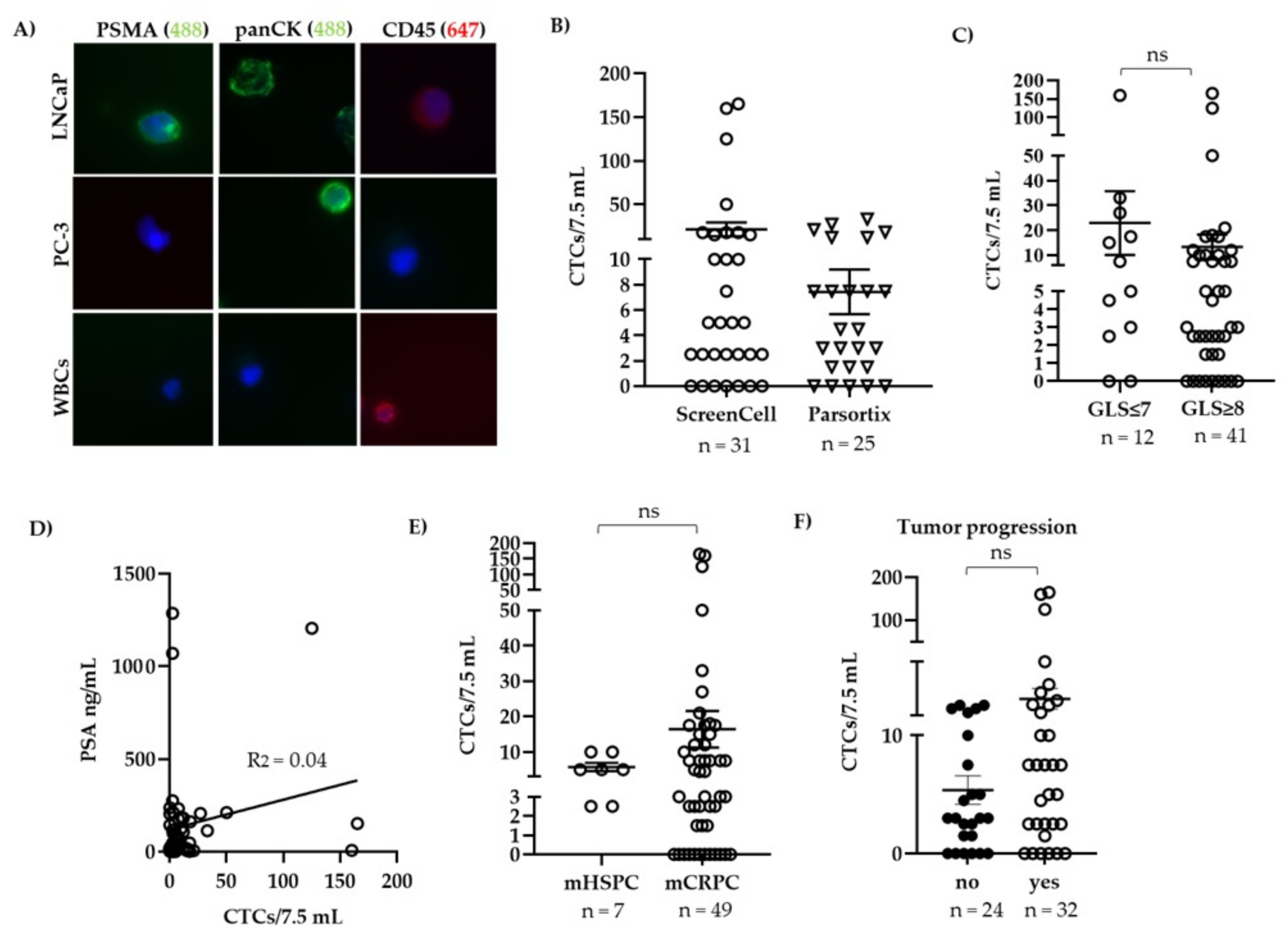
2.2. Enumeration and Counting of Patient Derived CTCs
2.3. Picking Single CTCs
2.4. RNA Isolation from CTCs and Plasma
2.5. cDNA Synthesis, Pre-Amplification, and Real-Time Quantitative RT-PCR (qPCR)
2.6. Cell Culture
2.7. Statistical Analysis
3. Results
3.1. CTC Count Did Not Correlate with Gleason Score or Serum PSA Levels
3.2. Gene Expression in Patient-Derived CTCs
3.3. Gene Expression in Plasma-Derived cfRNA
3.4. Follow-Up of a Patient with mCRPC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Heidegger, I.; Heidenreich, A.; Pfister, D. New Biomarkers for Selecting the Best Therapy Regimens in Metastatic Castration-Resistant Prostate Cancer. Target. Oncol. 2017, 12, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.B.; Rokhlin, O.W. Mechanisms of prostate cancer cell survival after inhibition of AR expression. J. Cell. Biochem. 2009, 106, 363–371. [Google Scholar] [CrossRef]
- Fizazi, K.; Tran, N.; Fein, L.; Matsubara, N.; Rodriguez-Antolin, A.; Alekseev, B.Y.; Özgüroğlu, M.; Ye, D.; Feyerabend, S.; Protheroe, A.; et al. Abiraterone plus Prednisone in Metastatic, Castration-Sensitive Prostate Cancer. N. Engl. J. Med. 2017, 377, 352–360. [Google Scholar] [CrossRef]
- Davis, I.D.; Martin, A.J.; Stockler, M.R.; Begbie, S.; Chi, K.N.; Chowdhury, S.; Coskinas, X.; Frydenberg, M.; Hague, W.E.; Horvath, L.G.; et al. Enzalutamide with Standard First-Line Therapy in Metastatic Prostate Cancer. N. Engl. J. Med. 2019, 381, 121–131. [Google Scholar] [CrossRef]
- Fizazi, K.; Shore, N.; Tammela, T.L.; Ulys, A.; Vjaters, E.; Polyakov, S.; Jievaltas, M.; Luz, M.; Alekseev, B.; Kuss, I.; et al. Nonmetastatic, Castration-Resistant Prostate Cancer and Survival with Darolutamide. N. Engl. J. Med. 2020, 383, 1040–1049. [Google Scholar] [CrossRef]
- Smith, M.R.; Saad, F.; Chowdhury, S.; Oudard, S.; Hadaschik, B.A.; Graff, J.N.; Olmos, D.; Mainwaring, P.N.; Lee, J.Y.; Uemura, H.; et al. Apalutamide Treatment and Metastasis-free Survival in Prostate Cancer. N. Engl. J. Med. 2018, 378, 1408–1418. [Google Scholar] [CrossRef]
- Parker, C.; Nilsson, S.; Heinrich, D.; Helle, S.I.; O’Sullivan, J.M.; Fosså, S.D.; Chodacki, A.; Wiechno, P.; Logue, J.; Seke, M.; et al. Alpha Emitter Radium-223 and Survival in Metastatic Prostate Cancer. N. Engl. J. Med. 2013, 369, 213–223. [Google Scholar] [CrossRef] [Green Version]
- De Bono, J.; Mateo, J.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Chi, K.N.; Sartor, O.; Agarwal, N.; Olmos, D.; et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2020, 382, 2091–2102. [Google Scholar] [CrossRef] [PubMed]
