Decreased MicroRNA-150 Exacerbates Neuronal Apoptosis in the Diabetic Retina
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Culture
2.3. Lipofectamine Transfection
2.4. Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL)
2.5. The 3-[4,5-Dimethylthiazol-2-yl]-2,5 Diphenyl Tetrazolium Bromide (MTT) Colorimetric Assay
2.6. Quantitative Real-Time RT-PCR (qPCR)
2.7. Western Blot
2.8. Immunofluorescent Staining (Retina and Cultured Cells)
2.9. Statistical Analysis
3. Results
3.1. MicroRNA-150 Knockout (miR-150−/−) Exacerbates Apoptosis in the Diabetic Retina
3.2. MicroRNA-150 Knockdown Exacerbates Palmitic Acid (PA)-Elicited Apoptosis in Cultured 661W Cells
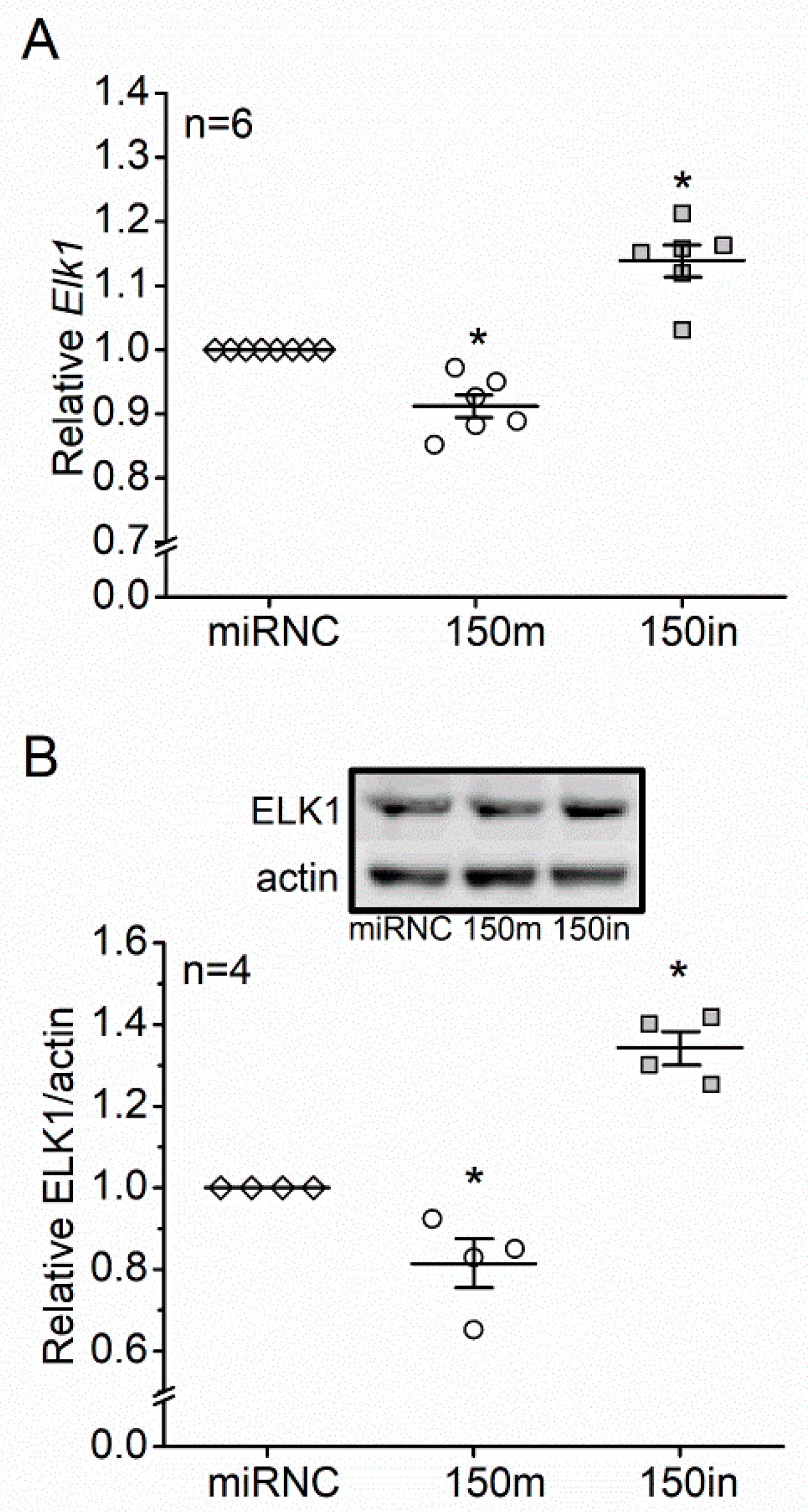
3.3. ETS-Domain Transcription Factor 1 (Elk1) Is a Direct Target of miR-150 and Contributes to T2D-Induced Apoptosis in Photoreceptors
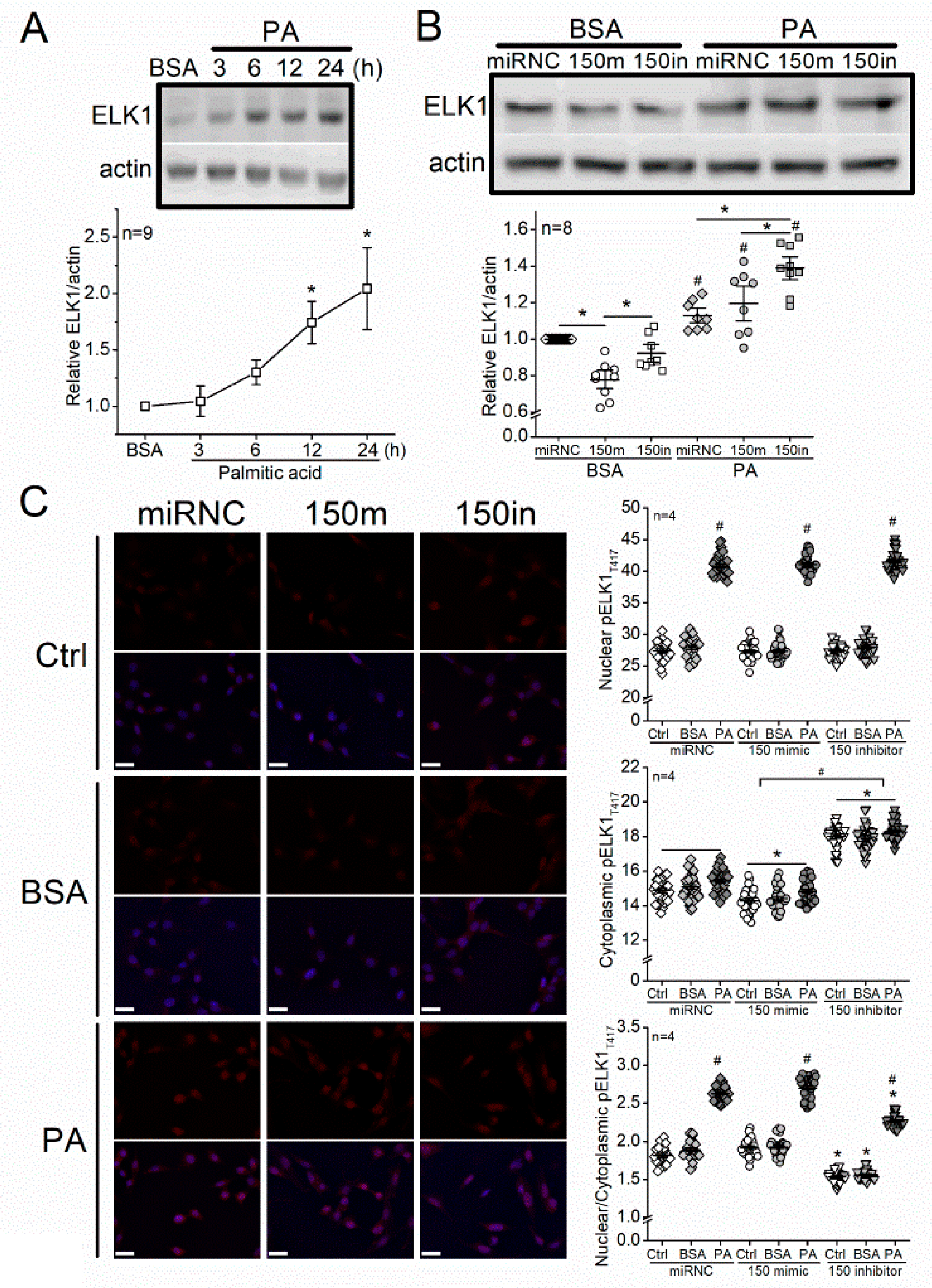
3.4. Treatment with PA Increases ELK1 and Nuclear pELK1T417, and Knocking Down miR-150 Upregulates ELK1 and Cytoplasmic pELK1T417 in 661W Cells
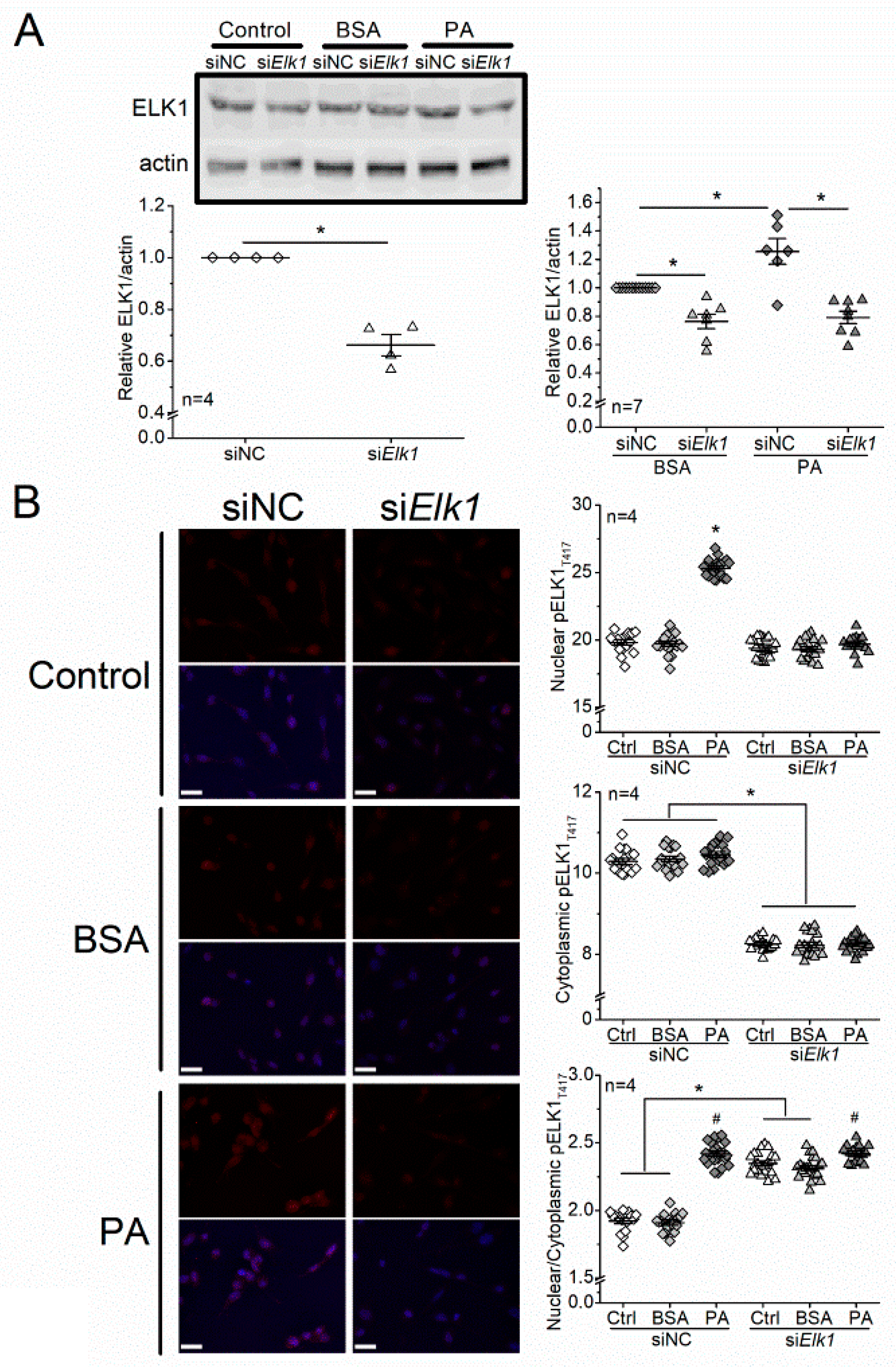
3.5. Knocking Down Elk1 Decreases PA-Elicited Increases in Total ELK1 and pELK1T417 but Does Not Alleviate PA-Elicited Apoptosis in 661W Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barsegian, A.; Kotlyar, B.; Lee, J.; Salifu, M.O.; McFarlane, S.I. Diabetic retinopathy: Focus on minority populations. Int. J. Clin. Endocrinol. Metab. 2017, 3, 034–045. [Google Scholar] [CrossRef] [Green Version]
- Wong, T.Y.; Cheung, C.M.; Larsen, M.; Sharma, S.; Simo, R. Diabetic retinopathy. Nat. Rev. Dis. Primers 2016, 2, 16012. [Google Scholar] [CrossRef]
- Moran, E.P.; Wang, Z.; Chen, J.; Sapieha, P.; Smith, L.E.; Ma, J.X. Neurovascular cross talk in diabetic retinopathy: Pathophysiological roles and therapeutic implications. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, H738–H749. [Google Scholar] [CrossRef] [Green Version]
