Self-Organization Provides Cell Fate Commitment in MSC Sheet Condensed Areas via ROCK-Dependent Mechanism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Reagents
2.2. Cell Sheet Assembly Procedure
2.3. Cell Culture Hematoxylin Staining in Cultures Dishes and Plates
2.4. Immunofluorescent Labeling of α-Smooth Muscle Actin (α-SMA)
2.5. Phalloidin Fluorescent Staining
2.6. PicoGreen Dye DNA Assay
2.7. Adipogenic, Osteogenic and Chondrogenic Differentiation of MSC
2.8. Alkaline Phosphatase Activity Assay
2.9. MitoTracker™ Probing for Mitochondrial Transmembrane Potential
2.10. Laser Microdissection Procedure
2.11. Protein Isolation, Electrophoresis and Western Blotting
2.12. Solid Phase Dot-ELISA
2.13. Lactate Dehydrogenase Activity Assay
2.14. AdipoQ Promoter Reporter Construct and AdipoQRep Cell Line
2.15. Adiponectin Reporter Assay and ROCK-1/2 Inhibitory Analysis
2.16. RNA-Seq Sample Preparation and Data Analysis
2.17. Statistical Analysis
3. Results
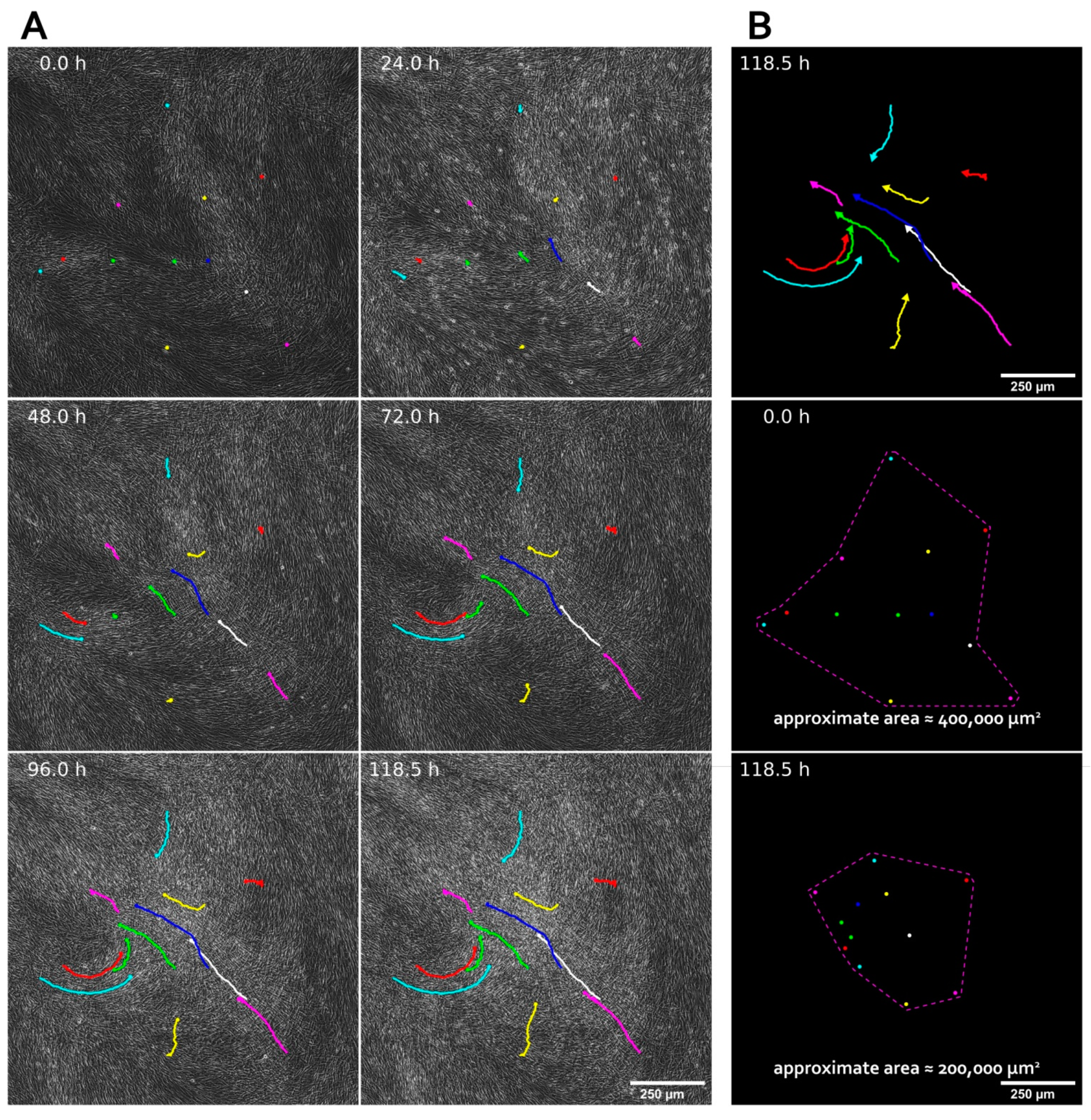
3.1. MSC Self-Organization in Cell Sheet Occurs through Condensation Mediated by Actin Cytoskeleton Rearrangement
3.2. MSC within CS Readily and More Effectively Undergo Osteogenic and Chondrogenic but Not Adipogenic Differentiation
3.3. MSC in Condensed Areas Demonstrate Early Signs of Commitment to Osteocytes Occurs Prior to the Induction of Differentiation
- mitochondrial transmembrane potential (MtMP);
- content of cytochrome C oxidase complex IV (COX-IV)—the terminal enzyme complex component of the respiratory chain;
- hypoxia inducible factor (HIF-1a);
- activity of lactate dehydrogenase (LDH)—the terminal enzyme of anaerobic glycolysis).