- Wadosky, K.M.; Koochekpour, S. Molecular mechanisms underlying resistance to androgen deprivation therapy in prostate cancer. Oncotarget 2016, 7, 64447–64470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, M.T.; Morris, M.J.; Heller, G.; I Scher, H.I. Post-therapy changes in PSA as an outcome measure in prostate cancer clinical trials. Nat. Clin. Pr. Oncol. 2006, 3, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Jia, X.; de Bono, J.S.; Fleisher, M.; Pienta, K.J.; Raghavan, D.; Heller, G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: A reanalysis of IMMC38 trial data. Lancet Oncol. 2009, 10, 233–239. [Google Scholar] [CrossRef] [Green Version]
- Goodman, O.B.; Symanowski, J.T.; Loudyi, A.; Fink, L.M.; Ward, D.C.; Vogelzang, N.J. Circulating Tumor Cells as a Predictive Biomarker in Patients with Hormone-sensitive Prostate Cancer. Clin. Genitourin. Cancer 2011, 9, 31–38. [Google Scholar] [CrossRef]
- Lorente, D.; Olmos, D.; Mateo, J.; Bianchini, D.; Seed, G.; Fleisher, M.; Danila, D.C.; Flohr, P.; Crespo, M.; Figueiredo, I.; et al. Decline in Circulating Tumor Cell Count and Treatment Outcome in Advanced Prostate Cancer. Eur. Urol. 2016, 70, 985–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, D.T.; Sequist, L.V.; Lee, R.J. Circulating tumour cells—Monitoring treatment response in prostate cancer. Nat. Rev. Clin. Oncol. 2014, 11, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, D.R.; Leversha, M.A.; Danila, D.C.; Lin, O.; Gonzalez-Espinoza, R.; Gu, B.; Anand, A.; Smith, K.; Maslak, P.; Doyle, G.V.; et al. Circulating Tumor Cell Analysis in Patients with Progressive Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2007, 13, 2023–2029. [Google Scholar] [CrossRef] [Green Version]
- Attard, G.; Swennenhuis, J.F.; Olmos, D.; Reid, A.H.; Vickers, E.; A’Hern, R.; Levink, R.; Coumans, F.; Moreira, J.; Riisnaes, R.; et al. Characterization of ERG, AR and PTEN Gene Status in Circulating Tumor Cells from Patients with Castration-Resistant Prostate Cancer. Cancer Res. 2009, 69, 2912–2918. [Google Scholar] [CrossRef] [Green Version]
- Antonarakis, E.S.; Armstrong, A.J.; Dehm, S.M.; Luo, J. Androgen receptor variant-driven prostate cancer: Clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis. 2016, 19, 231–241. [Google Scholar] [CrossRef] [Green Version]
- Antonarakis, E.S.; Lu, C.; Luber, B.; Wang, H.; Chen, Y.; Nakazawa, M.; Nadal, R.; Paller, C.J.; Denmeade, S.R.; Carducci, M.A.; et al. Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients with Metastatic Castration-Resistant Prostate Cancer. JAMA Oncol. 2015, 1, 582–591. [Google Scholar] [CrossRef] [Green Version]