- Stewart, M.W. Treatment of diabetic retinopathy: Recent advances and unresolved challenges. World J. Diabetes 2016, 7, 333–341. [Google Scholar] [CrossRef]
- Cheung, N.; Wong, I.Y.; Wong, T.Y. Ocular anti-vegf therapy for diabetic retinopathy: Overview of clinical efficacy and evolving applications. Diabetes Care 2014, 37, 900–905. [Google Scholar] [CrossRef] [Green Version]
- Cheung, N.; Mitchell, P.; Wong, T.Y. Diabetic retinopathy. Lancet 2010, 376, 124–136. [Google Scholar] [CrossRef]
- van Dijk, H.W.; Verbraak, F.D.; Stehouwer, M.; Kok, P.H.; Garvin, M.K.; Sonka, M.; DeVries, J.H.; Schlingemann, R.O.; Abramoff, M.D. Association of visual function and ganglion cell layer thickness in patients with diabetes mellitus type 1 and no or minimal diabetic retinopathy. Vis. Res. 2011, 51, 224–228. [Google Scholar] [CrossRef] [Green Version]
- Bronson-Castain, K.W.; Bearse, M.A., Jr.; Neuville, J.; Jonasdottir, S.; King-Hooper, B.; Barez, S.; Schneck, M.E.; Adams, A.J. Adolescents with type 2 diabetes: Early indications of focal retinal neuropathy, retinal thinning, and venular dilation. RETINA 2009, 29, 618–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, P.M.; Roon, P.; Van Ells, T.K.; Ganapathy, V.; Smith, S.B. Death of retinal neurons in streptozotocin-induced diabetic mice. Invest Ophthalmol. Vis. Sci. 2004, 45, 3330–3336. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Xu, Y.; Xie, P.; Cheng, H.; Song, Q.; Su, T.; Yuan, S.; Liu, Q. Retinal neurodegeneration in db/db mice at the early period of diabetes. J. Ophthalmol. 2015, 2015, 757412. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Park, J.W.; Park, S.J.; Kim, K.Y.; Chung, J.W.; Chun, M.H.; Oh, S.J. Apoptotic death of photoreceptors in the streptozotocin-induced diabetic rat retina. Diabetologia 2003, 46, 1260–1268. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Y.; Jin, Y.; Ji, F.; Sinclair, S.H.; Luo, Y.; Xu, G.; Lu, L.; Dai, W.; Yanoff, M.; et al. Intravitreal injection of erythropoietin protects both retinal vascular and neuronal cells in early diabetes. Invest Ophthalmol. Vis. Sci. 2008, 49, 732–742. [Google Scholar] [CrossRef]