3.4. Transcriptomic Analysis of MSC in Scattered and Condensed Areas Reveals Dramatic Changes in Expression Profiles
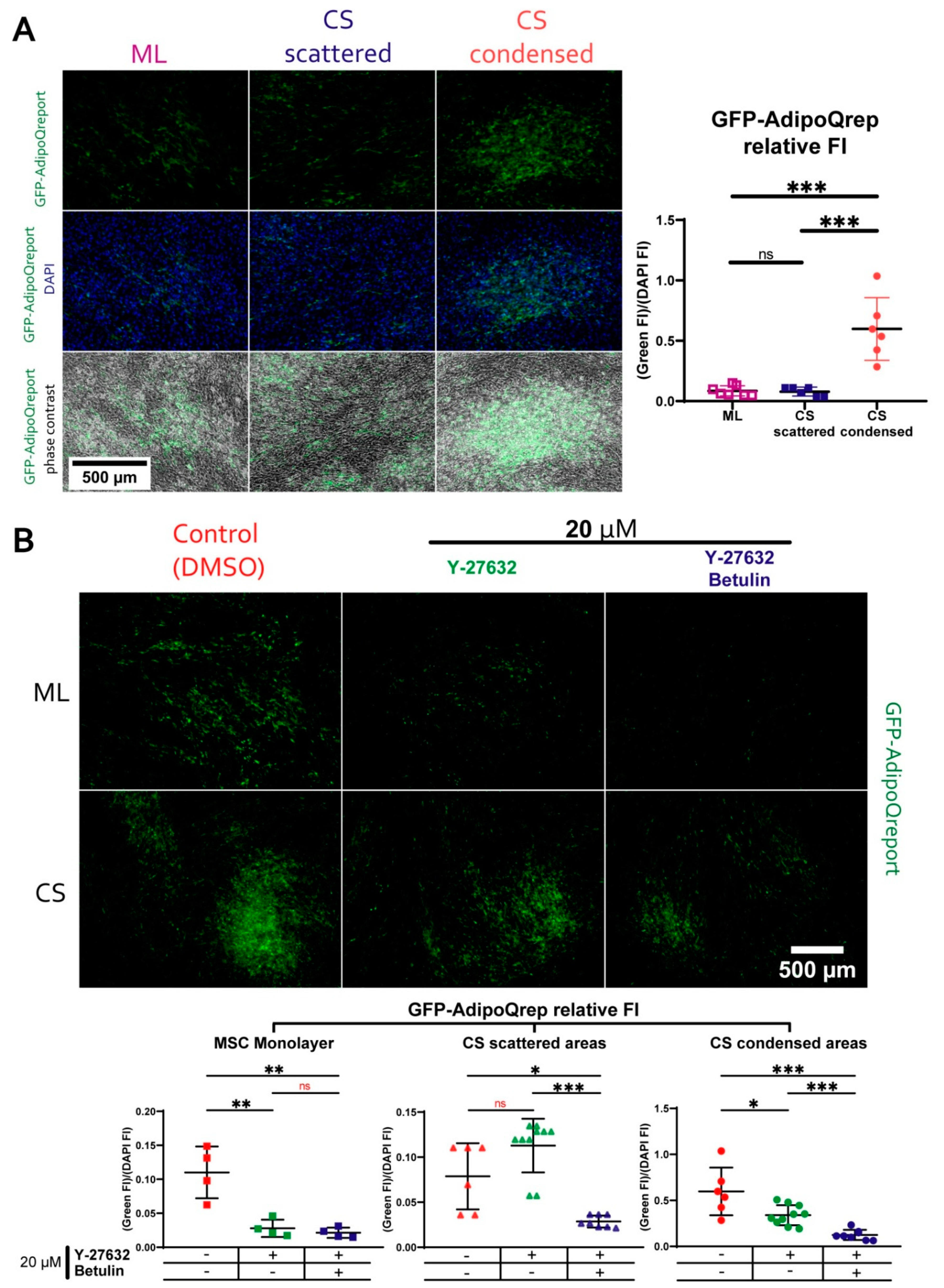
3.5. MSC Condensation in Cell Sheets Is Mediated by Activity of Rho GTPase Pathway
3.6. Reduced Adipogenic Potential of MSC in Condensed Areas Is Mediated by Local Activation of Rho GTPase Pathway
3.7. During Adipogenic Differentiation Adiponectin (AdipoQ) Expression in Condensed Areas Is Driven by Activation of Rho GTPase Pathway
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cohen, I.R.; Harel, D. Explaining a complex living system: Dynamics, multi-scaling and emergence. J. R. Soc. Interface 2006, 4, 175–182. [Google Scholar] [CrossRef] [Green Version]
- Forman, D.; Slack, J.M.W. Determination and cellular commitment in the embryonic amphibian mesoderm. Nature 1980, 286, 492–494. [Google Scholar] [CrossRef]
- Morales, J.S.; Raspopovic, J.; Marcon, L. From embryos to embryoids: How external signals and self-organization drive embryonic development. Stem Cell Rep. 2021, 16, 1039–1050. [Google Scholar] [CrossRef]
- Schweisguth, F.; Corson, F. Self-Organization in Pattern Formation. Dev. Cell 2019, 49, 659–677. [Google Scholar] [CrossRef] [PubMed]
- Sthijns, M.; Lapointe, V.L.; Van Blitterswijk, C.A. Building Complex Life Through Self-Organization. Tissue Eng. Part A 2019, 25, 1341–1346. [Google Scholar] [CrossRef] [Green Version]
- Kadoshima, T.; Sakaguchi, H.; Nakano, T.; Soen, M.; Ando, S.; Eiraku, M.; Sasai, Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA 2013, 110, 20284–20289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karus, M.; Blaess, S.; Brüstle, O. Self-organization of neural tissue architectures from pluripotent stem cells. J. Comp. Neurol. 2014, 522, 2831–2844. [Google Scholar] [CrossRef]
- Serra, D.; Mayr, U.; Boni, A.; Lukonin, I.; Rempfler, M.; Meylan, L.C.; Stadler, M.B.; Strnad, P.; Papasaikas, P.; Vischi, D.; et al. Self-organization and symmetry breaking in intestinal organoid development. Nature 2019, 569, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Sawada, Y.; Sugiyama, T. Minimum Tissue Size Required for Hydra Regeneration. Dev. Biol. 1993, 155, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Sasai, Y. Cytosystems dynamics in self-organization of tissue architecture. Nature 2013, 493, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Iismaa, S.E.; Kaidonis, X.; Nicks, A.M.; Bogush, N.; Kikuchi, K.; Naqvi, N.; Harvey, R.; Husain, A.; Graham, R.M. Comparative regenerative mechanisms across different mammalian tissues. NPJ Regen. Med. 2018, 3, 1–20. [Google Scholar] [CrossRef]
- Eremichev, R.; Kulebyakina, M.; Alexandrushkina, N.; Nimiritsky, P.; Basalova, N.; Grigorieva, O.; Egiazaryan, M.; Dyikanov, D.; Tkachuk, V.; Makarevich, P. Scar-Free Healing of Endometrium: Tissue-Specific Program of Stromal Cells and Its Induction by Soluble Factors Produced After Damage. Front. Cell Dev. Biol. 2021, 9, 212. [Google Scholar] [CrossRef]
- Pispa, J.; Thesleff, I. Mechanisms of ectodermal organogenesis. Dev. Biol. 2003, 262, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Olsen, B.R.; Reginato, A.M.; Wang, W. Bone Development. Annu. Rev. Cell Dev. Biol. 2000, 16, 191–220. [Google Scholar] [CrossRef]
- Ghosh, S.; Laha, M.; Mondal, S.; Sengupta, S.; Kaplan, D.L. In vitro model of mesenchymal condensation during chondrogenic development. Biomaterials 2009, 30, 6530–6540. [Google Scholar] [CrossRef] [Green Version]
- Mammoto, T.; Mammoto, A.; Torisawa, Y.-S.; Tat, T.; Gibbs, A.; Derda, R.; Mannix, R.; De Bruijn, M.; Yung, C.W.; Huh, D.; et al. Mechanochemical Control of Mesenchymal Condensation and Embryonic Tooth Organ Formation. Dev. Cell 2011, 21, 758–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Zhu, S.; Zhang, C.; Lu, P.; Hu, J.; Yin, Z.; Ma, Y.; Chen, X.; OuYang, H. Crucial transcription factors in tendon development and differentiation: Their potential for tendon regeneration. Cell Tissue Res. 2014, 356, 287–298. [Google Scholar] [CrossRef]
- Hilfer, S.; Rayner, R.M.; Brown, J.W. Mesenchymal control of branching pattern in the fetal mouse lung. Tissue Cell 1985, 17, 523–538. [Google Scholar] [CrossRef]
- Moore, K.A.; Polte, T.; Huang, S.; Shi, B.; Alsberg, E.; Sunday, M.E.; Ingber, D.E. Control of basement membrane remodeling and epithelial branching morphogenesis in embryonic lung by Rho and cytoskeletal tension. Dev. Dyn. 2005, 232, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Urban, A.E.; Zhou, X.; Ungos, J.M.; Raible, D.W.; Altmann, C.R.; Vize, P.D. FGF is essential for both condensation and mesenchymal–epithelial transition stages of pronephric kidney tubule development. Dev. Biol. 2006, 297, 103–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindström, N.O.; Hohenstein, P.; Davies, J.A. Nephrons require Rho-kinase for proximal-distal polarity development. Sci. Rep. 2013, 3, 2692. [Google Scholar] [CrossRef] [Green Version]