- Scher, H.I.; Morris, M.J.; Stadler, W.M.; Higano, C.; Basch, E.; Fizazi, K.; Antonarakis, E.S.; Beer, T.M.; Carducci, M.A.; Chi, K.N.; et al. Trial Design and Objectives for Castration-Resistant Prostate Cancer: Updated Recommendations from the Prostate Cancer Clinical Trials Working Group 3. J. Clin. Oncol. 2016, 34, 1402–1418. [Google Scholar] [CrossRef] [Green Version]
- Eslami-S, Z.; Cortés-Hernández, L.E.; Alix-Panabières, C. Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells. Cells 2020, 9, 1836. [Google Scholar] [CrossRef]
- Galletti, G.; Matov, A.; Beltran, H.; Fontugne, J.; Mosquera, J.M.; Cheung, C.; Macdonald, T.Y.; Sung, M.; O’Toole, S.; Kench, J.G.; et al. ERG induces taxane resistance in castration-resistant prostate cancer. Nat. Commun. 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pantel, K.; Speicher, M.R. The biology of circulating tumor cells. Oncogene 2016, 35, 1216–1224. [Google Scholar] [CrossRef]
- Xu, L.; Mao, X.; Grey, A.; Scandura, G.; Guo, T.; Burke, E.; Marzec, J.; Abdu, S.; Stankiewicz, E.; Davies, C.R.; et al. Noninvasive Detection of Clinically Significant Prostate Cancer Using Circulating Tumor Cells. J. Urol. 2020, 203, 73–82. [Google Scholar] [CrossRef]
- Hodara, E.; Morrison, G.; Cunha, A.; Zainfeld, D.; Xu, T.; Xu, Y.; Dempsey, P.W.; Pagano, P.C.; Bischoff, F.; Khurana, A.; et al. Multiparametric liquid biopsy analysis in metastatic prostate cancer. JCI Insight 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Luk, A.; Young, F.P.; Lynch, D.; Chua, W.; Balakrishnar, B.; De Souza, P.; Becker, T.M. Droplet Digital PCR Based Androgen Receptor Variant 7 (AR-V7) Detection from Prostate Cancer Patient Blood Biopsies. Int. J. Mol. Sci. 2016, 17, 1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todenhöfer, T.; Azad, A.; Stewart, C.; Gao, J.; Eigl, B.J.; Gleave, M.E.; Joshua, A.M.; Black, P.C.; Chi, K.N. AR-V7 Transcripts in Whole Blood RNA of Patients with Metastatic Castration Resistant Prostate Cancer Correlate with Response to Abiraterone Acetate. J. Urol. 2017, 197, 135–142. [Google Scholar] [CrossRef]
- Kohli, M.; Tan, W.; Zheng, T.; Wang, A.; Montesinos, C.; Wong, C.; Du, P.; Jia, S.; Yadav, S.; Horvath, L.G.; et al. Clinical and genomic insights into circulating tumor DNA-based alterations across the spectrum of metastatic hormone-sensitive and castrate-resistant prostate cancer. EBioMedicine 2020, 54, 102728. [Google Scholar] [CrossRef]