- Lv, J.; Bao, S.; Liu, T.; Wei, L.; Wang, D.; Ye, W.; Wang, N.; Song, S.; Li, J.; Chudhary, M.; et al. Sulforaphane delays diabetes-induced retinal photoreceptor cell degeneration. Cell Tissue Res. 2020, 382, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Arden, G.B. The absence of diabetic retinopathy in patients with retinitis pigmentosa: Implications for pathophysiology and possible treatment. Br. J. Ophthalm. 2001, 85, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahdenranta, J.; Pasqualini, R.; Schlingemann, R.O.; Hagedorn, M.; Stallcup, W.B.; Bucana, C.D.; Sidman, R.L.; Arap, W. An anti-angiogenic state in mice and humans with retinal photoreceptor cell degeneration. Proc. Natl. Acad. Sci. USA 2001, 98, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Tang, J.; Du, Y.; Saadane, A.; Tonade, D.; Samuels, I.; Veenstra, A.; Palczewski, K.; Kern, T.S. Photoreceptor cells influence retinal vascular degeneration in mouse models of retinal degeneration and diabetes. Invest Ophthalmol. Vis. Sci. 2016, 57, 4272–4281. [Google Scholar] [CrossRef] [Green Version]
- Assmann, T.S.; Recamonde-Mendoza, M.; De Souza, B.M.; Crispim, D. Microrna expression profiles and type 1 diabetes mellitus: Systematic review and bioinformatic analysis. Endocr. Connect. 2017, 6, 773–790. [Google Scholar] [CrossRef] [Green Version]
- Mazzeo, A.; Beltramo, E.; Lopatina, T.; Gai, C.; Trento, M.; Porta, M. Molecular and functional characterization of circulating extracellular vesicles from diabetic patients with and without retinopathy and healthy subjects. Exp. Eye Res. 2018, 176, 69–77. [Google Scholar] [CrossRef]