- Shinozawa, T.; Yoshikawa, H.Y.; Takebe, T. Reverse engineering liver buds through self-driven condensation and organization towards medical application. Dev. Biol. 2016, 420, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Takebe, T.; Enomura, M.; Yoshizawa, E.; Kimura, M.; Koike, H.; Ueno, Y.; Matsuzaki, T.; Yamazaki, T.; Toyohara, T.; Osafune, K.; et al. Vascularized and Complex Organ Buds from Diverse Tissues via Mesenchymal Cell-Driven Condensation. Cell Stem Cell 2015, 16, 556–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimitriou, R.; Jones, E.; McGonagle, D.; Giannoudis, P.V. Bone regeneration: Current concepts and future directions. BMC Med. 2011, 9, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, T.; Ugawa, Y.; Yamashiro, K.; Shimoe, M.; Tomikawa, K.; Hongo, S.; Kochi, S.; Ideguchi, H.; Maeda, H.; Takashiba, S. Osteogenic differentiation regulated by Rho-kinase in periodontal ligament cells. Differentiation 2014, 88, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.C.; Jacobs, C.R. Mechanically induced osteogenic lineage commitment of stem cells. Stem Cell Res. Ther. 2013, 4, 107. [Google Scholar] [CrossRef] [Green Version]
- Vervoort, M. Regeneration and Development in Animals. Biol. Theory 2011, 6, 25–35. [Google Scholar] [CrossRef]
- Goldman, D. Regeneration, morphogenesis and self-organization. Development 2014, 141, 2745–2749. [Google Scholar] [CrossRef] [Green Version]
- Laurent, J.; Blin, G.; Chatelain, F.; Vanneaux, V.; Fuchs, A.; Larghero, J.; Théry, M. Convergence of microengineering and cellular self-organization towards functional tissue manufacturing. Nat. Biomed. Eng. 2017, 1, 939–956. [Google Scholar] [CrossRef]
- Correia, C.R.; Bjørge, I.M.; Nadine, S.; Mano, J.F. Minimalist Tissue Engineering Approaches Using Low Material-Based Bioengineered Systems. Adv. Health Mater. 2021, 10, 2002110. [Google Scholar] [CrossRef] [PubMed]
- Toda, S.; Blauch, L.R.; Tang, S.K.Y.; Morsut, L.; Lim, W.A. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science 2018, 361, 156–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nimiritsky, P.P.; Eremichev, R.; Alexandrushkina, N.A.; Efimenko, A.Y.; Tkachuk, V.A.; Makarevich, P.I. Unveiling Mesenchymal Stromal Cells’ Organizing Function in Regeneration. Int. J. Mol. Sci. 2019, 20, 823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Chem. Biol. 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexandrushkina, N.; Nimiritsky, P.; Eremichev, R.; Popov, V.; Arbatskiy, M.; Danilova, N.; Malkov, P.; Akopyan, Z.; Tkachuk, V.; Makarevich, P. Cell Sheets from Adipose Tissue MSC Induce Healing of Pressure Ulcer and Prevent Fibrosis via Trigger Effects on Granulation Tissue Growth and Vascularization. Int. J. Mol. Sci. 2020, 21, 5567. [Google Scholar] [CrossRef]
- Podgorny, O.V.; Lazarev, V.N. Laser microdissection: A promising tool for exploring microorganisms and their interactions with hosts. J. Microbiol. Methods 2017, 138, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Goldring, J.P.D. Measuring Protein Concentration on Nitrocellulose and After the Electrophoretic Transfer of Protein to Nitrocellulose. Methods Mol. Biol. 2015, 1314, 19–25. [Google Scholar] [CrossRef]
- Longo, P.A.; Kavran, J.M.; Kim, M.-S.; Leahy, D.J. Transient Mammalian Cell Transfection with Polyethylenimine (PEI). Methods Enzymol. 2013, 529, 227–240. [Google Scholar] [CrossRef] [Green Version]
- Koshiishi, C.; Park, H.-M.; Uchiyama, H.; Tanaka, Y. Regulation of expression of the mouse adiponectin gene by the C/EBP family via a novel enhancer region. Gene 2008, 424, 141–146. [Google Scholar] [CrossRef]
- Uhrig-Schmidt, S.; Geiger, M.; Luippold, G.; Birk, G.; Mennerich, D.; Neubauer, H.; Grimm, D.; Wolfrum, C.; Kreuz, S. Gene Delivery to Adipose Tissue Using Transcriptionally Targeted rAAV8 Vectors. PLoS ONE 2014, 9, e116288. [Google Scholar] [CrossRef] [Green Version]
- Tyurin-Kuzmin, P.A.; Karagyaur, M.N.; Kulebyakin, K.Y.; Dyikanov, D.T.; Chechekhin, V.I.; Ivanova, A.M.; Skryabina, M.N.; Arbatskiy, M.S.; Sysoeva, V.Y.; Kalinina, N.I.; et al. Functional Heterogeneity of Protein Kinase A Activation in Multipotent Stromal Cells. Int. J. Mol. Sci. 2020, 21, 4442. [Google Scholar] [CrossRef]