- Fettke, H.; Kwan, E.M.; Docanto, M.M.; Bukczynska, P.; Ng, N.; Graham, L.-J.K.; Mahon, K.; Hauser, C.; Tan, W.; Wang, X.H.; et al. Combined Cell-free DNA and RNA Profiling of the Androgen Receptor: Clinical Utility of a Novel Multianalyte Liquid Biopsy Assay for Metastatic Prostate Cancer. Eur. Urol. 2020, 78, 173–180. [Google Scholar] [CrossRef]
- Gillessen, S.; Omlin, A.; Attard, G.; de Bono, J.S.; Efstathiou, E.; Fizazi, K.; Halabi, S.; Nelson, P.S.; Sartor, O.; Smith, M.R.; et al. Management of patients with advanced prostate cancer: Recommendations of the St Gallen Advanced Prostate Cancer Consensus Conference (APCCC) 2015. Ann. Oncol. 2015, 26, 1589–1604. [Google Scholar] [CrossRef]
- Neuwirt, H.; Bouchal, J.; Kharaishvili, G.; Ploner, C.; Jöhrer, K.; Pitterl, F.; Weber, A.; Klocker, H.; Eder, I.E. Cancer-associated fibroblasts promote prostate tumor growth and progression through upregulation of cholesterol and steroid biosynthesis. Cell Commun. Signal. 2020, 18, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoefer, J.; Akbor, M.; Handle, F.; Ofer, P.; Puhr, M.; Parson, W.; Culig, Z.; Klocker, H.; Heidegger, I. Critical role of androgen receptor level in prostate cancer cell resistance to new generation antiandrogen enzalutamide. Oncotarget 2016, 7, 59781–59794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.J.; Raghavan, D. Circulating Tumor Cells Predict Survival Benefit from Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [Green Version]
- Kafka, M.; Mayr, F.; Temml, V.; Möller, G.; Adamski, J.; Höfer, J.; Schwaiger, S.; Heidegger, I.; Matuszczak, B.; Schuster, D.; et al. Dual Inhibitory Action of a Novel AKR1C3 Inhibitor on Both Full-Length AR and the Variant AR-V7 in Enzalutamide Resistant Metastatic Castration Resistant Prostate Cancer. Cancers 2020, 12, 2092. [Google Scholar] [CrossRef] [PubMed]
- Danila, D.C.; Heller, G.; Gignac, G.A.; Gonzalez-Espinoza, R.; Anand, A.; Tanaka, E.; Lilja, H.; Schwartz, L.; Larson, S.; Fleisher, M.; et al. Circulating Tumor Cell Number and Prognosis in Progressive Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2007, 13, 7053–7058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folkersma, L.R.; Manso, L.S.J.; Romo, I.G.; Sierra, J.M.; Gómez, C.O. Prognostic Significance of Circulating Tumor Cell Count in Patients with Metastatic Hormone-sensitive Prostate Cancer. Urology 2012, 80, 1328–1332. [Google Scholar] [CrossRef]
- Schilling, D.; Todenhöfer, T.; Hennenlotter, J.; Schwentner, C.; Fehm, T.; Stenzl, A. Isolated, disseminated and circulating tumour cells in prostate cancer. Nat. Rev. Urol. 2012, 9, 448–463. [Google Scholar] [CrossRef]