- Yu, F.; Chapman, S.; Pham, D.L.; Ko, M.L.; Zhou, B.; Ko, G.Y. Decreased mir-150 in obesity-associated type 2 diabetic mice increases intraocular inflammation and exacerbates retinal dysfunction. BMJ Open Diabetes Res. Care 2020, 8, e001446. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Yao, K.; Guo, J.; Shi, H.; Ma, L.; Wang, Q.; Liu, H.; Gao, W.; Sun, A.; Zou, Y.; et al. Mir-181a and mir-150 regulate dendritic cell immune inflammatory responses and cardiomyocyte apoptosis via targeting jak1-stat1/c-fos pathway. J. Cell Mol. Med. 2017, 21, 2884–2895. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.L.; Guo, W.L.; Chen, X.M. Overexpressing microrna-150 attenuates hypoxia-induced human cardiomyocyte cell apoptosis by targeting glucose-regulated protein-94. Mol. Med. Rep. 2018, 17, 4181–4186. [Google Scholar] [CrossRef]
- Wright, W.S.; McElhatten, R.M.; Messina, J.E.; Harris, N.R. Hypoxia and the expression of hif-1alpha and hif-2alpha in the retina of streptozotocin-injected mice and rats. Exp. Eye Res. 2010, 90, 405–412. [Google Scholar] [CrossRef] [Green Version]
- Linsenmeier, R.A.; Zhang, H.F. Retinal oxygen: From animals to humans. Prog. Retin. Eye Res. 2017, 58, 115–151. [Google Scholar] [CrossRef] [Green Version]
- Barrett, L.E.; Bockstaele, E.J.V.; Sul, J.Y.; Takano, H.; Haydon, P.G.; Eberwine, J.H. Elk-1 associates with the mitochondrial permeablity transition pore complex in neurons. Proc. Natl. Acad. Sci. USA 2006, 103, 6. [Google Scholar] [CrossRef] [Green Version]
- Barrett, L.E.; Sul, J.Y.; Takano, H.; Van Bockstaele, E.J.; Haydon, P.G.; Eberwine, J.H. Region-directed phototransfection reveals the functional significance of a dendritically synthesized transcription factor. Nat. Methods 2006, 3, 455–460. [Google Scholar] [CrossRef]
- Lu, Z.; Miao, Z.; Zhu, J.; Zhu, G. Ets-domain containing protein (elk1) suppression protects cortical neurons against oxygen-glucose deprivation injury. Exp. Cell Res. 2018, 371, 42–49. [Google Scholar] [CrossRef]