- Babraham Bioinformatics. FastQC v. 0.11.2. 2014. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 January 2020).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17. [Google Scholar] [CrossRef]
- Joshi, N.; Fass, J. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33) [Software]. 2011. Available online: https://github.com/najoshi/sickle (accessed on 26 July 2021).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.; Smyth, G. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; He, Q.-Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. BioSyst. 2015, 12, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Majack, R.A. Beta-type transforming growth factor specifies organizational behavior in vascular smooth muscle cell cultures. J. Cell Biol. 1987, 105, 465–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faries, P.L.; Rohan, D.I.; Takahara, H.; Wyers, M.C.; Contreras, M.A.; Quist, W.C.; King, G.L.; LoGerfo, F.W. Human vascular smooth muscle cells of diabetic origin exhibit increased proliferation, adhesion, and migration. J. Vasc. Surg. 2001, 33, 601–607. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Wang, M.; Yan, Z.; Zhang, S.; Liu, H. Isolation and culture of vascular smooth muscle cells from rat placenta. J. Cell. Physiol. 2018, 234, 7675–7682. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.R.; Wang, C.J.; Xu, C.Y.; Wu, X.K.; Hong, D.; Shi, W.; Gong, Y.; Chen, H.X.; Long, F.; Wu, X.M. Signaling Cascades Governing Cdc42-Mediated Chondrogenic Differentiation and Mensenchymal Condensation. Genetics 2016, 202, 1055–1069. [Google Scholar] [CrossRef] [Green Version]
- Stefkova, K.; Prochazkova, J.; Pacherník, J. Alkaline Phosphatase in Stem Cells. Stem Cells Int. 2015, 2015, 628368. [Google Scholar] [CrossRef] [Green Version]
- Salasznyk, R.M.; Klees, R.F.; Hughlock, M.K.; Plopper, G.E. ERK Signaling Pathways Regulate the Osteogenic Differentiation of Human Mesenchymal Stem Cells on Collagen I and Vitronectin. Cell Commun. Adhes. 2004, 11, 137–153. [Google Scholar] [CrossRef] [PubMed]
- Novoseletskaya, E.; Grigorieva, O.; Nimiritsky, P.; Basalova, N.; Eremichev, R.; Milovskaya, I.; Kulebyakin, K.; Kulebyakina, M.; Rodionov, S.; Omelyanenko, N.; et al. Mesenchymal Stromal Cell-Produced Components of Extracellular Matrix Potentiate Multipotent Stem Cell Response to Differentiation Stimuli. Front. Cell Dev. Biol. 2020, 8, 983. [Google Scholar] [CrossRef]
- Li, Y.; Park, J.-S.; Deng, J.-H.; Bai, Y. Cytochrome c oxidase subunit IV is essential for assembly and respiratory function of the enzyme complex. J. Bioenerg. Biomembr. 2006, 38, 283–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernhardt, A.; Lode, A.; Mietrach, C.; Hempel, U.; Hänke, T.; Gelinsky, M. In vitroosteogenic potential of human bone marrow stromal cells cultivated in porous scaffolds from mineralized collagen. J. Biomed. Mater. Res. Part A 2009, 90, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.; Chapman, S.C. Cytoskeletal Reorganization Drives Mesenchymal Condensation and Regulates Downstream Molecular Signaling. PLoS ONE 2015, 10, e0134702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strzelecka-Kiliszek, A.; Mebarek, S.; Roszkowska, M.; Buchet, R.; Magne, D.; Pikula, S. Functions of Rho family of small GTPases and Rho-associated coiled-coil kinases in bone cells during differentiation and mineralization. Biochim. et Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 1009–1023. [Google Scholar] [CrossRef]
- Stanley, A.; Heo, S.; Mauck, R.L.; Mourkioti, F.; Shore, E.M. Elevated BMP and Mechanical Signaling Through YAP1/RhoA Poises FOP Mesenchymal Progenitors for Osteogenesis. J. Bone Miner. Res. 2019, 34, 1894–1909. [Google Scholar] [CrossRef]
- Gegg, C.; Yang, F. The Effects of ROCK Inhibition on Mesenchymal Stem Cell Chondrogenesis Are Culture Model Dependent. Tissue Eng. Part A 2020, 26, 130–139. [Google Scholar] [CrossRef]
- Schweizer, M.; Röder, K.; Zhang, L.; Wolf, S.S. Transcription factors acting on the promoter of the rat fatty acid synthase gene. Biochem. Soc. Trans. 2002, 30, 1070–1072. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Luo, J.; Xu, H.; Wang, M.; Zhu, J.; Shi, H.; Haile, A.B.; Wang, H.; Sun, Y. Fatty acid synthase promoter: Characterization, and transcriptional regulation by sterol regulatory element binding protein-1 in goat mammary epithelial cells. Gene 2015, 561, 157–164. [Google Scholar] [CrossRef]
- Bertolio, R.; Napoletano, F.; Mano, M.; Maurer-Stroh, S.; Fantuz, M.; Zannini, A.; Bicciato, S.; Sorrentino, G.; Del Sal, G. Sterol regulatory element binding protein 1 couples mechanical cues and lipid metabolism. Nat. Commun. 2019, 10, 1326. [Google Scholar] [CrossRef] [Green Version]