- Hofman, M.S.; Violet, J.; Hicks, R.J.; Ferdinandus, J.; Thang, S.P.; Akhurst, T.; Iravani, A.; Kong, G.; Kumar, A.R.; Murphy, D.G.; et al. [177 Lu]-PSMA-617 radionuclide treatment in patients with metastatic castration-resistant prostate cancer (LuPSMA trial): A single-centre, single-arm, phase 2 study. Lancet Oncol. 2018, 19, 825–833. [Google Scholar] [CrossRef]
- Ristau, B.T.; O’Keefe, D.S.; Bacich, D.J. The prostate-specific membrane antigen: Lessons and current clinical implications from 20 years of research. Urol. Oncol. Semin. Orig. Investig. 2014, 32, 272–279. [Google Scholar] [CrossRef] [Green Version]
- Uprimny, C.; Kroiss, A.S.; Decristoforo, C.; Fritz, J.; Warwitz, B.; Scarpa, L.; Roig, L.G.; Kendler, D.; Von Guggenberg, E.; Bektic, J.; et al. Early dynamic imaging in 68Ga- PSMA-11 PET/CT allows discrimination of urinary bladder activity and prostate cancer lesions. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 765–775. [Google Scholar] [CrossRef]
- Wright, G.L., Jr.; Haley, C.; Beckett, M.L.; Schellhammer, P.F. Expression of prostate-specific membrane antigen in normal, benign, and malignant prostate tissues. Urol. Oncol. Semin. Orig. Investig. 1995, 1, 18–28. [Google Scholar] [CrossRef]
- Ladurner, M.; Horninger, W.; Bektic, J. Lutetium-PSMA therapy—A new therapeutic option in metastatic castration-resistant prostate cancer? Memo Mag. Eur. Med. Oncol. 2018, 11, 301–304. [Google Scholar] [CrossRef]
- Gorges, T.M.; Kuske, A.; Röck, K.; Mauermann, O.; Müller, V.; Peine, S.; Verpoort, K.; Novosadova, V.; Kubista, M.; Riethdorf, S.; et al. Accession of Tumor Heterogeneity by Multiplex Transcriptome Profiling of Single Circulating Tumor Cells. Clin. Chem. 2016, 62, 1504–1515. [Google Scholar] [CrossRef] [PubMed]
- El-Heliebi, A.; Hille, C.; Laxman, N.; Svedlund, J.; Haudum, C.; Ercan, E.; Kroneis, T.; Chen, S.; Smolle, M.; Rossmann, C.; et al. In Situ Detection and Quantification of AR-V7, AR-FL, PSA, and KRAS Point Mutations in Circulating Tumor Cells. Clin. Chem. 2018, 64, 536–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, D.T.; Lee, R.J.; Stott, S.L.; Ting, D.T.; Wittner, B.S.; Ulman, M.; Smas, M.E.; Lord, J.B.; Brannigan, B.W.; Trautwein, J.; et al. Androgen Receptor Signaling in Circulating Tumor Cells as a Marker of Hormonally Responsive Prostate Cancer. Cancer Discov. 2012, 2, 995–1003. [Google Scholar] [CrossRef] [Green Version]
- Montgomery, R.B.; Mostaghel, E.A.; Vessella, R.; Hess, D.L.; Kalhorn, T.F.; Higano, C.S.; True, L.D.; Nelson, P.S. Maintenance of Intratumoral Androgens in Metastatic Prostate Cancer: A Mechanism for Castration-Resistant Tumor Growth. Cancer Res. 2008, 68, 4447–4454. [Google Scholar] [CrossRef] [Green Version]