- Sharma, A.; Callahan, L.M.; Sul, J.Y.; Kim, T.K.; Barrett, L.; Kim, M.; Powers, J.M.; Federoff, H.; Eberwine, J. A neurotoxic phosphoform of elk-1 associates with inclusions from multiple neurodegenerative diseases. PLoS ONE 2010, 5, e9002. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Kim, A.J.; Chang, R.C.; Chang, J.Y.; Ying, W.; Ko, M.L.; Zhou, B.; Ko, G.Y. Deletion of mir-150 exacerbates retinal vascular overgrowth in high-fat-diet induced diabetic mice. PLoS ONE 2016, 11, e0157543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, E.; Ding, X.; Saadi, A.; Agarwal, N.; Naash, M.I.; Al-Ubaidi, M.R. Expression of cone-photoreceptor-specific antigens in a cell line derived from retinal tumors in transgenic mice. Invest Ophthalmol. Vis. Sci. 2004, 45, 5. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Yu, F.; Shi, L.; Ko, M.L.; Ko, G.Y. Melatonin affects mitochondrial fission/fusion dynamics in the diabetic retina. J. Diabetes Res. 2019, 2019, 8463125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, L.; Ko, S.; Ko, M.L.; Kim, A.J.; Ko, G.Y. Peptide lv augments l-type voltage-gated calcium channels through vascular endothelial growth factor receptor 2 (vegfr2) signaling. Biochim. Biophys. Acta 2015, 1853, 1154–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, A.J.; Chang, J.Y.; Shi, L.; Chang, R.C.; Ko, M.L.; Ko, G.Y. The effects of metformin on obesity-induced dysfunctional retinas. Invest Ophthalmol. Vis. Sci. 2017, 58, 106–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajagopal, R.; Bligard, G.W.; Zhang, S.; Yin, L.; Peter, L.; Clay, F.S. Functional deficits precede structural lesions in mice with high-fat diet–induced diabetic retinopathy. Diabetes 2016, 65, 13. [Google Scholar] [CrossRef] [Green Version]
- Asare-Bediako, B.; Noothi, S.K.; Li Calzi, S.; Athmanathan, B.; Vieira, C.P.; Adu-Agyeiwaah, Y.; Dupont, M.; Jones, B.A.; Wang, X.X.; Chakraborty, D.; et al. Characterizing the retinal phenotype in the high-fat diet and western diet mouse models of prediabetes. Cells 2020, 9, 464. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. Micrornas: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Wong-Riley, M.T. Energy metabolism of the visual system. Eye Brain 2010, 2, 99–116. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Yang, J.; Huang, Y.; Deng, L. Microrna-150 inhibitors enhance cell apoptosis of melanoma by targeting pdcd4. Oncol. Lett. 2018, 15, 1475–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, H.; Teng, H.; Qin, Y.; Luo, X.; Yang, P.; Zhang, W.; Chen, W.; Lv, D.; Tang, H. Extracellular vesicles derived from microrna-150-5p-overexpressing mesenchymal stem cells protect rat hearts against ischemia/reperfusion. Aging 2020, 12, 15. [Google Scholar] [CrossRef]
- Ling, Z.; Fan, G.; Yao, D.; Zhao, J.; Zhou, Y.; Feng, J.; Zhou, G.; Chen, Y. Microrna-150 functions as a tumor suppressor and sensitizes osteosarcoma to doxorubicin-induced apoptosis by targeting runx2. Exp. Ther. Med. 2020, 19, 481–488. [Google Scholar] [CrossRef] [Green Version]
- Qin, B.; Shu, Y.; Xiao, L.; Lu, T.; Lin, Y.; Yang, H.; Lu, Z. Microrna-150 targets elk1 and modulates the apoptosis induced by ox-ldl in endothelial cells. Mol. Cell. Biochem. 2017, 429, 45–58. [Google Scholar] [CrossRef]
- Ying, W.; Tseng, A.; Chang, R.C.; Wang, H.; Lin, Y.L.; Kanameni, S.; Brehm, T.; Morin, A.; Jones, B.; Splawn, T.; et al. Mir-150 regulates obesity-associated insulin resistance by controlling b cell functions. Sci. Rep. 2016, 6, 20176. [Google Scholar] [CrossRef]
- Demir, O.; Aysit, N.; Onder, Z.; Turkel, N.; Ozturk, G.; Sharrocks, A.D.; Kurnaz, I.A. Ets-domain transcription factor elk-1 mediates neuronal survival: Smn as a potential target. Biochim. Biophys. Acta 2011, 1812, 652–662. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, T.; Shareef, H.K.; Aljarah, A.K.; Ide, H.; Li, Y.; Kashiwagi, E.; Netto, G.J.; Zheng, Y.; Miyamoto, H. Elk1 is up-regulated by androgen in bladder cancer cells and promotes tumor progression. Oncotarget 2015, 6, 17. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Chen, M.; Wang, J.; Cao, G.; Chen, W.; Xu, J. Pcna-associated factor kiaa0101 transcriptionally induced by elk1 controls cell proliferation and apoptosis in nasopharyngeal carcinoma: An integrated bioinformatics and experimental study. Aging 2020, 12, 26. [Google Scholar] [CrossRef]