- Yin, F.; Feng, F.; Wang, L.; Wang, X.; Li, Z.; Cao, Y. SREBP-1 inhibitor Betulin enhances the antitumor effect of Sorafenib on hepatocellular carcinoma via restricting cellular glycolytic activity. Cell Death Dis. 2019, 10, 672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Straub, L.G.; Scherer, P.E. Metabolic Messengers: Adiponectin. Nat. Metab. 2019, 1, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Bartosh, T.J.; Ylöstalo, J.H.; Mohammadipoor, A.; Bazhanov, N.; Coble, K.; Claypool, K.; Lee, R.H.; Choi, H.; Prockop, D.J. Aggregation of human mesenchymal stromal cells (MSCs) into 3D spheroids enhances their antiin fl ammatory properties. Proc. Natl. Acad. Sci. USA 2010, 107, 13724–13729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, P.-C.; Young, T.-H.; Wang, T.-M.; Peng, H.-W.; Hou, S.-M.; Yen, M.-L. Spontaneous osteogenesis of MSCs cultured on 3D microcarriers through alteration of cytoskeletal tension. Biomaterials 2012, 33, 556–564. [Google Scholar] [CrossRef]
- Imamura, A.; Kajiya, H.; Fujisaki, S.; Maeshiba, M.; Yanagi, T.; Kojima, H.; Ohno, J. Three-dimensional spheroids of mesenchymal stem/stromal cells promote osteogenesis by activating stemness and Wnt/β-catenin. Biochem. Biophys. Res. Commun. 2019, 523, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Laschke, M.W.; Menger, M.D. Life is 3D: Boosting Spheroid Function for Tissue Engineering. Trends Biotechnol. 2016, 35, 133–144. [Google Scholar] [CrossRef]
- Cesarz, Z.; Tamama, K. Spheroid Culture of Mesenchymal Stem Cells. Stem Cells Int. 2015, 2016, 9176357. [Google Scholar] [CrossRef] [Green Version]
- Saykali, B.; Mathiah, N.; Nahaboo, W.; Racu, M.-L.; Hammou, L.; Defrance, M.; Migeotte, I. Distinct mesoderm migration phenotypes in extra-embryonic and embryonic regions of the early mouse embryo. eLife 2019, 8, e42434. [Google Scholar] [CrossRef]
- Hughes, A.J.; Miyazaki, H.; Coyle, M.C.; Zhang, J.; Laurie, M.T.; Chu, D.; Vavrušová, Z.; Schneider, R.A.; Klein, O.D.; Gartner, Z.J. Engineered Tissue Folding by Mechanical Compaction of the Mesenchyme. Dev. Cell 2017, 44, 165–178.e6. [Google Scholar] [CrossRef] [Green Version]
- Ghone, N.V.; Grayson, W.L. Recapitulation of mesenchymal condensation enhances in vitro chondrogenesis of human mesenchymal stem cells. J. Cell. Physiol. 2012, 227, 3701–3708. [Google Scholar] [CrossRef]
- McDermott, A.M.; Herberg, S.; Mason, D.E.; Collins, J.M.; Pearson, H.B.; Dawahare, J.H.; Tang, R.; Patwa, A.N.; Grinstaff, M.W.; Kelly, D.J.; et al. Recapitulating bone development through engineered mesenchymal condensations and mechanical cues for tissue regeneration. Sci. Transl. Med. 2019, 11, eaav7756. [Google Scholar] [CrossRef]
- Sarem, M.; Otto, O.; Tanaka, S.; Shastri, V.P. Cell number in mesenchymal stem cell aggregates dictates cell stiffness and chondrogenesis. Stem Cell Res. Ther. 2019, 10, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, B.K.; Miyake, T. All for one and one for all: Condensations and the initiation of skeletal development. Bioessays 2000, 22, 138–147. [Google Scholar] [CrossRef]
- Lam, J.; Bellayr, I.H.; Marklein, R.A.; Bauer, S.R.; Puri, R.K.; Sung, K.E. Functional Profiling of Chondrogenically Induced Multipotent Stromal Cell Aggregates Reveals Transcriptomic and Emergent Morphological Phenotypes Predictive of Differentiation Capacity. Stem Cells Transl. Med. 2018, 7, 664–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldring, M.B.; Tsuchimochi, K.; Ijiri, K. The control of chondrogenesis. J. Cell. Biochem. 2005, 97, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Chatzeli, L.; Simons, B.D. Tracing the Dynamics of Stem Cell Fate. Cold Spring Harb. Perspect. Biol. 2020, 12, a036202. [Google Scholar] [CrossRef]
- Mendes, S.C.; Tibbe, J.M.; Veenhof, M.; Both, S.; Oner, F.; van Blitterswijk, C.; De Bruijn, J.D. Relation between in vitro and in vivo osteogenic potential of cultured human bone marrow stromal cells. J. Mater. Sci. Mater. Med. 2004, 15, 1123–1128. [Google Scholar] [CrossRef]
- Kadenbach, B. Regulation of mitochondrial respiration and ATP synthesis via cytochrome c oxidase. Rend. Lince 2018, 29, 421–435. [Google Scholar] [CrossRef]
- Barilani, M.; Palorini, R.; Votta, G.; Piras, R.; Buono, G.; Grassi, M.; Bollati, V.; Chiaradonna, F.; Lazzari, L. Central metabolism of functionally heterogeneous mesenchymal stromal cells. Sci. Rep. 2019, 9, 15420. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-T.; Shih, Y.-R.V.; Kuo, T.K.; Lee, O.K.; Wei, Y.-H. Coordinated Changes of Mitochondrial Biogenesis and Antioxidant Enzymes During Osteogenic Differentiation of Human Mesenchymal Stem Cells. Stem Cells 2008, 26, 960–968. [Google Scholar] [CrossRef]