- Stanbrough, M.; Bubley, G.J.; Ross, K.; Golub, T.R.; Rubin, M.A.; Penning, T.M.; Febbo, P.G.; Balk, S.P. Increased Expression of Genes Converting Adrenal Androgens to Testosterone in Androgen-Independent Prostate Cancer. Cancer Res. 2006, 66, 2815–2825. [Google Scholar] [CrossRef] [Green Version]
- Steckelbroeck, S.; Jin, Y.; Gopishetty, S.; Oyesanmi, B.; Penning, T.M. Human Cytosolic 3α-Hydroxysteroid Dehydrogenases of the Aldo-keto Reductase Superfamily Display Significant 3β-Hydroxysteroid Dehydrogenase Activity: Implications for steroid hormone metabolism and action. J. Biol. Chem. 2004, 279, 10784–10795. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Armstrong, C.M.; Lou, W.; Lombard, A.; Evans, C.P.; Gao, A.C. Inhibition of AKR1C3 Activation Overcomes Resistance to Abiraterone in Advanced Prostate Cancer. Mol. Cancer Ther. 2017, 16, 35–44. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Lou, W.; Zhu, Y.; Yang, J.C.; Nadiminty, N.; Gaikwad, N.W.; Evans, C.P.; Gao, A.C. Intracrine Androgens and AKR1C3 Activation Confer Resistance to Enzalutamide in Prostate Cancer. Cancer Res. 2015, 75, 1413–1422. [Google Scholar] [CrossRef] [Green Version]
- Verma, K.; Gupta, N.; Zang, T.; Wangtrakluldee, P.; Srivastava, S.K.; Penning, T.M.; Trippier, P.C. AKR1C3 Inhibitor KV-37 Exhibits Antineoplastic Effects and Potentiates Enzalutamide in Combination Therapy in Prostate Adenocarcinoma Cells. Mol. Cancer Ther. 2018, 17, 1833–1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poeppel, T.D.; Handkiewicz-Junak, D.; Andreeff, M.; Becherer, A.; Bockisch, A.; Fricke, E.; Geworski, L.; Heinzel, A.; Krause, B.J.; Krause, T.; et al. EANM guideline for radionuclide therapy with radium-223 of metastatic castration-resistant prostate cancer. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 824–845. [Google Scholar] [CrossRef] [PubMed]
- Carles, J.; Castellano, D.; Méndez-Vidal, M.-J.; Mellado, B.; Saez, M.-I.; Del Alba, A.G.; Perez-Gracia, J.-L.; Jimenez, J.; Suárez, C.; Sepúlveda, J.-M.; et al. Circulating Tumor Cells as a Biomarker of Survival and Response to Radium-223 Therapy: Experience in a Cohort of Patients with Metastatic Castration-Resistant Prostate Cancer. Clin. Genitourin. Cancer 2018, 16, e1133–e1139. [Google Scholar] [CrossRef] [PubMed]







| Total Number of Patient Samples | 62 |
| Median age, years (range) | 71 (56–88) |
| ISUP grade (number of samples) | |
| 1–3 | 13 |
| 4 | 13 |
| 5 | 36 |