- Halestrap, A.P.; McStay, G.P.; Clarke, S.J. The permeability transition pore complex: Another view. Biochimie 2002, 84, 14. [Google Scholar] [CrossRef]
- Lavaur, J.; Bernard, F.; Trifilieff, P.; Pascoli, V.; Kappes, V.; Pages, C.; Vanhoutte, P.; Caboche, J. A tat-def-elk-1 peptide regulates the cytonuclear trafficking of elk-1 and controls cytoskeleton dynamics. J. Neurosci. 2007, 27, 14448–14458. [Google Scholar] [CrossRef]
- Wu, W.; Mosteller, R.D.; Broek, D. Sphingosine kinase protects lipopolysaccharide-activated macrophages from apoptosis. Mol. Cell Biol. 2004, 24, 7359–7369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, D.M.; Zhang, Y.T.; Lu, J.; Zheng, Y.L. Effects of microrna-129 and its target gene c-fos on proliferation and apoptosis of hippocampal neurons in rats with epilepsy via the mapk signaling pathway. J. Cell Physiol. 2018, 233, 6632–6643. [Google Scholar] [CrossRef]
- Fan, L.; Jiang, L.; Du, Z. Myeloid cell leukemia 1 (mcl(-1)) protects against 1-methyl-4-phenylpyridinium ion (mpp+) induced apoptosis in parkinson’s disease. Metab. Brain Dis. 2015, 30, 1269–1274. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, F.; Ko, M.L.; Ko, G.Y.-P. Decreased MicroRNA-150 Exacerbates Neuronal Apoptosis in the Diabetic Retina. Biomedicines 2021, 9, 1135. https://doi.org/10.3390/biomedicines9091135
Yu F, Ko ML, Ko GY-P. Decreased MicroRNA-150 Exacerbates Neuronal Apoptosis in the Diabetic Retina. Biomedicines. 2021; 9(9):1135. https://doi.org/10.3390/biomedicines9091135
Chicago/Turabian StyleYu, Fei, Michael L. Ko, and Gladys Y.-P. Ko. 2021. "Decreased MicroRNA-150 Exacerbates Neuronal Apoptosis in the Diabetic Retina" Biomedicines 9, no. 9: 1135. https://doi.org/10.3390/biomedicines9091135