- Esen, E.; Long, F. Aerobic Glycolysis in Osteoblasts. Curr. Osteoporos. Rep. 2014, 12, 433–438. [Google Scholar] [CrossRef] [Green Version]
- Hong, M.; Zhang, X.-B.; Xiang, F.; Fei, X.; Ouyang, X.-L.; Peng, X.-C. MiR-34a suppresses osteoblast differentiation through glycolysis inhibition by targeting lactate dehydrogenase-A (LDHA). Vitr. Cell. Dev. Biol. Anim. 2020, 56, 480–487. [Google Scholar] [CrossRef]
- Karner, C.; Long, F. Wnt signaling and cellular metabolism in osteoblasts. Cell. Mol. Life Sci. 2016, 74, 1649–1657. [Google Scholar] [CrossRef] [PubMed]
- Esen, E.; Chen, J.; Karner, C.; Okunade, A.L.; Patterson, B.W.; Long, F. WNT-LRP5 Signaling Induces Warburg Effect through mTORC2 Activation during Osteoblast Differentiation. Cell Metab. 2013, 17, 745–755. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Wang, M.; Feng, H.; Peng, Y.; Sun, J.; Qu, X.; Li, C. Lactate induces osteoblast differentiation by stabilization of HIF1α. Mol. Cell. Endocrinol. 2017, 452, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, N.; Kajiya, M.; Motoike, S.; Takewaki, M.; Horikoshi, S.; Iwata, T.; Ouhara, K.; Takeda, K.; Matsuda, S.; Fujita, T.; et al. Type I collagen deposition via osteoinduction ameliorates YAP/TAZ activity in 3D floating culture clumps of mesenchymal stem cell/extracellular matrix complexes. Stem Cell Res. Ther. 2018, 9, 342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghaemi, S.R.; Delalat, B.; Cetó, X.; Harding, F.J.; Tuke, J.; Voelcker, N.H. Synergistic influence of collagen I and BMP 2 drives osteogenic differentiation of mesenchymal stem cells: A cell microarray analysis. Acta Biomater. 2016, 34, 41–52. [Google Scholar] [CrossRef]
- Oosterlaken, B.M.; Vena, M.P.; de With, G. In Vitro Mineralization of Collagen. Adv. Mater. 2021, 33, 2004418. [Google Scholar] [CrossRef]
- Lee, M.J.; Agrahari, G.; Kim, H.-Y.; An, E.-J.; Chun, K.-H.; Kang, H.; Kim, Y.-S.; Bang, C.W.; Tak, L.-J.; Kim, T.-Y. Extracellular Superoxide Dismutase Prevents Skin Aging by Promoting Collagen Production through the Activation of AMPK and Nrf2/HO-1 Cascades. J. Investig. Dermatol. 2021, in press. [Google Scholar] [CrossRef]
- Wheeler, A.P.; Ridley, A. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp. Cell Res. 2004, 301, 43–49. [Google Scholar] [CrossRef]
- Hodge, R.; Ridley, A. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 2016, 17, 496–510. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.M.; Gleghorn, J. Sculpting Organs: Mechanical Regulation of Tissue Development. Annu. Rev. Biomed. Eng. 2012, 14, 129–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffin, M.J.; Wong, R.H.F.; Pandya, N.; Sul, H.S. Direct Interaction between USF and SREBP-1c Mediates Synergistic Activation of the Fatty-acid Synthase Promoter. J. Biol. Chem. 2007, 282, 5453–5467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBeath, R.; Pirone, D.M.; Nelson, C.M.; Bhadriraju, K.; Chen, C.S. Cell Shape, Cytoskeletal Tension, and RhoA Regulate Stem Cell Lineage Commitment. Dev. Cell 2004, 6, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Barnea, M.; Chapnik, N.; Genzer, Y.; Froy, O. The circadian clock machinery controls adiponectin expression. Mol. Cell. Endocrinol. 2015, 399, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, J.; Lerin, C.; Gerhart-Hines, Z.; Puigserver, P. Metabolic adaptations through the PGC-1α and SIRT1 pathways. FEBS Lett. 2007, 582, 46–53. [Google Scholar] [CrossRef] [Green Version]
- Imran, K.M.; Rahman, N.; Yoon, D.; Jeon, M.; Lee, B.-T.; Kim, Y.-S. Cryptotanshinone promotes commitment to the brown adipocyte lineage and mitochondrial biogenesis in C3H10T1/2 mesenchymal stem cells via AMPK and p38-MAPK signaling. Biochim. et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.N.; Clark, J.P.; Anderson, R.M. Mitochondrial regulator PGC-1a—Modulating the modulator. Curr. Opin. Endocr. Metab. Res. 2019, 5, 37–44. [Google Scholar] [CrossRef]
- Qiao, L.; Lee, B.; Kinney, B.; Yoo, H.S.; Shao, J. Energy intake and adiponectin gene expression. Am. J. Physiol. Metab. 2011, 300, E809–E816. [Google Scholar] [CrossRef]