| Tumor stage (number of samples) | |
| mHSPC | 8 |
| High volume | 7 |
| High risk | 7 |
| mCRPC | 54 |
| Primary M1 PCa | 36 |
| Metastatic sites (number of samples) | |
| Lymph nodes | 53 |
| Bone | 58 |
| Liver | 5 |
| Lung | 7 |
| Other visceral mets | 3 |
| Blood analytes | |
| Median PSA, ng/mL (range) | 46 (0.6–7014) |
| Median alkaline phosphatase, U/L (range) | 95 (38–783) |
| Median LDH, U/L (range) | 226 (133–1173) |
| Median CRP, mg/dL (range) | 0.6 (0.1–13.3) |
| Median hemoglobin, g/dL (range) | 12.3 (7.15.8) |
| Local treatment (number of samples) | |
| Radical prostatectomy | 19 |
| EBRT (external beam radiation) | 5 |
| LDR (low dose rate) brachytherapy | 2 |
| Therapies prior to sample collection (number of samples) | |
| ADT | 61 |
| Enzalutamide | 22 |
| Abiraterone | 26 |
| Docetaxel | 41 |
| Cabazitaxel | 12 |
| Radium-223 | 17 |
| Lutetium177-PSMA | 6 |
| PrimePCR PreAmp and PrimePCR Probe Assay (Biorad) | ||
|---|---|---|
| Gene symbol | Gene name | Unique Assay ID |
| AKR1C3 | Aldo-keto reductase family 1 member C3 | qHsaCEP0040990 |
| AR-FL | Androgen receptor (full-length) | qHsaCIP0026366 |
| CD45 | Tyrosine phosphatase receptor type C | qHsaCEP0041630 |
| ERG | ETS-related gene | qHsaCEP0041582 |
| FASN | Fatty acid synthase | qHsaCIP0026813 |
| KRT18 | Keratin 18 | qHsaCEP0035862 |
| PSMA | Prostate-specific membrane antigen | qHsaCEP0049804 |
| TMPRSS2 | Transmembrane Serine Protease 2 | qHsaCIP0028919 |
| TP53 | Tumor protein P53 | qHsaCEP0052284 |
| PreAmp Primer | ||
| Gene | Primer forward (F), Primer reverse (R) | |
| AR-FL | F: ACATCAAGGAACTCGATCGTATCA, R: GGGCACTTGCACAGAGATGA | |
| AR-V7 | F: AAGAGCCGCTGAAGGGAAAC, R: TCCAGACTATCCACTAGAGCCC | |
| HPRT1 | F: ACACTGGCAAAACAATGCAGA, R: AGTCAAGGGCATATCCTACAACAA | |
| KLK-2 | F: TCAGAGCCTGCCAAGATCAC, R: TTTACCACCTGTCCAGAGCC | |
| PSA | F: AGTGCGAGAAGCATTCCCAA, R: AAGCTGTGGCTGACCTGAAA | |
| TBP | F: GCCGAATATAATCCCAAGCGG, R: TTAGCTGGAAAACCCAACTTCTG | |
| PCR Primer | ||
| AR-FL | F: CTGCTCAAGACGCTTCTA, R: ATCATTTCCGGAAAGTCCA, P (Probe): TCCGTGCAGCCTATTGCGAG | |
| AR-V7 | F: GTCCATCTTGTCGTCTTC, R: GCAAGTCAGCCTTTCTTCA, P: GGGAGAAAAATTCCGGGTTGGC | |
| HPRT1 | F: GCTTTCCTTGGTCAGGCAGTA, R: GTCTGGCTTATATCCAACACTTCGT, P: TCAAGGTCGCAAGCTTGCTGGTGAAAAGAA | |
| KLK-2 | F: GACCACCTGCTACGCCTCAG, R: GGACAGGAGATGGAGGCTCA, P: ACCAGAGGAGTTCTTGCGCCCCA | |
| PSA | F: GTCTGCGGCGGTGTTCTG, R: TGCCGACCCACGAAGATC, P: CACAGCTGCCCACTGCATCAGGA | |
| TBP | F: CACGAACCACGGCACTGATT, R: TTTTCTTGCTGCCAGTCTGGAC, P: TCTTCACTCTTGGCTCCTGTGCACA | |
| Total | ScreenCell | ParsortixTM | |
|---|---|---|---|
| Samples analyzed, n | 56 | 31 | 25 |
| Mean CTC count/7.5 mL (SEM) | 15.1 (±4.5) | 21.2 (±7.6) | 7.4 (±1.8) |
| CTC count ≥ 1/7.5 mL, n (%) | 44 (78.6%) | 24 (77.4%) | 20 (80.0%) |
| CTC count ≥ 5/7.5 mL, n (%) | 28 (50.0%) | 17 (54.8%) | 11 (44.0%) |
| Patient | Method | PSA (ng/mL) | CTCs/7.5 mL | Number of CTCs Picked |
|---|---|---|---|---|
| 47 | Parsortix+ALS | 144.8 | 0 | 0 |
| 51 | Parsortix+ALS | 2.7 | 0 | 0 |
| 56 | Parsortix+ALS | 26.8 | 1.5 | 0 |
| 58 | Parsortix+ALS | 10.9 | 1.5 | 0 |
| 59 | Parsortix+ALS | 33.8 | 0 | 0 |
| 61 | Parsortix+ALS | 9.0 | 0 | 0 |
| 62 | Parsortix+ALS | 237.5 | 0 | 0 |
| 64 | Parsortix+ALS | 32.9 | 1.5 | 0 |
| 66 | Parsortix+ALS | 67.91 | 7.5 | 0 |
| 67 | Parsortix+ALS | 232.6 | 7.5 | 0 |
| 55 | Parsortix+ALS | 212.3 | 3.0 | 1 |
| 63 | Parsortix+ALS | 23.2 | 3.0 | 2 |
| 50 | Parsortix+ALS | 22.3 | 7.5 | 3 |
| 57 | Parsortix+ALS | 162.3 | 18.0 | 4 |
| 46 | Parsortix+ALS | 178.1 | 7.5 | 5 |
| 65 | Parsortix+ALS | 182.1 | 12.0 | 8 |
| 49 | Parsortix+ALS | 6.0 | 21.0 | 12 |
| 60 | Parsortix+ALS | 207.4 | 27.0 | 18 |
| 43b | Parsortix+ALS | 113.6 | 33.0 | 22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ladurner, M.; Wieser, M.; Eigentler, A.; Seewald, M.; Dobler, G.; Neuwirt, H.; Kafka, M.; Heidegger, I.; Horninger, W.; Bektic, J.; et al. Validation of Cell-Free RNA and Circulating Tumor Cells for Molecular Marker Analysis in Metastatic Prostate Cancer. Biomedicines 2021, 9, 1004. https://doi.org/10.3390/biomedicines9081004
Ladurner M, Wieser M, Eigentler A, Seewald M, Dobler G, Neuwirt H, Kafka M, Heidegger I, Horninger W, Bektic J, et al. Validation of Cell-Free RNA and Circulating Tumor Cells for Molecular Marker Analysis in Metastatic Prostate Cancer. Biomedicines. 2021; 9(8):1004. https://doi.org/10.3390/biomedicines9081004
Chicago/Turabian StyleLadurner, Michael, Manuel Wieser, Andrea Eigentler, Martin Seewald, Gabriele Dobler, Hannes Neuwirt, Mona Kafka, Isabel Heidegger, Wolfgang Horninger, Jasmin Bektic, and et al. 2021. "Validation of Cell-Free RNA and Circulating Tumor Cells for Molecular Marker Analysis in Metastatic Prostate Cancer" Biomedicines 9, no. 8: 1004. https://doi.org/10.3390/biomedicines9081004
APA StyleLadurner, M., Wieser, M., Eigentler, A., Seewald, M., Dobler, G., Neuwirt, H., Kafka, M., Heidegger, I., Horninger, W., Bektic, J., Klocker, H., Obrist, P., & Eder, I. E. (2021). Validation of Cell-Free RNA and Circulating Tumor Cells for Molecular Marker Analysis in Metastatic Prostate Cancer. Biomedicines, 9(8), 1004. https://doi.org/10.3390/biomedicines9081004