- Almabrouk, T.A.M.; Ugusman, A.B.; Katwan, O.J.; Salt, I.P.; Kennedy, S. Deletion of AMPKα1 attenuates the anticontractile effect of perivascular adipose tissue (PVAT) and reduces adiponectin release. Br. J. Pharmacol. 2016, 174, 3398–3410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almabrouk, T.A.M.; White, A.D.; Ugusman, A.B.; Skiba, D.; Katwan, O.J.; Alganga, H.; Guzik, T.J.; Touyz, R.M.; Salt, I.; Kennedy, S. High Fat Diet Attenuates the Anticontractile Activity of Aortic PVAT via a Mechanism Involving AMPK and Reduced Adiponectin Secretion. Front. Physiol. 2018, 9, 51. [Google Scholar] [CrossRef]
- Viswakarma, N.; Jia, Y.; Bai, L.; Vluggens, A.; Borensztajn, J.; Xu, J.; Reddy, J.K. Coactivators in PPAR-Regulated Gene Expression. PPAR Res. 2010, 2010, 250126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daval, M.; Foufelle, F.; Ferré, P. Functions of AMP-activated protein kinase in adipose tissue. J. Physiol. 2006, 574, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Zulian, A.; Cancello, R.; Girola, A.; Gilardini, L.; Alberti, L.; Croci, M.; Micheletto, G.; Danelli, P.; Invitti, C. In vitro and in vivo Effects of Metformin on Human Adipose Tissue Adiponectin. Obes. Facts 2011, 4, 27–33. [Google Scholar] [CrossRef]
- Shimada, T.; Nakayama, Y.; Harasawa, Y.; Matsui, H.; Kobayashi, H.; Sai, Y.; Miyamoto, K.-I.; Tomatsu, S.; Aburada, M. Salacia reticulata has therapeutic effects on obesity. J. Nat. Med. 2014, 68, 668–676. [Google Scholar] [CrossRef]
- Ke, R.; Xu, Q.; Li, C.; Luo, L.; Huang, D. Mechanisms of AMPK in the maintenance of ATP balance during energy metabolism. Cell Biol. Int. 2018, 42, 384–392. [Google Scholar] [CrossRef] [Green Version]
- Viscomi, C.; Bottani, E.; Civiletto, G.; Cerutti, R.; Moggio, M.; Fagiolari, G.; Schon, E.A.; Lamperti, C.; Zeviani, M. In Vivo Correction of COX Deficiency by Activation of the AMPK/PGC-1α Axis. Cell Metab. 2011, 14, 80–90. [Google Scholar] [CrossRef] [Green Version]
- Summermatter, S.; Santos, G.; Pérez-Schindler, J.; Handschin, C. Skeletal muscle PGC-1 controls whole-body lactate homeostasis through estrogen-related receptor -dependent activation of LDH B and repression of LDH A. Proc. Natl. Acad. Sci. USA 2013, 110, 8738–8743. [Google Scholar] [CrossRef] [Green Version]
- de Diego, C.S.; Artigas, N.; Lopes, C.I.P.C.d.S.; Valer, J.A.; Torrejon, B.; Gama-Pérez, P.; A Villena, J.; Garcia-Roves, P.M.; Rosa, J.L.; Ventura, F. Glucose Restriction Promotes Osteocyte Specification by Activating a PGC-1α-Dependent Transcriptional Program. iScience 2019, 15, 79–94. [Google Scholar] [CrossRef]
- Jang, W.G.; Kim, E.J.; Lee, K.-N.; Son, H.-J.; Koh, J.-T. AMP-activated protein kinase (AMPK) positively regulates osteoblast differentiation via induction of Dlx5-dependent Runx2 expression in MC3T3E1 cells. Biochem. Biophys. Res. Commun. 2011, 404, 1004–1009. [Google Scholar] [CrossRef]
- Wang, Y.-G.; Qu, X.-H.; Yang, Y.; Han, X.-G.; Wang, L.; Qiao, H.; Fan, Q.-M.; Tang, T.-T.; Dai, K.-R. AMPK promotes osteogenesis and inhibits adipogenesis through AMPK-Gfi1-OPN axis. Cell. Signal. 2016, 28, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Pantovic, A.; Krstic, J.; Janjetovic, K.; Kocic, J.; Harhaji-Trajkovic, L.; Bugarski, D.; Trajkovic, V. Coordinated time-dependent modulation of AMPK/Akt/mTOR signaling and autophagy controls osteogenic differentiation of human mesenchymal stem cells. Bone 2013, 52, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, I.; Yamaguchi, T.; Yano, S.; Yamauchi, M.; Sugimoto, T. Metformin enhances the differentiation and mineralization of osteoblastic MC3T3-E1 cells via AMP kinase activation as well as eNOS and BMP-2 expression. Biochem. Biophys. Res. Commun. 2008, 375, 414–419. [Google Scholar] [CrossRef]
- Bahrambeigi, S.; Yousefi, B.; Rahimi, M.; Shafiei-Irannejad, V. Metformin; an old antidiabetic drug with new potentials in bone disorders. Biomed. Pharmacother. 2018, 109, 1593–1601. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-G.; Han, X.-G.; Yang, Y.; Qiao, H.; Dai, K.-R.; Fan, Q.-M.; Tang, T.-T. Functional differences between AMPK α1 and α2 subunits in osteogenesis, osteoblast-associated induction of osteoclastogenesis, and adipogenesis. Sci. Rep. 2016, 6, 32771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.-Y.; Kim, E.-J.; Jang, W.-G. Piperine induces osteoblast differentiation through AMPK-dependent Runx2 expression. Biochem. Biophys. Res. Commun. 2017, 495, 1497–1502. [Google Scholar] [CrossRef]
- Sedlinsky, C.; Molinuevo, M.S.; Cortizo, A.M.; Tolosa, M.J.; Felice, J.I.; Sbaraglini, M.L.; Schurman, L.; McCarthy, A.D. Metformin prevents anti-osteogenic in vivo and ex vivo effects of rosiglitazone in rats. Eur. J. Pharmacol. 2011, 668, 477–485. [Google Scholar] [CrossRef]
- Liu, X.; Chen, T.; Wu, Y.; Tang, Z. Role and mechanism of PTEN in adiponectin-induced osteogenesis in human bone marrow mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2017, 483, 712–717. [Google Scholar] [CrossRef]
- Oshima, K.; Nampei, A.; Matsuda, M.; Iwaki, M.; Fukuhara, A.; Hashimoto, J.; Yoshikawa, H.; Shimomura, I. Adiponectin increases bone mass by suppressing osteoclast and activating osteoblast. Biochem. Biophys. Res. Commun. 2005, 331, 520–526. [Google Scholar] [CrossRef]
- Kanazawa, I.; Yamaguchi, T.; Yano, S.; Yamauchi, M.; Yamamoto, M.; Sugimoto, T. Adiponectin and AMP kinase activator stimulate proliferation, differentiation, and mineralization of osteoblastic MC3T3-E1 cells. BMC Cell Biol. 2007, 8, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, X.-H.; Guo, L.-J.; Yuan, L.-Q.; Xie, H.; Zhou, H.-D.; Wu, X.-P.; Liao, E.-Y. Adiponectin stimulates human osteoblasts proliferation and differentiation via the MAPK signaling pathway. Exp. Cell Res. 2005, 309, 99–109. [Google Scholar] [CrossRef]









| Method or Assay | Procedure | Equipment | Notes |
|---|---|---|---|
| Cell sheet assembly procedure; Adiponectin reporter assay and ROCK-1/2 inhibitory analysis | Long-term microscopic observations of the cell sheet assembly and fluorescence assays (coculture with endothelium or differentiation experiments) | Nikon ECLIPSE Ti microscope (Nikon, Tokyo, Japan) | Monitoring of live cells in culture gas mixture, temperature and humidity conditions suitable for long-term growth. Photocapture was carried out for 12–16 days, with a frequency of shooting each 40 min using the control software Nikon program NIS—Elements Advanced (Nikon, Tokyo, Japan) |
| Images or videos processing in all cases | Image and videos processing in all experiments | Fiji plugin collection for NIH ImageJ (1.53c version, NIH, Bethesda, MD, USA) [33] | |
| Cell culture hematoxylin staining in cultures dishes and plates; Adipogenic, osteogenic, and chondrogenic differentiation of MSC; Alkaline phosphatase activity assay | Acquisition and counts of condensed areas; assessment of the differentiation at the endpoint; visualization of ALP activity after incubation with chromogenic substrates | Nikon SMZ18 (Nikon, Tokyo, Japan) stereomicroscope | Surface and number of condensed areas in each well were measured using unified thresholding and binarization procedures in Fiji plugin collection for NIH ImageJ (1.53c version, NIH, Bethesda, MD, USA) |
| Immunofluorescent labeling of α-smooth muscle actin (α-SMA) and Phalloidin fluorescent staining; Adipogenic, osteogenic and chondrogenic differentiation of MSC; Alkaline phosphatase activity assay; MitoTracker™ probing for mitochondrial transmembrane potential; Adiponectin reporter assay and ROCK-1/2 inhibitory analysis | Visualization and image acquisition | Leica DMi8 inverted fluorescence microscope with a DFC7000T camera (Leica Microsystems, Wetzlar, Germany). | Additional image processing and analysis was carried out in the Fiji plugin collection for NIH ImageJ (1.53c version, NIH, Bethesda, MD, USA) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nimiritsky, P.; Novoseletskaya, E.; Eremichev, R.; Alexandrushkina, N.; Karagyaur, M.; Vetrovoy, O.; Basalova, N.; Khrustaleva, A.; Tyakht, A.; Efimenko, A.; et al. Self-Organization Provides Cell Fate Commitment in MSC Sheet Condensed Areas via ROCK-Dependent Mechanism. Biomedicines 2021, 9, 1192. https://doi.org/10.3390/biomedicines9091192
Nimiritsky P, Novoseletskaya E, Eremichev R, Alexandrushkina N, Karagyaur M, Vetrovoy O, Basalova N, Khrustaleva A, Tyakht A, Efimenko A, et al. Self-Organization Provides Cell Fate Commitment in MSC Sheet Condensed Areas via ROCK-Dependent Mechanism. Biomedicines. 2021; 9(9):1192. https://doi.org/10.3390/biomedicines9091192
Chicago/Turabian StyleNimiritsky, Peter, Ekaterina Novoseletskaya, Roman Eremichev, Natalia Alexandrushkina, Maxim Karagyaur, Oleg Vetrovoy, Nataliya Basalova, Anastasia Khrustaleva, Alexander Tyakht, Anastasia Efimenko, and et al. 2021. "Self-Organization Provides Cell Fate Commitment in MSC Sheet Condensed Areas via ROCK-Dependent Mechanism" Biomedicines 9, no. 9: 1192. https://doi.org/10.3390/biomedicines9091192
APA StyleNimiritsky, P., Novoseletskaya, E., Eremichev, R., Alexandrushkina, N., Karagyaur, M., Vetrovoy, O., Basalova, N., Khrustaleva, A., Tyakht, A., Efimenko, A., Tkachuk, V., & Makarevich, P. (2021). Self-Organization Provides Cell Fate Commitment in MSC Sheet Condensed Areas via ROCK-Dependent Mechanism. Biomedicines, 9(9), 1192. https://doi.org/10.3390/biomedicines9091192









